PDK4 Constitutes a Novel Prognostic Biomarker and Therapeutic Target in Gastric Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Public Data Acquisition and Preprocessing
2.2. Analysis of PDK Mutations and Prognosis
2.3. Survival Analysis
2.4. Multivariate Cox Regression Analysis
2.5. Construction and Evaluation of the Nomogram
2.6. Analysis of DEGs between the PDK4-High- and -Low-Expression Groups
2.7. Enrichment Analysis
2.8. Protein–Protein Interaction (PPI) Network Construction and Hub Gene Identification
2.9. Immune Infiltration Analysis
2.10. Cell Culture
2.11. Gastric Tissue Specimens
2.12. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.13. Protein Extraction and Western Blot Analysis
2.14. Lentivirus Construction and Infection
2.15. Cell Proliferation Assay
2.16. Colony-Formation Assay
2.17. Flow Cytometric Detection of Cell-Cycle Distribution
2.18. Transwell Assay
2.19. Wound Healing Assay
2.20. Statistical Analysis
3. Results
3.1. Aberrant PDK Expression in Various Cancers
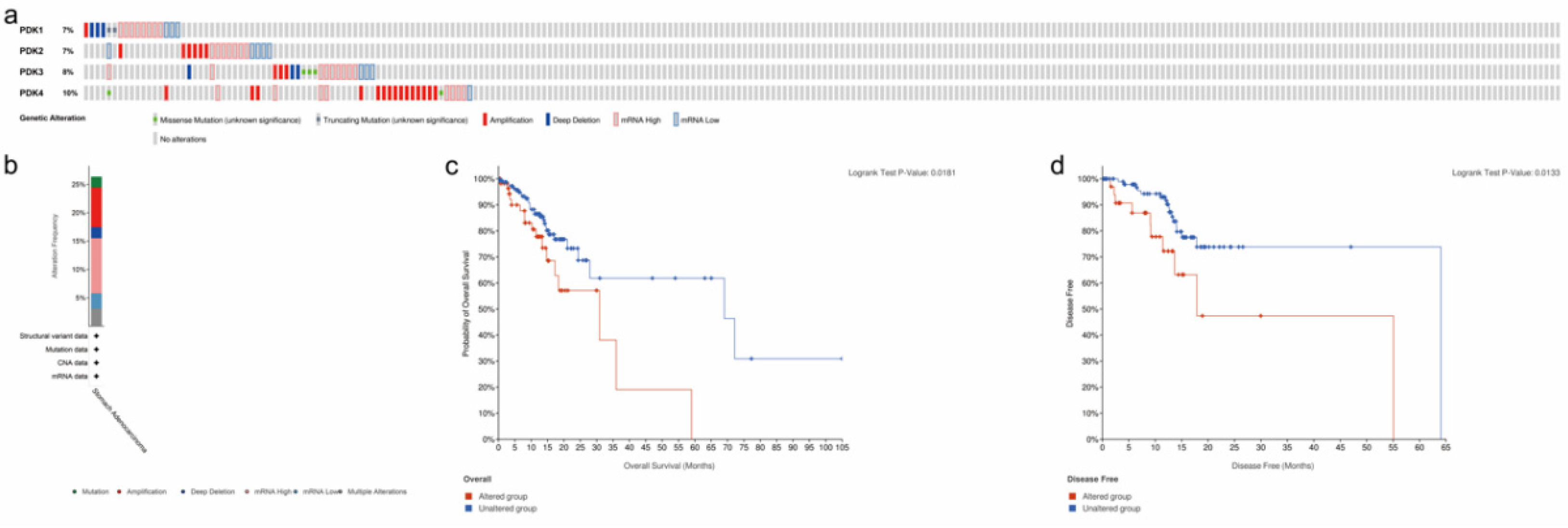
3.2. Analysis of PDK Genetic Alteration and Association with Prognosis in GC
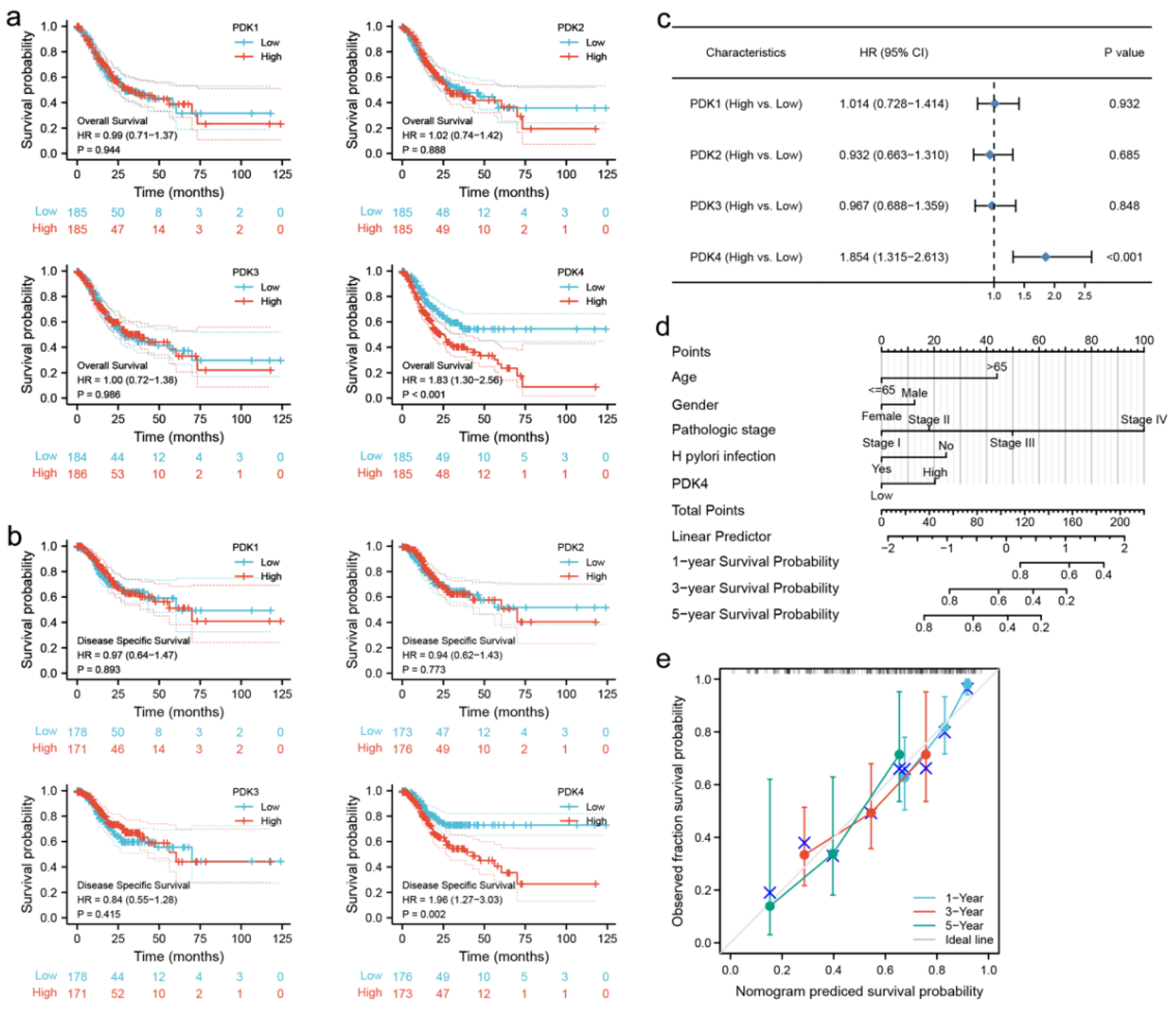
3.3. Prognostic Value of PDK Expression in GC
3.4. Identification of DEGs of PDK4
3.5. Enrichment Analyses of PDK4 in GC
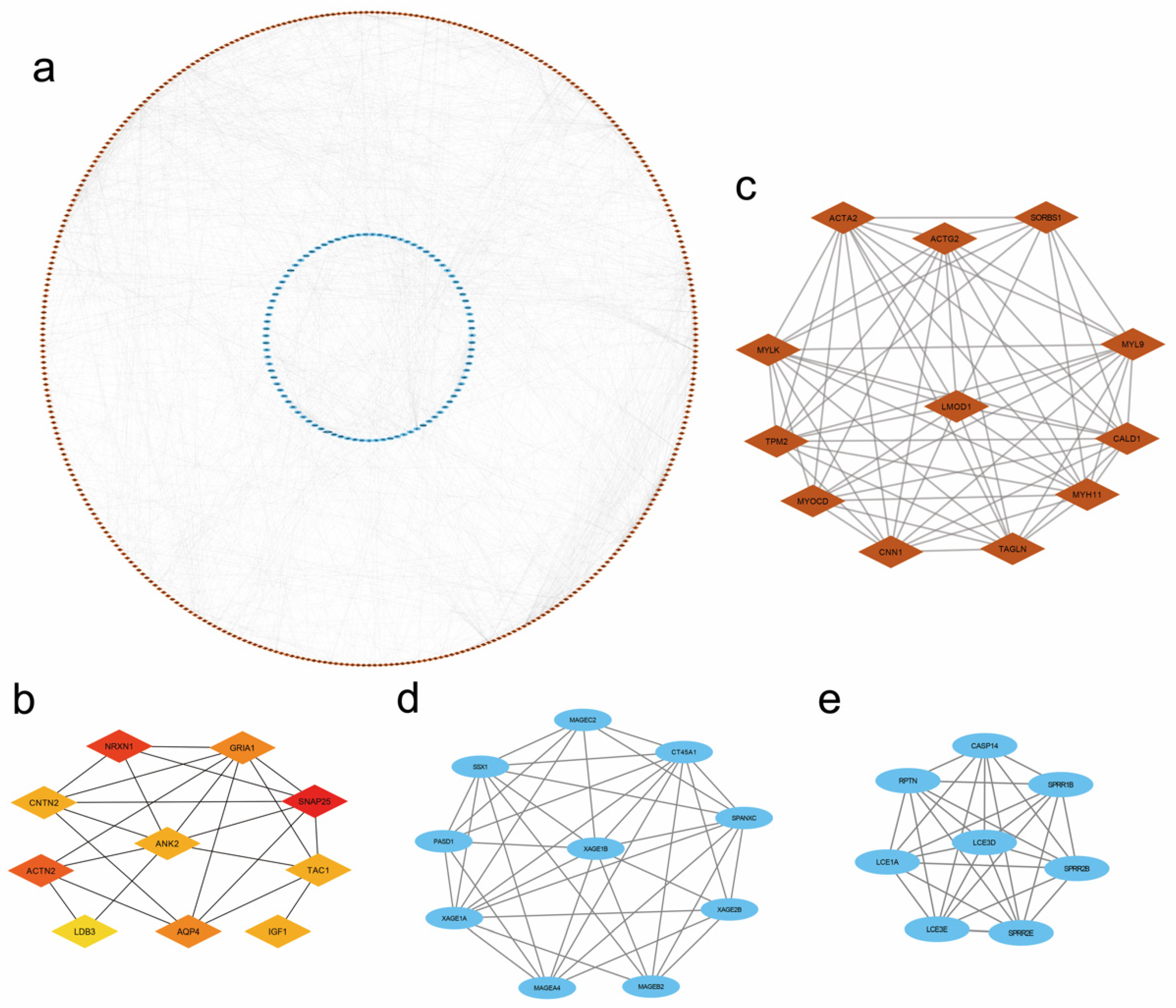
3.6. PPI Network and Module Analysis
3.7. Immune Infiltration Analysis of PDK4 Expression in GC
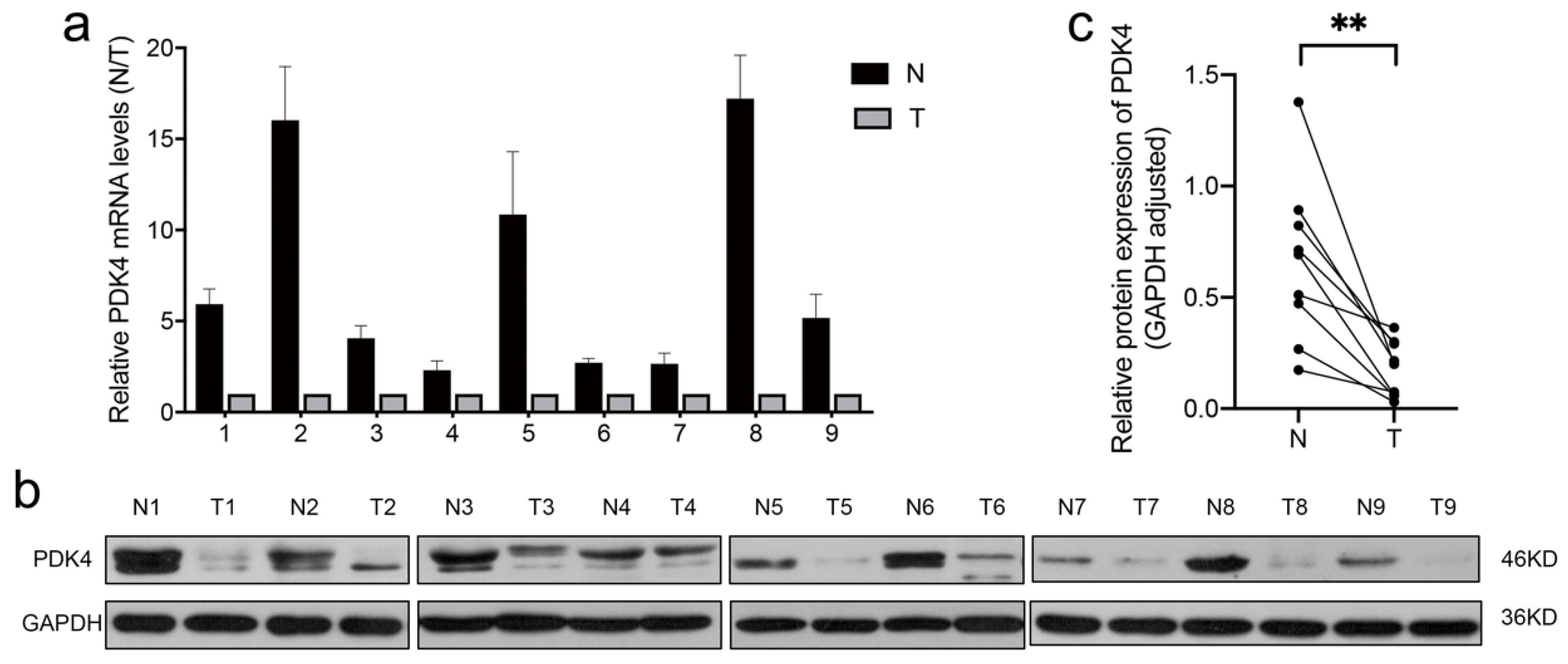
3.8. PDK4 Expression Is Downregulated in GC Tissues
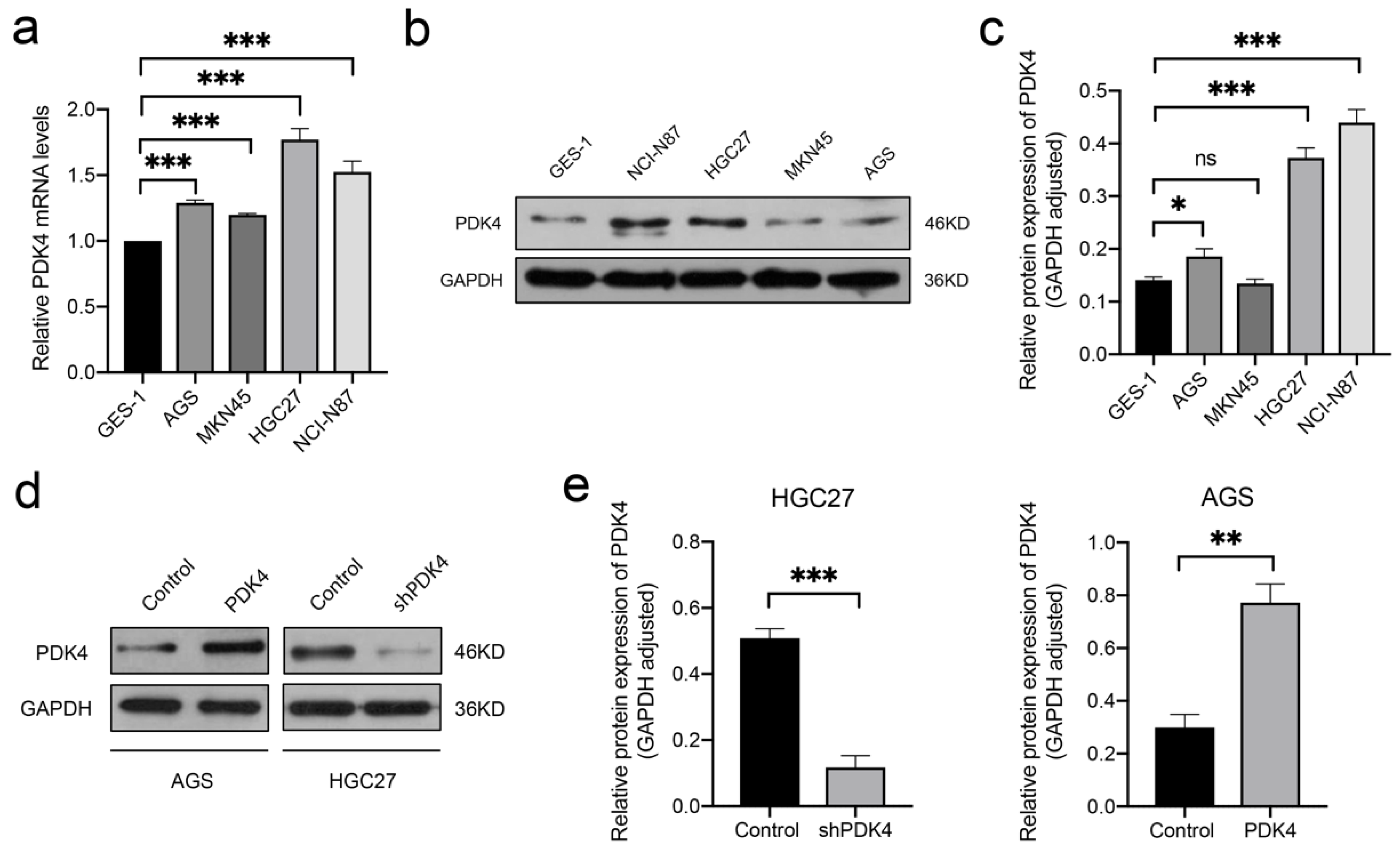
3.9. High PDK4 Expression in GC Cells and Validation of the Intervention
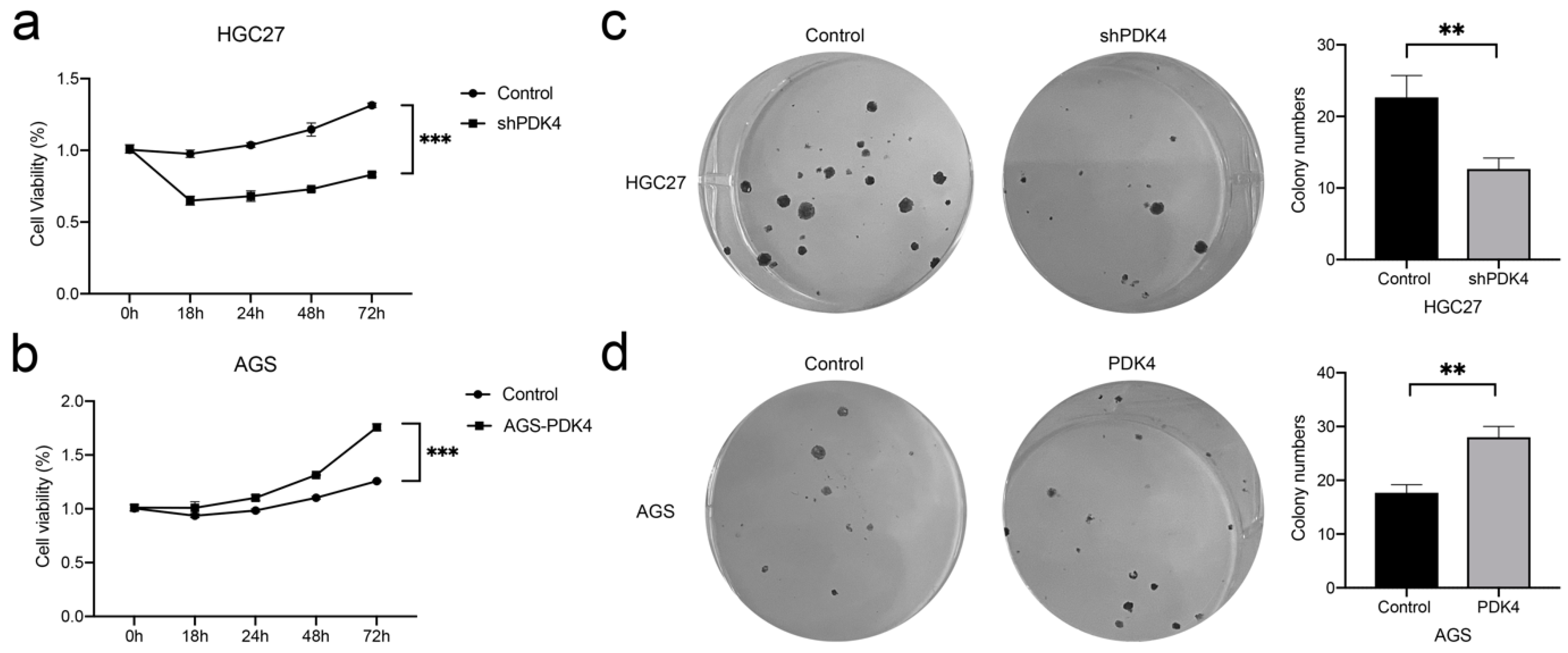
3.10. PDK4 Expression Regulates the Proliferation and Colony Formation of GC Cells
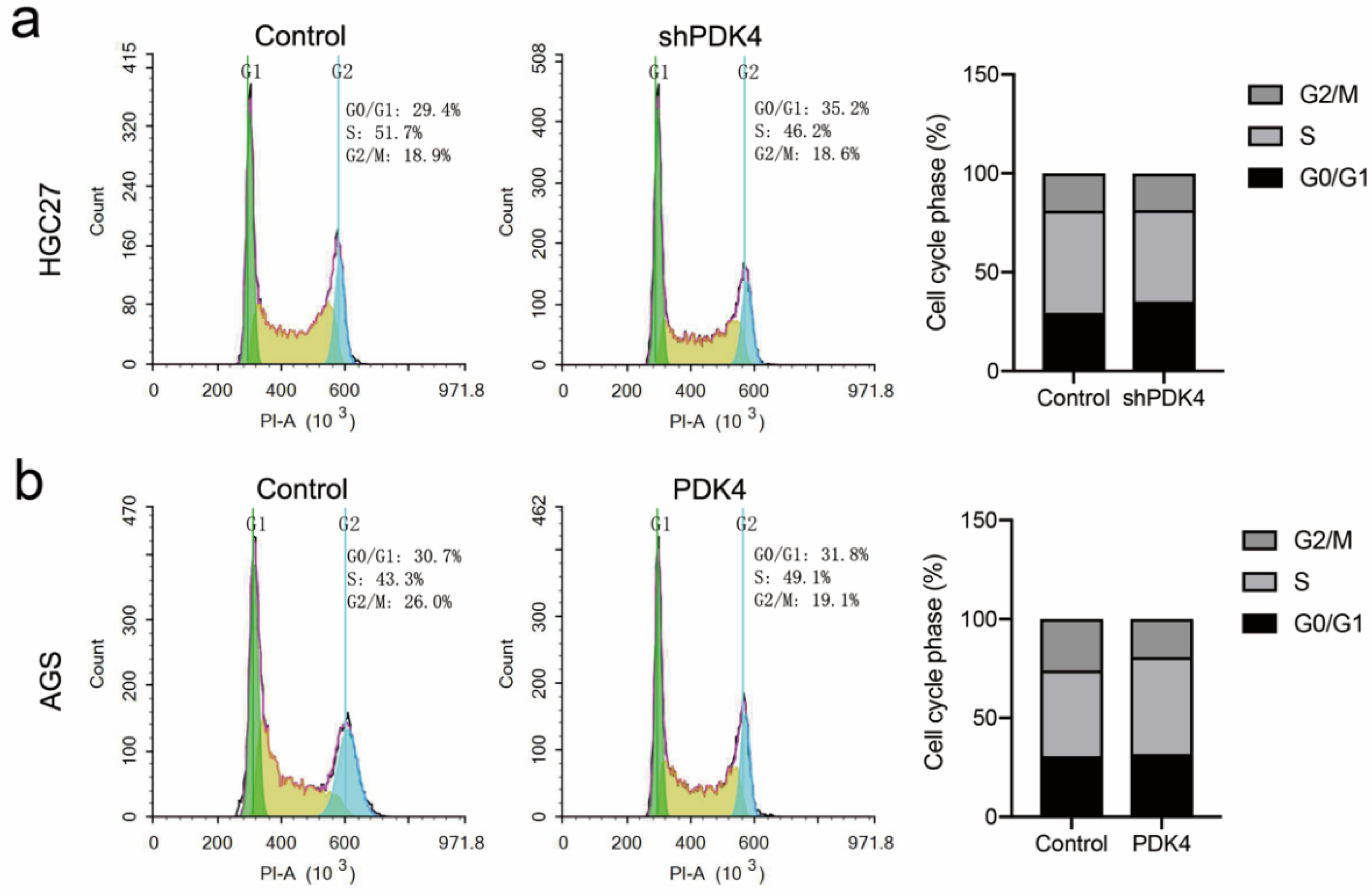
3.11. Suppression or Overexpression of PDK4 Could Affect Cell Cycle in GC Cells
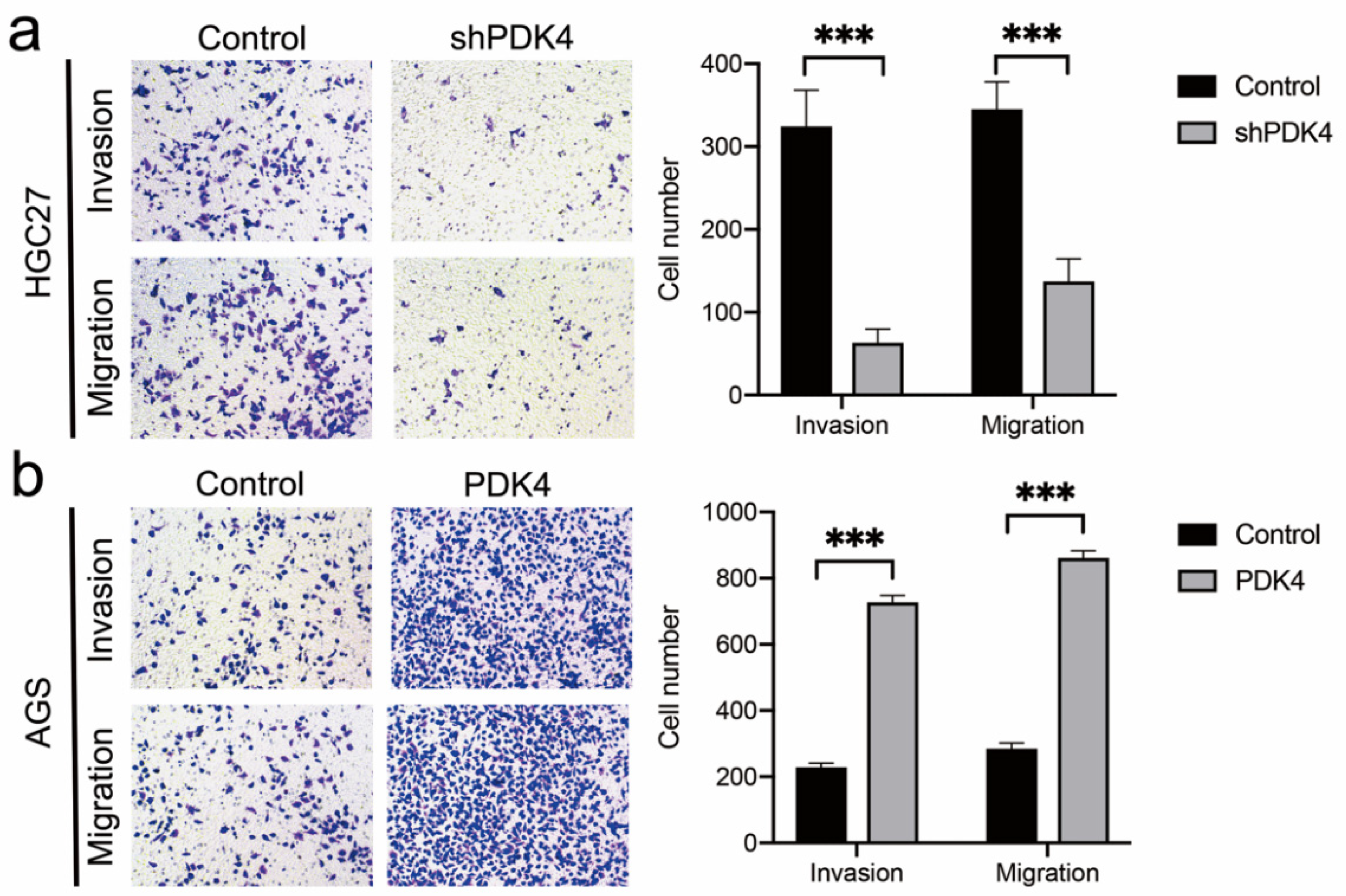
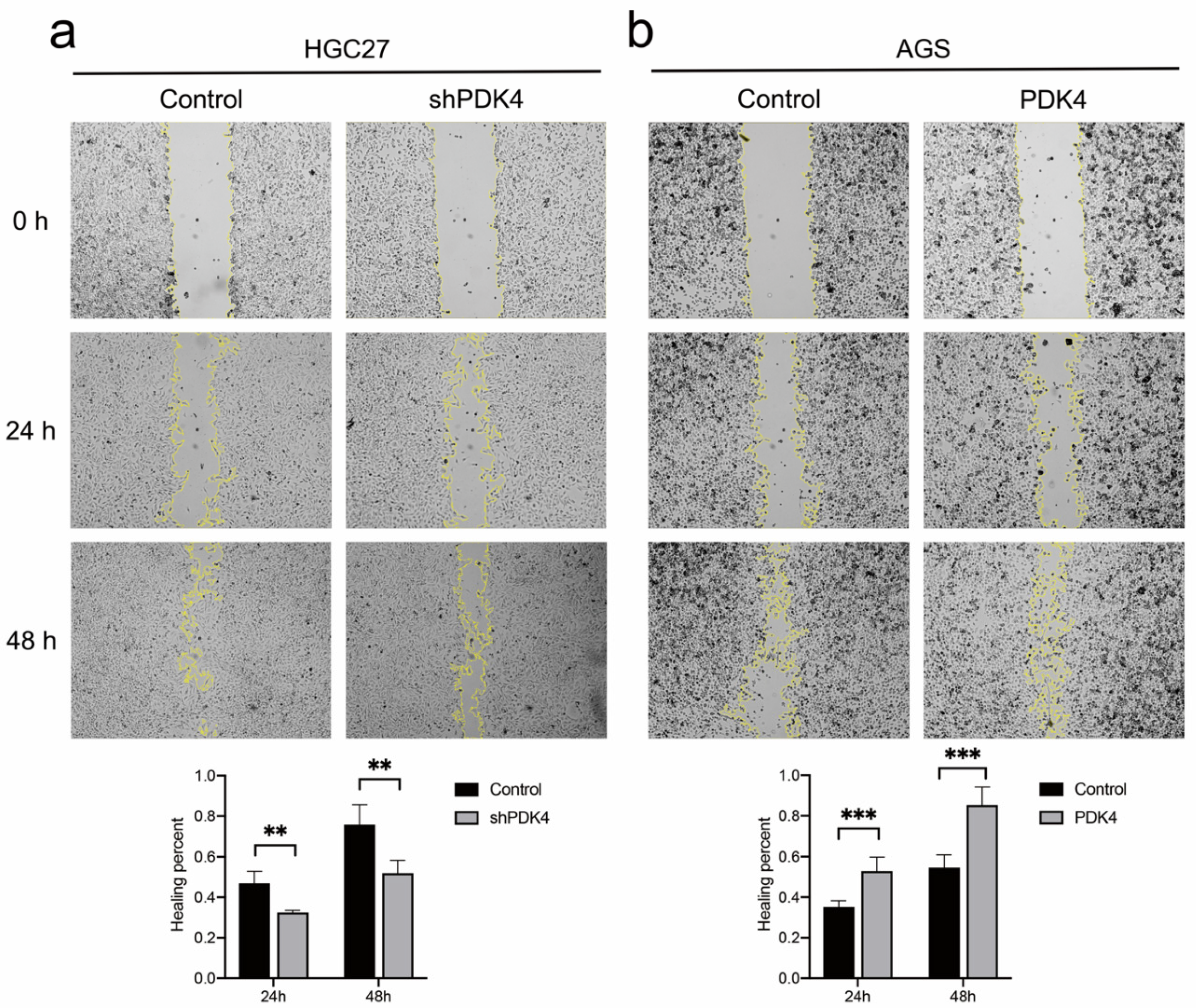
3.12. PDK4 Expression Exerted an Effect on GC Invasion and Migration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zheng, R.; Baade, P.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thrift, A.P.; El-Serag, H.B. Burden of Gastric Cancer. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2020, 18, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- San-Millán, I.; Brooks, G.A. Reexamining cancer metabolism: Lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 2017, 38, 119–133. [Google Scholar] [CrossRef]
- Ganapathy-Kanniappan, S.; Geschwind, J.-F.H. Tumor glycolysis as a target for cancer therapy: Progress and prospects. Mol. Cancer 2013, 12, 152. [Google Scholar] [CrossRef] [Green Version]
- Stacpoole, P.W. Therapeutic Targeting of the Pyruvate Dehydrogenase Complex/Pyruvate Dehydrogenase Kinase (PDC/PDK) Axis in Cancer. JNCI J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [Green Version]
- Woolbright, B.L.; Rajendran, G.; Harris, R.A.; Taylor, J.A. Metabolic Flexibility in Cancer: Targeting the Pyruvate Dehydrogenase Kinase:Pyruvate Dehydrogenase Axis. Mol. Cancer Ther. 2019, 18, 1673–1681. [Google Scholar] [CrossRef] [Green Version]
- Leclerc, D.; Pham, D.N.T.; Lévesque, N.; Truongcao, M.; Foulkes, W.D.; Sapienza, C.; Rozen, R. Oncogenic role of PDK4 in human colon cancer cells. Br. J. Cancer 2017, 116, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Woolbright, B.L.; Choudhary, D.; Mikhalyuk, A.; Trammel, C.; Shanmugam, S.; Abbott, E.; Pilbeam, C.C.; Taylor, J.A. The Role of Pyruvate Dehydrogenase Kinase-4 (PDK4) in Bladder Cancer and Chemoresistance. Mol. Cancer Ther. 2018, 17, 2004–2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Qian, Y.; Gao, M. Overexpression of PDK4 is associated with cell proliferation, drug resistance and poor prognosis in ovarian cancer. Cancer Manag. Res. 2018, 11, 251–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guda, M.R.; Asuthkar, S.; Labak, C.M.; Tsung, A.J.; Alexandrov, I.; Mackenzie, M.J.; Prasad, D.V.; Velpula, K.K. Targeting PDK4 inhibits breast cancer metabolism. Am. J. Cancer Res. 2018, 8, 1725–1738. [Google Scholar]
- Atas, E.; Oberhuber, M.; Kenner, L. The Implications of PDK1–4 on Tumor Energy Metabolism, Aggressiveness and Therapy Resistance. Front. Oncol. 2020, 10, 583217. [Google Scholar] [CrossRef]
- Wu, N.; He, C.; Zhu, B.; Jiang, J.; Chen, Y.; Ma, T. 3-Phosphoinositide Dependent Protein Kinase-1 (PDK-1) Promotes Migration and Invasion in Gastric Cancer Cells Through Activating the NF-κB Pathway. Oncol. Res. 2017, 25, 1153–1159. [Google Scholar] [CrossRef]
- Zahalka, A.H.; Frenette, P.S. Nerves in cancer. Nat. Rev. Cancer 2020, 20, 143–157. [Google Scholar] [CrossRef]
- Vyas, S.; Zaganjor, E.; Haigis, M.C. Mitochondria and Cancer. Cell 2016, 166, 555–566. [Google Scholar] [CrossRef]
- Xiao, S.; Zhou, L. Gastric cancer: Metabolic and metabolomics perspectives (Review). Int. J. Oncol. 2017, 51, 5–17. [Google Scholar] [CrossRef] [Green Version]
- Gatenby, R.A.; Gillies, R. Why do cancers have high aerobic glycolysis? Nat. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Li, Z.; Peng, Y.; Li, J.; Chen, Z.; Chen, F.; Tu, J.; Lin, S.; Wang, H. N6-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat. Commun. 2020, 11, 2578. [Google Scholar] [CrossRef] [PubMed]
- Rabben, H.-L.; Andersen, G.T.; Olsen, M.K.; Øverby, A.; Ianevski, A.; Kainov, D.; Wang, T.C.; Lundgren, S.; Grønbech, J.E.; Chen, D.; et al. Neural signaling modulates metabolism of gastric cancer. iScience 2021, 24, 102091. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, X.; Chen, X.; Yang, X.; Song, Q.; Wu, H. High SALM3 Expression in Tumor Cells and Fibroblasts Is Correlated with Poor Prognosis in Gastric Cancer Patients. Dis. Markers 2019, 2019, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.-M.; Hayakawa, Y.; Kodama, Y.; Muthupalani, S.; Westphalen, C.B.; Andersen, G.T.; Flatberg, A.; Johannessen, H.; Friedman, R.A.; Renz, B.W.; et al. Denervation suppresses gastric tumorigenesis. Sci. Transl. Med. 2014, 6, 250ra115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Zhang, M.; Ma, Z.; Yuan, D.; Zhu, J.; Tuo, B.; Li, T.; Liu, X. Alteration and dysfunction of ion channels/transporters in a hypoxic microenvironment results in the development and progression of gastric cancer. Cell. Oncol. 2021, 44, 739–749. [Google Scholar] [CrossRef]
- Tian, S.; Zhang, R.; Tian, P.; Chi, Q.; Wang, J.; Wang, Y.; Sun, L.; Liu, Y.; Zhang, Q. Human ether-à-go-go-related gene expression is essential for cisplatin to induce apoptosis in human gastric cancer. Oncol. Rep. 2012, 27, 433–440. [Google Scholar] [CrossRef]
- Crociani, O.; Lastraioli, E.; Boni, L.; Pillozzi, S.; Romoli, M.R.; D’Amico, M.; Stefanini, M.; Crescioli, S.; Taddei, A.; Bencini, L.; et al. hERG1 Channels Regulate VEGF-A Secretion in Human Gastric Cancer: Clinicopathological Correlations and Therapeutical Implications. Clin. Cancer Res. 2014, 20, 1502–1512. [Google Scholar] [CrossRef] [Green Version]
- Kamarajugadda, S.; Stemboroski, L.; Cai, Q.; Simpson, N.E.; Nayak, S.; Tan, M.; Lu, J. Glucose Oxidation Modulates Anoikis and Tumor Metastasis. Mol. Cell. Biol. 2012, 32, 1893–1907. [Google Scholar] [CrossRef] [Green Version]
- Oya, Y.; Hayakawa, Y.; Koike, K. Tumor microenvironment in gastric cancers. Cancer Sci. 2020, 111, 2696–2707. [Google Scholar] [CrossRef]
- Varricchi, G.; Galdiero, M.R.; Loffredo, S.; Marone, G.; Iannone, R.; Marone, G.; Granata, F. Are Mast Cells MASTers in Cancer? Front. Immunol. 2017, 8, 424. [Google Scholar] [CrossRef] [Green Version]
- Teng, L.; Pereira, B.; Keerthikumar, S.; Huang, C.; Niranjan, B.; Lee, S.; Richards, M.; Schittenhelm, R.; Furic, L.; Goode, D.; et al. Mast Cell-Derived SAMD14 Is a Novel Regulator of the Human Prostate Tumor Microenvironment. Cancers 2021, 13, 1237. [Google Scholar] [CrossRef] [PubMed]
- Aller, M.A.; Arias, A.; Arias, J.I.; Arias, J. Carcinogenesis: The cancer cell-mast cell connection. Inflamm. Res. Off. J. Eur. Histamine Res. Soc. 2019, 68, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Sammarco, G.; Varricchi, G.; Ferraro, V.; Ammendola, M.; De Fazio, M.; Altomare, D.F.; Luposella, M.; Maltese, L.; Currò, G.; Marone, G.; et al. Mast Cells, Angiogenesis and Lymphangiogenesis in Human Gastric Cancer. Int. J. Mol. Sci. 2019, 20, 2106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komi, D.E.A.; Redegeld, F.A. Role of Mast Cells in Shaping the Tumor Microenvironment. Clin. Rev. Allergy Immunol. 2020, 58, 313–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aponte-López, A.; Muñoz-Cruz, S. Mast Cells in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1273, 159–173. [Google Scholar] [CrossRef]
- Schreiber, S.; Hammers, C.M.; Kaasch, A.J.; Schraven, B.; Dudeck, A.; Kahlfuss, S. Metabolic Interdependency of Th2 Cell-Mediated Type 2 Immunity and the Tumor Microenvironment. Front. Immunol. 2021, 12, 632581. [Google Scholar] [CrossRef]
- Rui, X.; Shao, S.; Wang, L.; Leng, J. Identification of recurrence marker associated with immune infiltration in prostate cancer with radical resection and build prognostic nomogram. BMC Cancer 2019, 19, 1179. [Google Scholar] [CrossRef]
- Guéry, L.; Hugues, S. Th17 Cell Plasticity and Functions in Cancer Immunity. BioMed Res. Int. 2015, 2015, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Jiang, Y.; Li, G.; Fisher, G.A.; Li, R. Natural killer cell and stroma abundance are independently prognostic and predict gastric cancer chemotherapy benefit. JCI Insight 2020, 5, e136570. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wu, X.; Chen, Y. Stromal-Immune Score-Based Gene Signature: A Prognosis Stratification Tool in Gastric Cancer. Front. Oncol. 2019, 9, 1212. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Heinz, A.; Jahn, D.; Hiller, K. Complexity of macrophage metabolism in infection. Curr. Opin. Biotechnol. 2021, 68, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-H.; Curtis, J.D.; Maggi, L.B.; Faubert, B.; Villarino, A.V.; O’Sullivan, D.; Huang, S.C.-C.; van der Windt, G.J.; Blagih, J.; Qiu, J.; et al. Posttranscriptional Control of T Cell Effector Function by Aerobic Glycolysis. Cell 2013, 153, 1239–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckermann, K.E.; Dudzinski, S.O.; Rathmell, J.C. Dysfunctional T cell metabolism in the tumor microenvironment. Cytokine Growth Factor Rev. 2017, 35, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Viola, A.; Munari, F.; Sánchez-Rodríguez, R.; Scolaro, T.; Castegna, A. The Metabolic Signature of Macrophage Responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.; Ni, D.; Wali, J.A.; Cox, D.A.; Pinget, G.V.; Taitz, J.; Daïen, C.I.; Senior, A.; Read, M.N.; Simpson, S.J.; et al. Dietary carbohydrate, particularly glucose, drives B cell lymphopoiesis and function. iScience 2021, 24, 102835. [Google Scholar] [CrossRef]
- Mendoza, R.; Fudge, D.; Brown, J. Cellular Energetics of Mast Cell Development and Activation. Cells 2021, 10, 524. [Google Scholar] [CrossRef]
- Gerriets, V.A.; Kishton, R.J.; Nichols, A.G.; Macintyre, A.; Inoue, M.; Ilkayeva, O.; Winter, P.S.; Liu, X.; Priyadharshini, B.; Slawinska, M.E.; et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Investig. 2015, 125, 194–207. [Google Scholar] [CrossRef]
- Böttcher, M.; DHofmann, A.; Bruns, H.; Haibach, M.; Loschinski, R.; Saul, D.; Mougiakakos, D. Mesenchymal Stromal Cells Disrupt mTOR-Signaling and Aerobic Glycolysis During T-Cell Activation. Stem cells (Dayton, OH, USA). Stem Cells 2016, 34, 516–521. [Google Scholar] [CrossRef] [Green Version]



| Characteristic | Low Expression of PDK4 | High Expression of PDK4 | p Value |
|---|---|---|---|
| n | 187 | 188 | |
| Age, n (%) | 0.232 | ||
| ≤65 | 76 (20.5%) | 88 (23.7%) | |
| >65 | 110 (29.6%) | 97 (26.1%) | |
| Gender, n (%) | 0.352 | ||
| Female | 62 (16.5%) | 72 (19.2%) | |
| Male | 125 (33.3%) | 116 (30.9%) | |
| H pylori infection, n (%) | 1.000 | ||
| No | 80 (49.1%) | 65 (39.9%) | |
| Yes | 10 (6.1%) | 8 (4.9%) | |
| T stage, n (%) | 0.211 | ||
| T1 | 12 (3.3%) | 7 (1.9%) | |
| T2 | 41 (11.2%) | 39 (10.6%) | |
| T3 | 89 (24.3%) | 79 (21.5%) | |
| T4 | 42 (11.4%) | 58 (15.8%) | |
| N stage, n (%) | 0.056 | ||
| N0 | 63 (17.6%) | 48 (13.4%) | |
| N1 | 49 (13.7%) | 48 (13.4%) | |
| N2 | 39 (10.9%) | 36 (10.1%) | |
| N3 | 27 (7.6%) | 47 (13.2%) | |
| M stage, n (%) | 0.605 | ||
| M0 | 170 (47.9%) | 160 (45.1%) | |
| M1 | 11 (3.1%) | 14 (3.9%) | |
| Pathologic stage, n (%) | 0.277 | ||
| Stage I | 30 (8.5%) | 23 (6.5%) | |
| Stage II | 63 (17.9%) | 48 (13.6%) | |
| Stage III | 70 (19.9%) | 80 (22.7%) | |
| Stage IV | 17 (4.8%) | 21 (6%) | |
| OS event, n (%) | <0.001 | ||
| Alive | 131 (34.9%) | 97 (25.9%) | |
| Dead | 56 (14.9%) | 91 (24.3%) | |
| DSS event, n (%) | 0.003 | ||
| Alive | 145 (41%) | 118 (33.3%) | |
| Dead | 33 (9.3%) | 58 (16.4%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Han, S.; Ouyang, S.; Zeng, Z.; Liu, Z.; Sun, J.; Kang, W. PDK4 Constitutes a Novel Prognostic Biomarker and Therapeutic Target in Gastric Cancer. Diagnostics 2022, 12, 1101. https://doi.org/10.3390/diagnostics12051101
Zhang Z, Han S, Ouyang S, Zeng Z, Liu Z, Sun J, Kang W. PDK4 Constitutes a Novel Prognostic Biomarker and Therapeutic Target in Gastric Cancer. Diagnostics. 2022; 12(5):1101. https://doi.org/10.3390/diagnostics12051101
Chicago/Turabian StyleZhang, Zimu, Shiyuan Han, Siwen Ouyang, Ziyang Zeng, Zhen Liu, Juan Sun, and Weiming Kang. 2022. "PDK4 Constitutes a Novel Prognostic Biomarker and Therapeutic Target in Gastric Cancer" Diagnostics 12, no. 5: 1101. https://doi.org/10.3390/diagnostics12051101
APA StyleZhang, Z., Han, S., Ouyang, S., Zeng, Z., Liu, Z., Sun, J., & Kang, W. (2022). PDK4 Constitutes a Novel Prognostic Biomarker and Therapeutic Target in Gastric Cancer. Diagnostics, 12(5), 1101. https://doi.org/10.3390/diagnostics12051101





