Endometriosis Associated-miRNome Analysis of Blood Samples: A Prospective Study
Abstract
:1. Introduction
2. Material and Methods
2.1. Study Population
2.2. Sample Collection
2.3. RNA Sample Extraction, Preparation and Quality Control
3. Bioinformatics
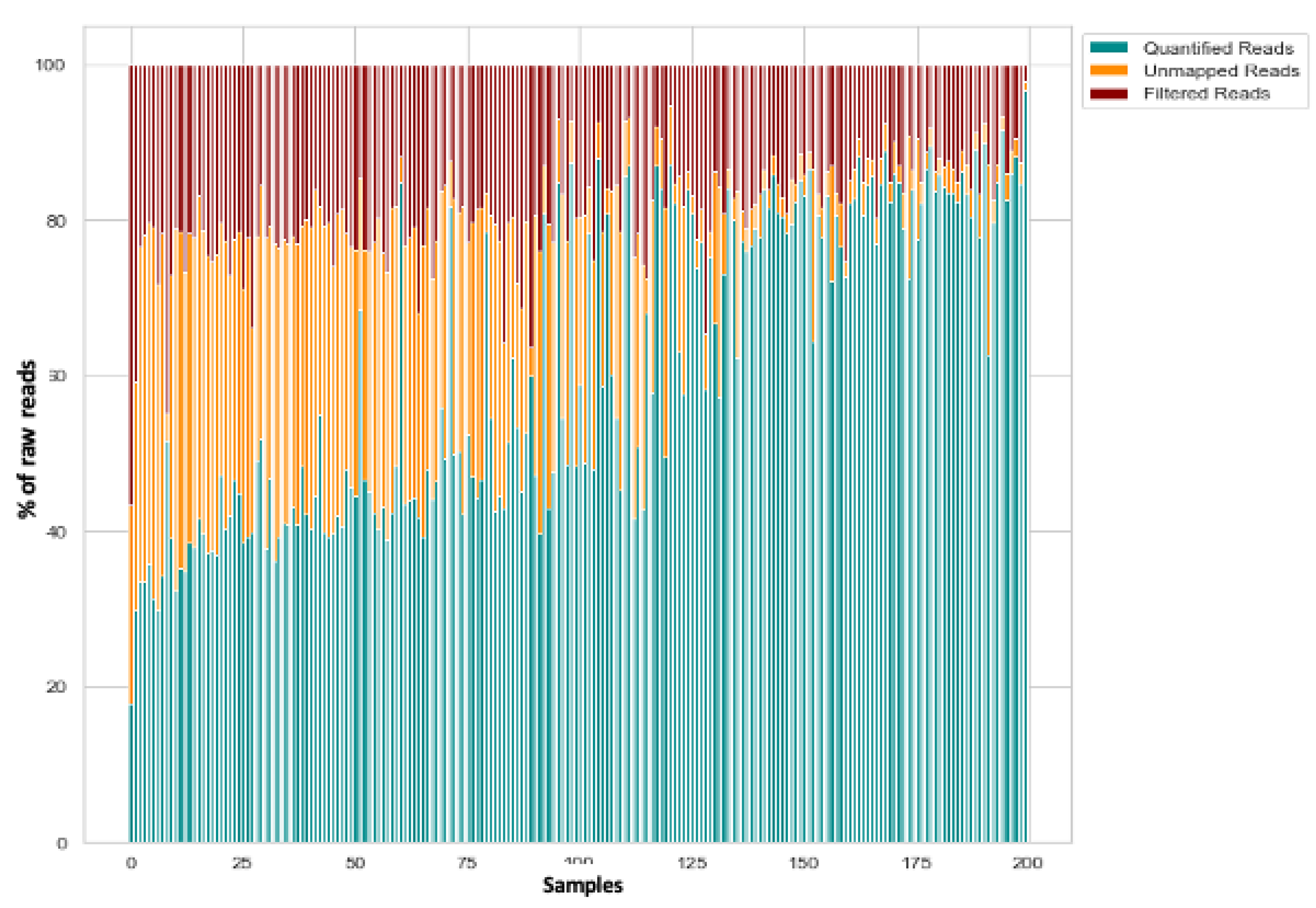
3.1. Raw Data Preprocessing (Raw, Filtered, Aligned Reads) and Quality Control
3.2. Differential Expression Analysis of miRNA
3.3. miRNome Accuracy
4. Results
4.1. Description of the ENDO-miRNA Cohort
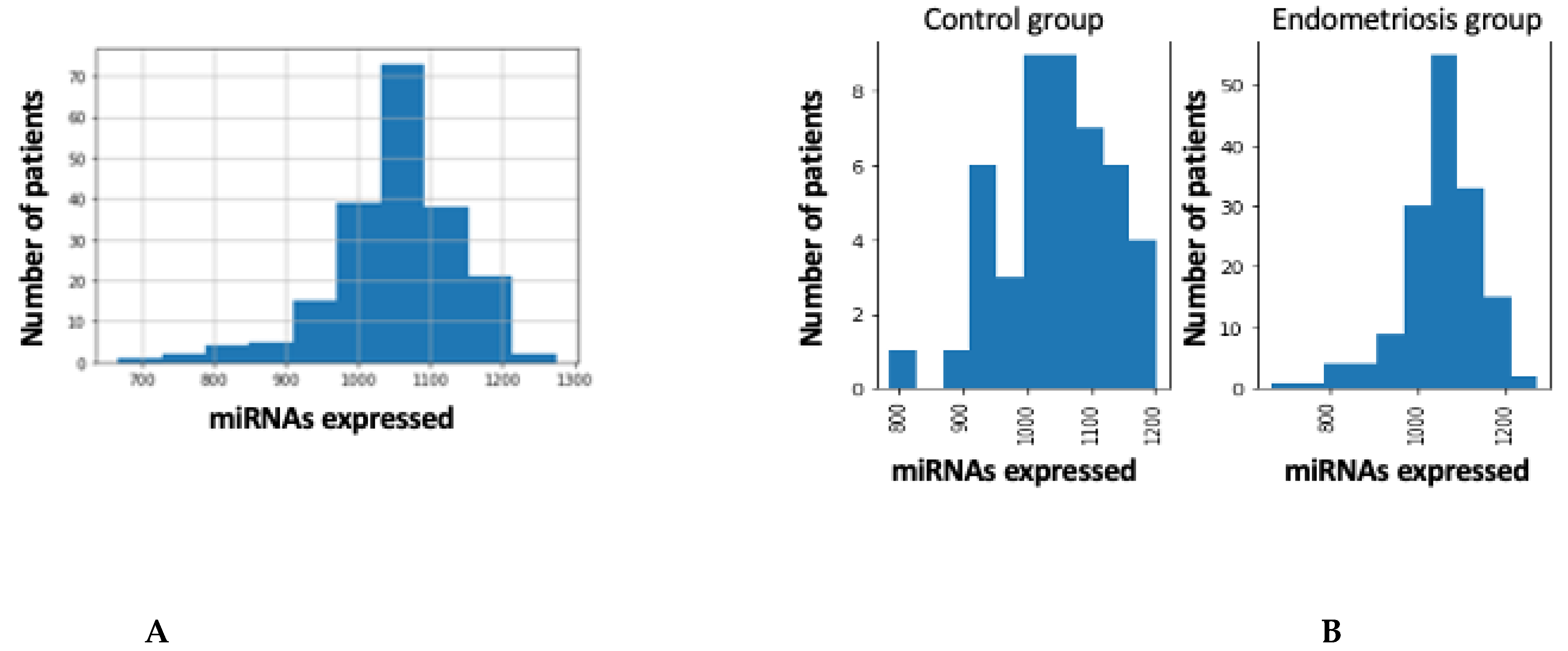
4.2. Global Overview of miRNA Transcriptome
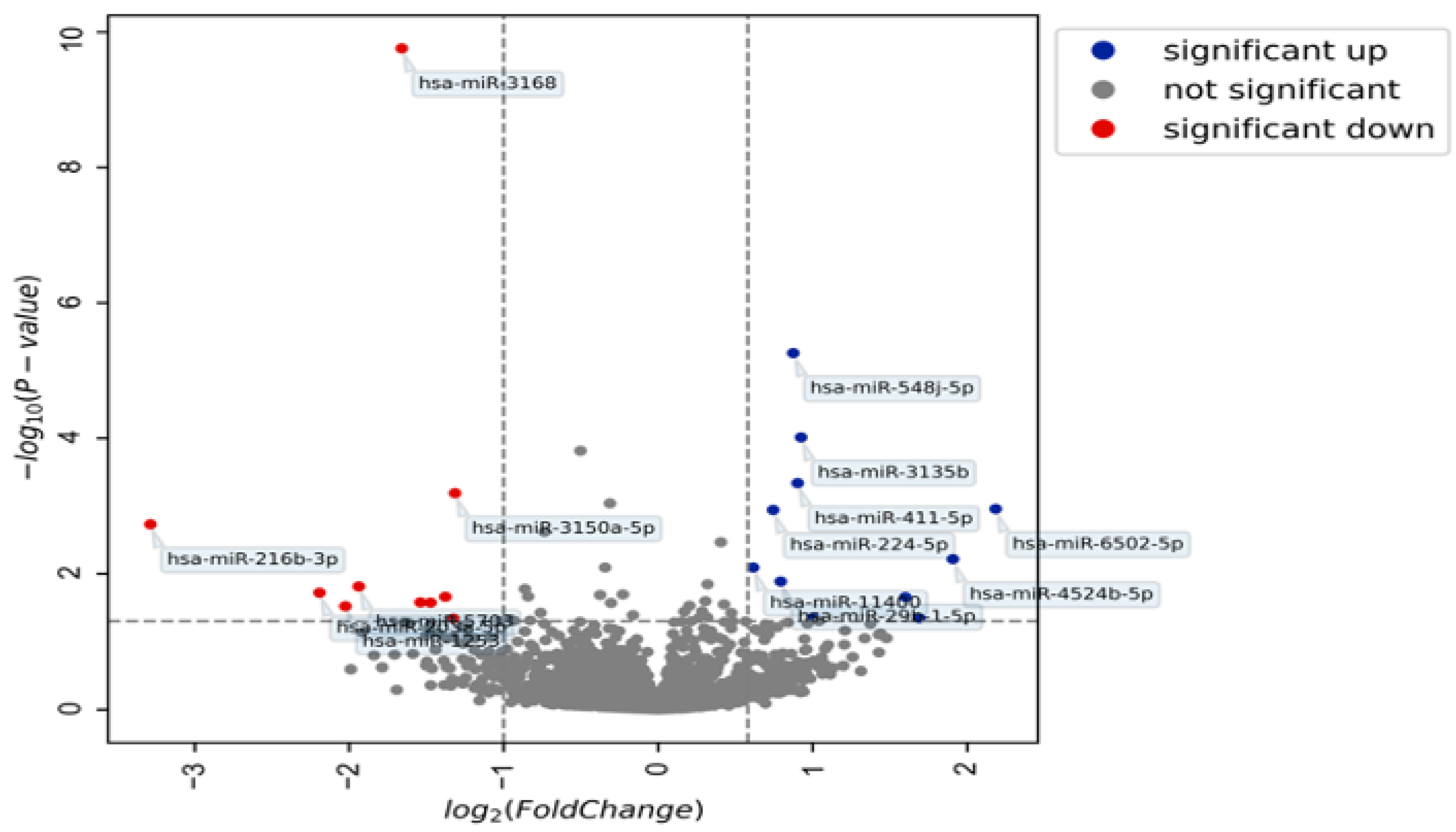
4.3. miRNA Expression in Patients with and without Endometriosis
4.4. Diagnostic Accuracy of Regulated miRNAs
5. Discussion
6. Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shahrouki, P.; Larsson, E. The Non-Coding Oncogene: A Case of Missing DNA Evidence? Front. Genet. 2012, 3, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, D.; Fraser, W.D.; Dalmay, T. Transfer RNA-Derived Small RNAs in the Cancer Transcriptome. Pflugers Arch. 2016, 468, 1041–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibb, E.A.; Brown, C.J.; Lam, W.L. The Functional Role of Long Non-Coding RNA in Human Carcinomas. Mol. Cancer 2011, 10, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, T.; Huang, X.; Woodcock, M.; Du, M.; Dittmar, R.; Wang, Y.; Tsai, S.; Kohli, M.; Boardman, L.; Patel, T.; et al. Plasma Extracellular RNA Profiles in Healthy and Cancer Patients. Sci. Rep. 2016, 6, 19413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, S.; Tapmeier, T.; Rahmioglu, N.; Kirtley, S.; Zondervan, K.; Becker, C. The MiRNA Mirage: How Close Are We to Finding a Non-Invasive Diagnostic Biomarker in Endometriosis? A Systematic Review. Int. J. Mol. Sci. 2018, 19, 599. [Google Scholar] [CrossRef] [Green Version]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. MiRBase: MicroRNA Sequences, Targets and Gene Nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. MiRBase: Tools for MicroRNA Genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef] [Green Version]
- Griffiths-Jones, S. The MicroRNA Registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Kang, J.-W.; Eun, Y.-G.; Lee, Y.-C. Diagnostic Value of Salivary MiRNA in Head and Neck Squamous Cell Cancer: Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2021, 22, 7026. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Rincon, A.; Mendoza-Maldonado, L.; Martinez-Archundia, M.; Schönhuth, A.; Kraneveld, A.D.; Garssen, J.; Tonda, A. Machine Learning-Based Ensemble Recursive Feature Selection of Circulating MiRNAs for Cancer Tumor Classification. Cancers 2020, 12, 1785. [Google Scholar] [CrossRef] [PubMed]
- Vanhie, A.; O, D.; Peterse, D.; Beckers, A.; Cuéllar, A.; Fassbender, A.; Meuleman, C.; Mestdagh, P.; D’Hooghe, T. Plasma MiRNAs as Biomarkers for Endometriosis. Hum. Reprod. 2019, 34, 1650–1660. [Google Scholar] [CrossRef]
- Courts, C.; Madea, B. Specific Micro-RNA Signatures for the Detection of Saliva and Blood in Forensic Body-Fluid Identification. J. Forensic Sci. 2011, 56, 1464–1470. [Google Scholar] [CrossRef]
- Monnaka, V.U.; Hernandes, C.; Heller, D.; Podgaec, S. Overview of MiRNAs for the Non-Invasive Diagnosis of Endometriosis: Evidence, Challenges and Strategies. A Systematic Review. Einstein 2021, 19, eRW5704. [Google Scholar] [CrossRef]
- Zondervan, K.T.; Becker, C.M.; Missmer, S.A. Endometriosis. N. Engl. J. Med. 2020, 382, 1244–1256. [Google Scholar] [CrossRef]
- Zondervan, K.T.; Becker, C.M.; Koga, K.; Missmer, S.A.; Taylor, R.N.; Viganò, P. Endometriosis. Nat. Rev. Dis. Primers 2018, 4, 9. [Google Scholar] [CrossRef]
- As-Sanie, S.; Soliman, A.M.; Evans, K.; Erpelding, N.; Lanier, R.; Katz, N.P. Healthcare Utilization and Cost Burden among Women with Endometriosis by Opioid Prescription Status in the First Year after Diagnosis: A Retrospective Claims Database Analysis. J. Med. Econ. 2020, 23, 371–377. [Google Scholar] [CrossRef]
- Surrey, E.; Soliman, A.M.; Trenz, H.; Blauer-Peterson, C.; Sluis, A. Impact of Endometriosis Diagnostic Delays on Healthcare Resource Utilization and Costs. Adv. Ther. 2020, 37, 1087–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dabi, Y.; Suisse, S.; Jornea, L.; Bouteiller, D.; Touboul, C.; Puchar, A.; Daraï, E.; Bendifallah, S. Clues for Improving the Pathophysiology Knowledge for Endometriosis Using Serum Micro-RNA Expression. Diagnostics 2022, 12, 175. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, S.; Burn, M.; Mamillapalli, R.; Nematian, S.; Flores, V.; Taylor, H.S. Accurate Diagnosis of Endometriosis Using Serum MicroRNAs. Am. J. Obstet. Gynecol. 2020, 223, e1–e557. [Google Scholar] [CrossRef]
- Bendifallah, S. Evaluation of MiRNAs in Endometriosis. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04728152 (accessed on 25 April 2022).
- Bendifallah, S.; Dabi, Y.; Suisse, S.; Jornea, L.; Bouteiller, D.; Touboul, C.; Puchar, A.; Daraï, E. MicroRNome Analysis Generates a Blood-Based Signature for Endometriosis. Sci. Rep. 2022, 12, 4051. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.E.; Abi Khalil, E.D.; Taffel, M.; Moawad, G.N. Magnetic Resonance Imaging Correlation to Intraoperative Findings of Deeply Infiltrative Endometriosis. Fertil. Steril. 2017, 107, e11–e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazot, M.; Daraï, E. Diagnosis of Deep Endometriosis: Clinical Examination, Ultrasonography, Magnetic Resonance Imaging, and Other Techniques. Fertil. Steril. 2017, 108, 886–894. [Google Scholar] [CrossRef] [Green Version]
- Bazot, M.; Darai, E.; Hourani, R.; Thomassin, I.; Cortez, A.; Uzan, S.; Buy, J.-N. Deep Pelvic Endometriosis: MR Imaging for Diagnosis and Prediction of Extension of Disease. Radiology 2004, 232, 379–389. [Google Scholar] [CrossRef] [Green Version]
- Bendifallah, S.; Suisse, S.; Puchar, A.; Delbos, L.; Poilblanc, M.; Descamps, P.; Golfier, F.; Jornea, L.; Bouteiller, D.; Touboul, C.; et al. Salivary MicroRNA Signature for Diagnosis of Endometriosis. J. Clin. Med. 2022, 11, 612. [Google Scholar] [CrossRef]
- Canis, M.; Donnez, J.G.; Guzick, D.S.; Halme, J.K.; Rock, J.A.; Schenken, R.S.; Vernon, M.W. Revised American Society for Reproductive Medicine Classification of Endometriosis: 1996. Fertil. Steril. 1997, 67, 817–821. [Google Scholar] [CrossRef]
- Bendifallah, S.; Puchar, A.; Suisse, S.; Delbos, L.; Poilblanc, M.; Descamps, P.; Golfier, F.; Touboul, C.; Dabi, Y.; Daraï, E. Machine Learning Algorithms as New Screening Approach for Patients with Endometriosis. Sci. Rep. 2022, 12, 639. [Google Scholar] [CrossRef]
- Canlorbe, G.; Castela, M.; Bendifallah, S.; Wang, Z.; Lefevre, M.; Chabbert-Buffet, N.; Aractingi, S.; DaraÏ, E.; Méhats, C.; Ballester, M. Micro-RNA Signature of Lymphovascular Space Involvement in Type 1 Endometrial Cancer. Histol. Histopathol. 2017, 32, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Canlorbe, G.; Wang, Z.; Laas, E.; Bendifallah, S.; Castela, M.; Lefevre, M.; Chabbert-Buffet, N.; Daraï, E.; Aractingi, S.; Méhats, C.; et al. Identification of MicroRNA Expression Profile Related to Lymph Node Status in Women with Early-Stage Grade 1-2 Endometrial Cancer. Mod. Pathol. 2016, 29, 391–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Foucher, T.; Sbeih, M.; Uzan, J.; Bendifallah, S.; Lefevre, M.; Chabbert-Buffet, N.; Aractingi, S.; Uzan, C.; Abd Alsalam, I.; Mitri, R.; et al. Identification of Micro-RNA Expression Profile Related to Recurrence in Women with ESMO Low-Risk Endometrial Cancer. J. Transl. Med. 2018, 16, 131. [Google Scholar] [CrossRef] [PubMed]
- Gyvyte, U.; Juzenas, S.; Salteniene, V.; Kupcinskas, J.; Poskiene, L.; Kucinskas, L.; Jarmalaite, S.; Stuopelyte, K.; Steponaitiene, R.; Hemmrich-Stanisak, G.; et al. MiRNA Profiling of Gastrointestinal Stromal Tumors by Next-Generation Sequencing. Oncotarget 2017, 8, 37225–37238. [Google Scholar] [CrossRef]
- Gyvyte, U.; Kupcinskas, J.; Juzenas, S.; Inciuraite, R.; Poskiene, L.; Salteniene, V.; Link, A.; Fassan, M.; Franke, A.; Kupcinskas, L.; et al. Identification of Long Intergenic Non-Coding RNAs (LincRNAs) Deregulated in Gastrointestinal Stromal Tumors (GISTs). PLoS ONE 2018, 13, e0209342. [Google Scholar] [CrossRef]
- Potla, P.; Ali, S.A.; Kapoor, M. A Bioinformatics Approach to MicroRNA-Sequencing Analysis. Osteoarthr. Cartil. Open 2021, 3, 100131. [Google Scholar] [CrossRef]
- Li, Q.; Liu, G.; Bao, Y.; Wu, Y.; You, Q. Evaluation and Application of Tools for the Identification of Known MicroRNAs in Plants. Appl. Plant Sci. 2021, 9, e11414. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Zhang, L. Construction and Comprehensive Analysis of a CeRNA Network to Reveal Potential Prognostic Biomarkers for Lung Adenocarcinoma. BMC Cancer 2021, 21, 849. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Bargaje, R.; Hariharan, M.; Scaria, V.; Pillai, B. Consensus MiRNA Expression Profiles Derived from Interplatform Normalization of Microarray Data. RNA 2010, 16, 16–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrell, F.E.; Lee, K.L.; Mark, D.B. Multivariable Prognostic Models: Issues in Developing Models, Evaluating Assumptions and Adequacy, and Measuring and Reducing Errors. Stat. Med. 1996, 15, 361–387. [Google Scholar] [CrossRef]
- Steyerberg, E.W.; Eijkemans, M.J.; Harrell, F.E.; Habbema, J.D. Prognostic Modelling with Logistic Regression Analysis: A Comparison of Selection and Estimation Methods in Small Data Sets. Stat. Med. 2000, 19, 1059–1079. [Google Scholar] [CrossRef]
- Panir, K.; Schjenken, J.E.; Robertson, S.A.; Hull, M.L. Non-Coding RNAs in Endometriosis: A Narrative Review. Hum. Reprod. Update 2018, 24, 497–515. [Google Scholar] [CrossRef] [Green Version]
- Ghafouri-Fard, S.; Shoorei, H.; Taheri, M. Role of Non-Coding RNAs in the Pathogenesis of Endometriosis. Front. Oncol. 2020, 10, 1370. [Google Scholar] [CrossRef]
- Maged, A.M.; Deeb, W.S.; El Amir, A.; Zaki, S.S.; El Sawah, H.; Al Mohamady, M.; Metwally, A.A.; Katta, M.A. Diagnostic accuracy of serum miR-122 and miR-199a in women with endometriosis. Int. J. Gynaecol. Obstet. 2018, 141, 14–19. [Google Scholar] [CrossRef]
- Wang, W.-T.; Zhao, Y.-N.; Han, B.-W.; Hong, S.-J.; Chen, Y.-Q. Circulating MicroRNAs Identified in a Genome-Wide Serum MicroRNA Expression Analysis as Noninvasive Biomarkers for Endometriosis. J. Clin. Endocrinol. Metab. 2013, 98, 281–289. [Google Scholar] [CrossRef] [Green Version]
- Rekker, K.; Saare, M.; Roost, A.M.; Kaart, T.; Sõritsa, D.; Karro, H.; Sõritsa, A.; Simón, C.; Salumets, A.; Peters, M. Circulating MiR-200-Family Micro-RNAs Have Altered Plasma Levels in Patients with Endometriosis and Vary with Blood Collection Time. Fertil. Steril. 2015, 104, 938–946.e2. [Google Scholar] [CrossRef]
- Fernandes, J.; Acuña, S.; Aoki, J.; Floeter-Winter, L.; Muxel, S. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. ncRNA 2019, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Zubrzycka, A.; Zubrzycki, M.; Perdas, E.; Zubrzycka, M. Genetic, Epigenetic, and Steroidogenic Modulation Mechanisms in Endometriosis. J. Clin. Med. 2020, 9, 1309. [Google Scholar] [CrossRef]
- Angioni, S.; D’Alterio, M.N.; Coiana, A.; Anni, F.; Gessa, S.; Deiana, D. Genetic Characterization of Endometriosis Patients: Review of the Literature and a Prospective Cohort Study on a Mediterranean Population. Int. J. Mol. Sci. 2020, 21, 1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deiana, D.; Gessa, S.; Anardu, M.; Daniilidis, A.; Nappi, L.; D’Alterio, M.N.; Pontis, A.; Angioni, S. Genetics of Endometriosis: A Comprehensive Review. Gynecol. Endocrinol. 2019, 35, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Rincon, A.; Martinez-Archundia, M.; Martinez-Ruiz, G.U.; Schoenhuth, A.; Tonda, A. Automatic Discovery of 100-MiRNA Signature for Cancer Classification Using Ensemble Feature Selection. BMC Bioinform. 2019, 20, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akter, S.; Xu, D.; Nagel, S.C.; Bromfield, J.J.; Pelch, K.E.; Wilshire, G.B.; Joshi, T. GenomeForest: An Ensemble Machine Learning Classifier for Endometriosis. AMIA Jt. Summits Transl. Sci. Proc. 2020, 2020, 33–42. [Google Scholar] [PubMed]
- Jang, H.-J.; Lee, H.-S.; Burt, B.M.; Lee, G.K.; Yoon, K.-A.; Park, Y.-Y.; Sohn, B.H.; Kim, S.B.; Kim, M.S.; Lee, J.M.; et al. Integrated Genomic Analysis of Recurrence-Associated Small Non-Coding RNAs in Oesophageal Cancer. Gut 2017, 66, 215–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akter, S.; Xu, D.; Nagel, S.C.; Bromfield, J.J.; Pelch, K.; Wilshire, G.B.; Joshi, T. Machine Learning Classifiers for Endometriosis Using Transcriptomics and Methylomics Data. Front. Genet. 2019, 10, 766. [Google Scholar] [CrossRef] [Green Version]
- Kleczyk, E.J.; Peri, A.; Yadav, T.; Komera, R.; Peri, M.; Guduru, V.; Amirtharaj, S.; Huang, M. Predicting Endometriosis Onset Using Machine Learning Algorithms. 2020; in review. [Google Scholar]
- Zondervan, K.T.; Cardon, L.R.; Kennedy, S.H. What Makes a Good Case-Control Study? Design Issues for Complex Traits Such as Endometriosis. Hum. Reprod. 2002, 17, 1415–1423. [Google Scholar] [CrossRef] [Green Version]
- Kolanska, K.; Bendifallah, S.; Canlorbe, G.; Mekinian, A.; Touboul, C.; Aractingi, S.; Chabbert-Buffet, N.; Daraï, E. Role of MiRNAs in Normal Endometrium and in Endometrial Disorders: Comprehensive Review. J. Clin. Med. 2021, 10, 3457. [Google Scholar] [CrossRef]
- Kuokkanen, S.; Chen, B.; Ojalvo, L.; Benard, L.; Santoro, N.; Pollard, J.W. Genomic Profiling of MicroRNAs and Messenger RNAs Reveals Hormonal Regulation in MicroRNA Expression in Human Endometrium. Biol. Reprod. 2010, 82, 791–801. [Google Scholar] [CrossRef] [Green Version]
- Sha, A.-G.; Liu, J.-L.; Jiang, X.-M.; Ren, J.-Z.; Ma, C.-H.; Lei, W.; Su, R.-W.; Yang, Z.-M. Genome-Wide Identification of Micro-Ribonucleic Acids Associated with Human Endometrial Receptivity in Natural and Stimulated Cycles by Deep Sequencing. Fertil. Steril. 2011, 96, 150–155.e5. [Google Scholar] [CrossRef]
- Altmäe, S.; Martinez-Conejero, J.A.; Esteban, F.J.; Ruiz-Alonso, M.; Stavreus-Evers, A.; Horcajadas, J.A.; Salumets, A. MicroRNAs MiR-30b, MiR-30d, and MiR-494 Regulate Human Endometrial Receptivity. Reprod. Sci. 2013, 20, 308–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafrir, A.L.; Farland, L.V.; Shah, D.K.; Harris, H.R.; Kvaskoff, M.; Zondervan, K.; Missmer, S.A. Risk for and Consequences of Endometriosis: A Critical Epidemiologic Review. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
| Controls N = 47 | Endometriosis N = 153 | p Value | |
|---|---|---|---|
| Age years (mean ± SD) | 30.92 ± 13.79 | 31.17 ± 10.78 | 0.19 |
| BMI (body mass index) (mean ± SD) | 24.84 ± 11.10 | 24.36 ± 8.38 | 0.52 |
| rASRM classification | |||
- I–II | - | 80 (52%) | |
- III–IV | - | 73 (48%) | |
| Control diagnoses | |||
- No abnormality | 24 (51%) | - | - |
- Leiomyoma | 1 (2%) | ||
- Cystadenoma | 5 (11%) | ||
- Teratoma | 11 (23%) | ||
- Others gynecological disorders | 6 (13%) | ||
| Dysmenorrhea | 100% | 100% | |
| Abdominal pain outside menstruation | |||
- Yes | 21 (66%) | 89 (71%) | 0.69 |
| Patients with pain suggesting sciatica | 10 (31%) | 70 (56%) | 0.02 |
| Dyspareunia intensity at VAS (mean ± SD) | 4.95 ± 3.52 | 5.28 ± 3.95 | <0.001 |
| Patients with lower back pain outside menstruation | 20 (62%) | 101 (81%) | 0.049 |
| Intensity of pain during defecation at VAS (mean ± SD) | 2.84 ± 2.76 | 4.35 ± 3.47 | <0.001 |
| Patient with right shoulder pain during menstruation | 3 (9%) | 26 (21%) | 0.21 |
| Intensity of urinary pain during menstruation at VAS (mean ± SD) | 2.84 ± 2.76 | 4.35 ± 3.36 | <0.001 |
| Patient with blood in the stools during menstruation | 4 (12%) | 30 (24%) | 0.24 |
| Patient with blood in urine during menstruation | 8 (25%) | 21 (17%) | 0.41 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bendifallah, S.; Dabi, Y.; Suisse, S.; Delbos, L.; Poilblanc, M.; Descamps, P.; Golfier, F.; Jornea, L.; Bouteiller, D.; Touboul, C.; et al. Endometriosis Associated-miRNome Analysis of Blood Samples: A Prospective Study. Diagnostics 2022, 12, 1150. https://doi.org/10.3390/diagnostics12051150
Bendifallah S, Dabi Y, Suisse S, Delbos L, Poilblanc M, Descamps P, Golfier F, Jornea L, Bouteiller D, Touboul C, et al. Endometriosis Associated-miRNome Analysis of Blood Samples: A Prospective Study. Diagnostics. 2022; 12(5):1150. https://doi.org/10.3390/diagnostics12051150
Chicago/Turabian StyleBendifallah, Sofiane, Yohann Dabi, Stéphane Suisse, Léa Delbos, Mathieu Poilblanc, Philippe Descamps, Francois Golfier, Ludmila Jornea, Delphine Bouteiller, Cyril Touboul, and et al. 2022. "Endometriosis Associated-miRNome Analysis of Blood Samples: A Prospective Study" Diagnostics 12, no. 5: 1150. https://doi.org/10.3390/diagnostics12051150
APA StyleBendifallah, S., Dabi, Y., Suisse, S., Delbos, L., Poilblanc, M., Descamps, P., Golfier, F., Jornea, L., Bouteiller, D., Touboul, C., Puchar, A., & Daraï, E. (2022). Endometriosis Associated-miRNome Analysis of Blood Samples: A Prospective Study. Diagnostics, 12(5), 1150. https://doi.org/10.3390/diagnostics12051150







