MRI-Based Radiomics Models to Discriminate Hepatocellular Carcinoma and Non-Hepatocellular Carcinoma in LR-M According to LI-RADS Version 2018
Abstract
:1. Introduction
2. Materials and Methods
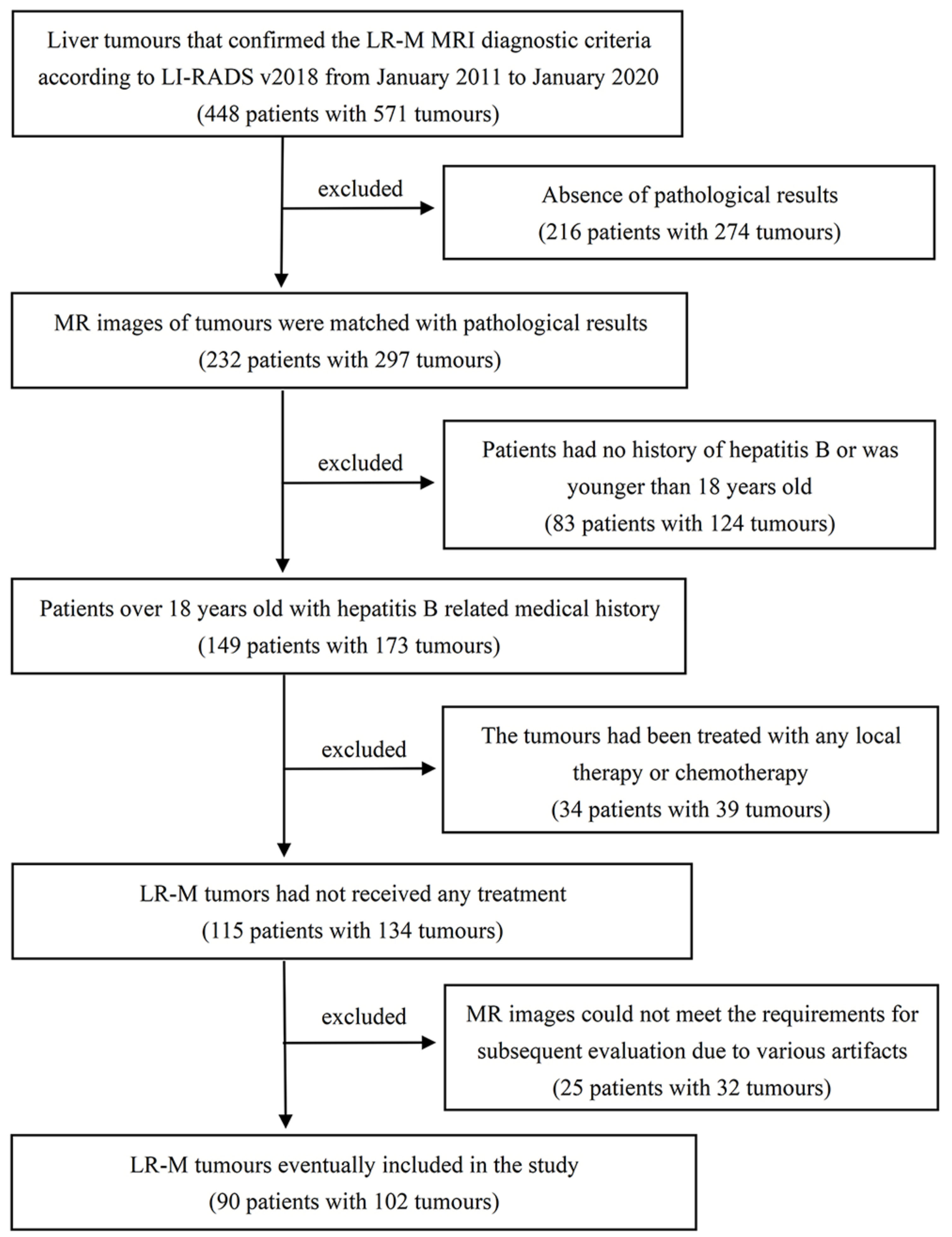
2.1. Study Population
2.2. MR Examination
2.3. Image Analysis
2.4. Histopathologic Evaluation
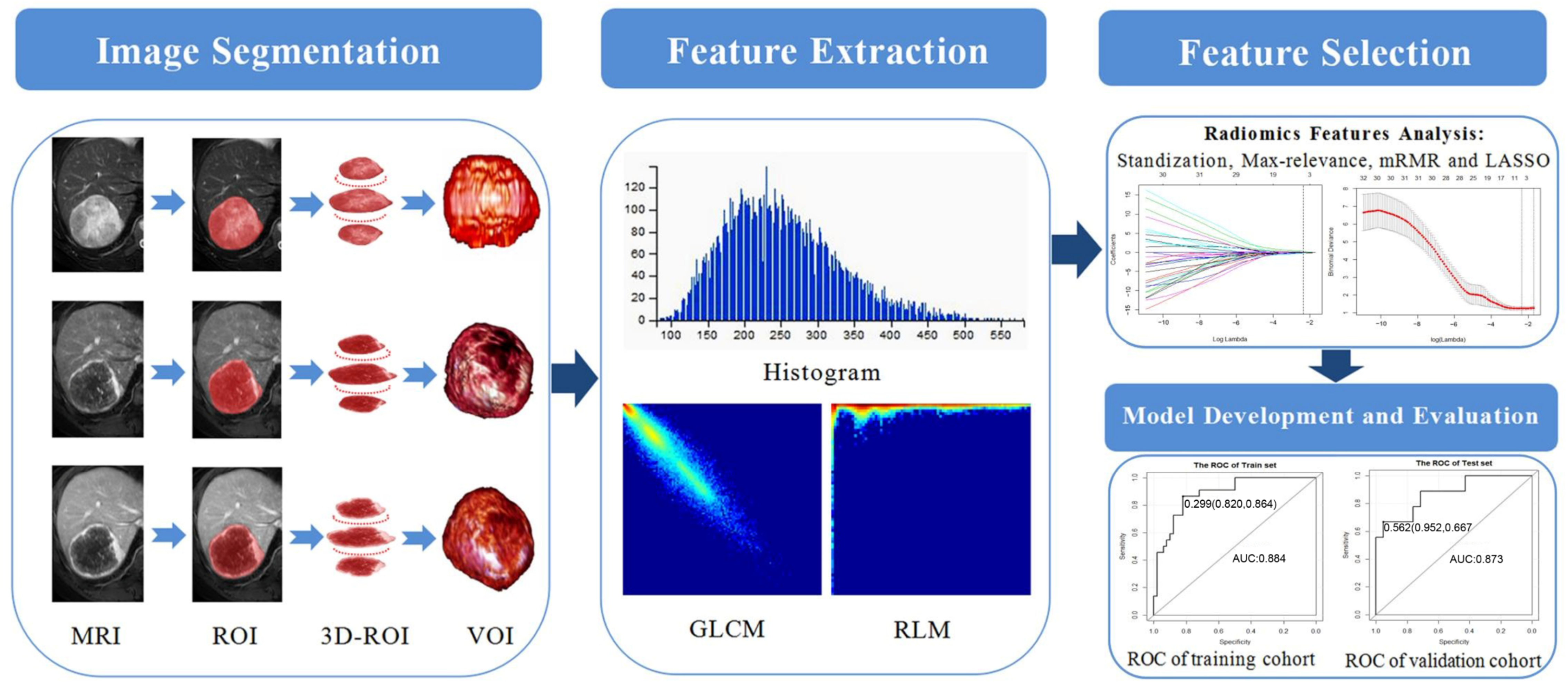
2.5. Image Segmentation and Radiomics Feature Extraction
2.6. Radiomics Features Analysis
2.7. Model Construction and Validation
2.8. Statistical Analysis
3. Results
3.1. Tumour Characteristics
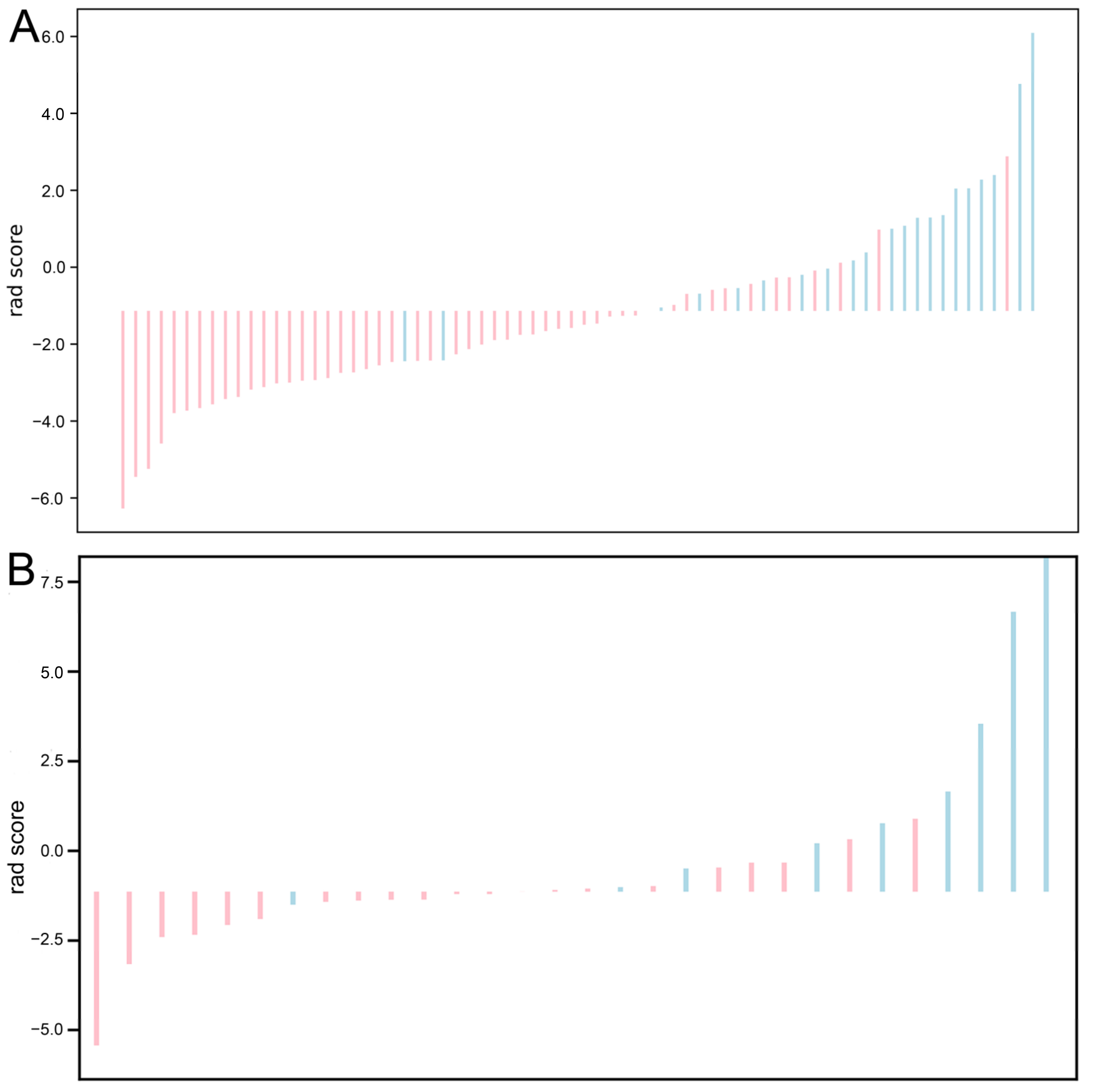
3.2. Feature Selection and Radiomics Signature Construction
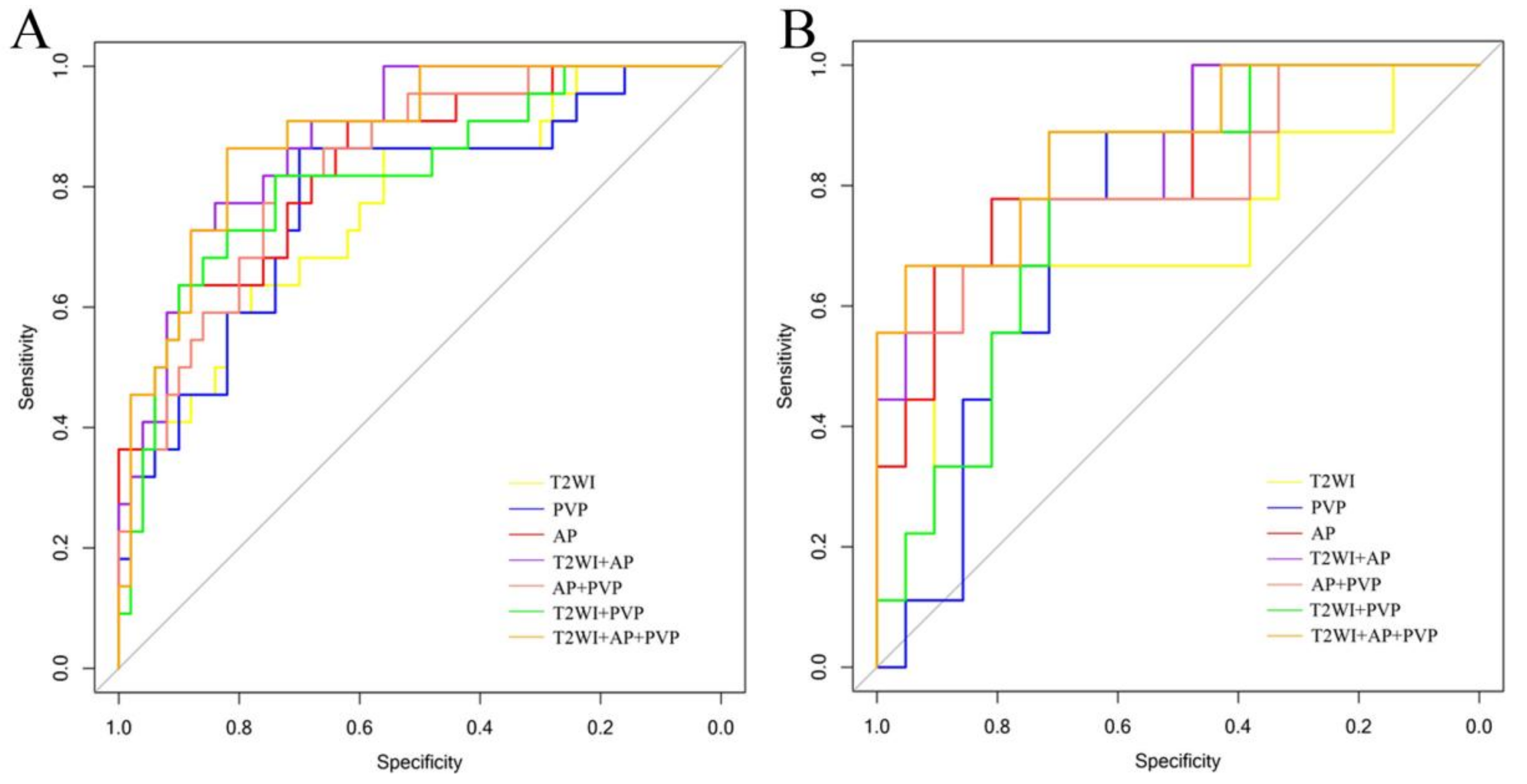
3.3. Performance of Radiomics Models and Verification
3.4. Classification Performance Verification of Visual Evaluation and Imaging Features
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liver Imaging Reporting and Data System Version 2018. American College of Radiology. 2018. Available online: https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/LI-RADS/CT-MRI-LI-RADS-v2018 (accessed on 30 May 2021).
- Centonze, L.; de Carlis, R.; Vella, I.; Carbonaro, L.; Incarbone, N.; Palmieri, L.; Sgrazzutti, C.; Ficarelli, A.; Valsecchi, M.G.; Dello Iacono, U.; et al. From LI-RADS Classification to HCC Pathology: A Retrospective Single-Institution Analysis of Clinico-Pathological Features Affecting Oncological Outcomes after Curative Surgery. Diagnostics 2022, 12, 160. [Google Scholar] [CrossRef] [PubMed]
- Centonze, L.; Di Sandro, S.; Lauterio, A.; de Carlis, R.; Frassoni, S.; Rampoldi, A.; Tuscano, B.; Bagnardi, V.; Vanzulli, A.; de Carlis, L. Surgical Resection vs. Percutaneous Ablation for Single Hepatocellular Carcinoma: Exploring the Impact of Li-RADS Classification on Oncological Outcomes. Cancers 2021, 13, 1671. [Google Scholar] [CrossRef] [PubMed]
- Piñero, F.; Thompson, M.A.; Diaz Telli, F.; Trentacoste, J.; Padín, C.; Mendizabal, M.; Colaci, C.; Gonzalez Campaña, A.; Pages, J.; Montal, S.; et al. LI-RADS 4 or 5 categorization may not be clinically relevant for decision-making processes: A prospective cohort study. Ann. Hepatol. 2020, 19, 662–667. [Google Scholar] [CrossRef]
- Centonze, L.; Di Sandro, S.; Lauterio, A.; de Carlis, R.; Sgrazzutti, C.; Ciulli, C.; Vella, I.; Vicentin, I.; Incarbone, N.; Bagnardi, V.; et al. A retrospective single-centre analysis of the oncological impact of LI-RADS classification applied to Metroticket 2.0 calculator in liver transplantation: Every nodule matters. Transpl. Int. 2021, 34, 1712–1721. [Google Scholar] [CrossRef] [PubMed]
- Van der Pol, C.B.; Lim, C.S.; Sirlin, C.B.; McGrath, T.A.; Salameh, J.P.; Bashir, M.R.; Tang, A.; Singal, A.G.; Costa, A.F.; Fowler, K.; et al. Accuracy of the Liver Imaging Reporting and Data System in Computed Tomography and Magnetic Resonance Image Analysis of Hepatocellular Carcinoma or Overall Malignancy-A Systematic Review. Gastroenterology 2019, 156, 976–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forner, A.; Reig, M.E.; de Lope, C.R.; Bruix, J. Current strategy for staging and treatment: The BCLC update and future prospects. Semin. Liver Dis. 2010, 30, 61–74. [Google Scholar] [CrossRef]
- Yin, X.; Zhang, B.H.; Qiu, S.J.; Ren, Z.G.; Zhou, J.; Chen, X.H.; Zhou, Y.; Fan, J. Combined hepatocellular carcinoma and cholangiocarcinoma: Clinical features, treatment modalities, and prognosis. Ann. Surg. Oncol. 2012, 19, 2869–2876. [Google Scholar] [CrossRef]
- Shao, S.; Liang, Y.; Kuang, S.; Chen, J.; Shan, Q.; Yang, H.; Zhang, Y.; Wang, B.; J Fowler, K.; Wang, J.; et al. Diagnostic performance of LI-RADS version 2018 in differentiating hepatocellular carcinoma from other hepatic malignancies in patients with hepatitis B virus infection. Bosn. J. Basic. Med. Sci. 2020, 20, 401–410. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.; Kwon, H.; Cho, J. Inter-reader agreement and imaging-pathology correlation of the LI-RADS M on gadoxetic acid-enhanced magnetic resonance imaging: Efforts to improve diagnostic performance. Abdom. Radiol. 2020, 45, 2430–2439. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, Y.K.; Cha, D.I.; Ko, S.E.; Kim, S.; Lee, E.S.; Ahn, S. Targetoid hepatic observations on gadoxetic acid-enhanced MRI using LI-RADS version 2018: Emphasis on hepatocellular carcinomas assigned to the LR-M category. Clin. Radiol. 2020, 75, 478.e13–478.e23. [Google Scholar] [CrossRef]
- Horva, N.; Nikolovski, I.; Long, N.; Gerst, S.; Zheng, J.; Pak, L.M.; Simpson, A.; Zheng, J.; Capanu, M.; Jarnagin, W.R.; et al. Imaging features of hepatocellular carcinoma compared to intrahepatic cholangiocarcinoma and combined tumor on MRI using liver imaging and data system (LI-RADS) version 2014. Abdom. Radiol. 2018, 43, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Ronot, M.; Fouque, O.; Esvan, M.; Lebigot, J.; Aubé, C.; Vilgrain, V. Comparison of the accuracy of AASLD and LI-RADS criteria for the non-invasive diagnosis of HCC smaller than 3 cm. J. Hepatol. 2018, 68, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiessling, F. The changing face of cancer diagnosis: From computational image analysis to systems biology. Eur. Radiol. 2018, 28, 3160–3164. [Google Scholar] [CrossRef] [Green Version]
- Zheng, B.H.; Liu, L.Z.; Zhang, Z.Z.; Shi, J.Y.; Dong, L.Q.; Tian, L.Y.; Ding, Z.B.; Ji, Y.; Rao, S.X.; Zhou, J.; et al. Radiomics score: A potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer 2018, 18, 1148. [Google Scholar] [CrossRef]
- Jiang, H.Y.; Liu, X.J.; Chen, J.; Wei, Y.; Lee, J.M.; Cao, L.; Wu, Y.; Duan, T.; Li, X.; Ma, L.; et al. Man or machine? Prospective comparison of the version 2018 EASL, LI-RADS criteria and a radiomics model to diagnose hepatocellular carcinoma. Cancer Imaging 2019, 19, 84. [Google Scholar] [CrossRef] [Green Version]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Terzi, E.; Iavarone, M.; Pompili, M.; Veronese, L.; Cabibbo, G.; Fraquelli, M.; Riccardi, L.; de Bonis, L.; Sangiovanni, A.; Leoni, S.; et al. Contrast ultrasound LI-RADS LR-5 identifies hepatocellular carcinoma in cirrhosis in a multicenter restropective study of 1,006 nodules. J. Hepatol. 2018, 68, 485–492. [Google Scholar] [CrossRef]
- Wilson, S.R.; Lyshchik, A.; Piscaglia, F.; Cosgrove, D.; Jang, H.J.; Sirlin, C.; Dietrich, C.F.; Kim, T.K.; Willmann, J.K.; Kono, Y. CEUS LI-RADS: Algorithm, implementation, and key differences from CT/MRI. Abdom. Radiol. 2018, 43, 127–142. [Google Scholar] [CrossRef]
- Kierans, A.S.; Makkar, J.; Guniganti, P.; Cornman-Homonoff, J.; Lee, M.J.; Pittman, M.; Askin, G.; Hecht, E.M. Validation of Liver Imaging Reporting and Data System 2017 (LI-RADS) Criteria for Imaging Diagnosis of Hepatocellular Carcinoma. J. Magn. Reason. Imaging 2019, 49, e205–e215. [Google Scholar] [CrossRef]
- Lee, S.M.; Lee, J.M.; Ahn, S.J.; Kang, H.J.; Yang, H.K.; Yoon, J.H. LI-RADS Version 2017 versus Version 2018: Diagnosis of Hepatocellular Carcinoma on Gadoxetate Disodium-enhanced MRI. Radiology 2019, 292, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Fowler, K.J.; Sheybani, A.; Parker, R.A.; Doherty, S.; MBrunt, E.; Chapman, W.C.; Menias, C.O. Combined hepatocellular and cholangiocarcinoma (biphenotypic) tumors: Imaging features and diagnostic accuracy of contrast-enhanced CT and MRI. Am. J. Roentgenol. 2013, 201, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parma, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Wu, J.J.; Liu, A.L.; Cui, J.J.; Chen, A.L.; Song, Q.W.; Xie, L.Z. Radiomics-based classification of hepatocellular carcinoma and hepatic haemangioma on precontrast magnetic resonance images. BMC Med. Imaging 2019, 19, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, B.; Zhang, L.Z.; Ning, P.G.; Ding, F.; Wu, F.T.; Lu, G.; Geng, Y.; Ma, J. Preoperative prediction for pathological grade of hepatocellular carcinoma via machine learning–based radiomics. Eur. Radiol. 2020, 30, 6924–6932. [Google Scholar] [CrossRef]
- Song, W.L.; Yu, X.L.; Guo, D.J.; Liu, H.; Tang, Z.Y.; Liu, X.J.; Zhou, J.; Zhang, H.P.; Liu, Y.Y.; Liu, X. MRI-Based Radiomics: Associations With the Recurrence-Free Survival of Patients With Hepatocellular Carcinoma Treated With Conventional Transcatheter Arterial Chemoembolization. J. Magn. Reason. Imaging 2020, 52, 461–473. [Google Scholar] [CrossRef]
- Wu, M.H.; Tan, H.N.; Gao, F.; Hai, J.J.; Ning, P.G.; Chen, J.; Zhu, S.C.; Wang, M.Y.; Dou, S.W.; Shi, D.P. Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur. Radiol. 2019, 29, 2802–2811. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, G.Z.; Duan, C.F.; Zhou, X.M.; Wang, X.; Yu, H.Y.; Yu, L.; Li, Z.M.; Gao, Y.X.; Zhao, R.R.; et al. Radiomics Analysis of MR Imaging with Gd-EOB-DTPA for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma: Investigation and Comparison of Different Hepatobiliary Phase Delay Times. Biomed. Res. Int. 2021, 2021, 6685723. [Google Scholar] [CrossRef]
- Feng, S.T.; Jia, Y.M.; Liao, B.; Huang, B.S.; Zhou, Q.; Li, X.; Wei, K.K.; Chen, L.L.; Li, B.; Wang, W.; et al. Preoperative prediction of microvascular invasion in hepatocellular cancer: A radiomics model using Gd-EOB-DTPA-enhanced MRI. Eur Radiol. 2019, 29, 4648–4659. [Google Scholar] [CrossRef]
- Zhong, X.; Guan, T.P.; Tang, D.R.; Li, J.S.; Lu, B.G.; Cui, S.Z.; Tang, H.S. Differentiation of small (≤3 cm) hepatocellular carcinomas from benign nodules in cirrhotic liver: The added additive value of MRI-based radiomics analysis to LI-RADS version 2018 algorithm. BMC Gastroenterol. 2021, 21, 155. [Google Scholar] [CrossRef]
- Liu, D.F.; Zhang, X.H.; Zheng, T.; Shi, Q.L.; Cui, Y.J.; Wang, Y.J.; Liu, L.X. Optimisation and evaluation of the random forest model in the efficacy prediction of chemoradiotherapy for advanced cervical cancer based on radiomics signature from high-resolution T2 weighted images. Arch. Gynecol. Obstet. 2021, 303, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Shen, F.; Jia, Y.; Xia, Y.; Li, Q.; Lu, J. MRI-based radiomics of rectal cancer: Preoperative assessment of the pathological features. BMC Med. Imaging. 2019, 19, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Xu, L.; Wen, X.; Zhang, J.H.; Yang, P.F.; Zhang, L.X.; Xue, X.; Wang, X.L.; Huang, Q.; Guo, C.G.; et al. A nomogram based on bi-regional radiomics features from multimodal magnetic resonance imaging for preoperative prediction of microvascular invasion in hepatocellular carcinoma. Quant. Imaging Med. Surg. 2019, 9, 1503–1515. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Y.; Franc, B.L.; Harnish, R.J.; Liu, G.; Mitra, D.; Copeland, T.P.; Arasu, V.A.; Kornak, J.; Jones, E.F.; Behr, S.C.; et al. Exploration of PET and MRI radiomic features for decoding breast cancer phenotypes and prognosis. NPJ Breast Cancer 2018, 4, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.; Nam, Y.; Jang, J.; Shin, N.Y.; Ahn, K.J.; Kim, B.S.; Lee, Y.S.; Kim, M.S. Prediction of Human Papillomavirus Status and Overall Survival in Patients with Untreated Oropharyngeal Squamous Cell Carcinoma: Development and Validation of CT-Based Radiomics. Am. J. Neuroradiol. 2020, 41, 1897–1904. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Zhang, X.Y.; Shi, Y.J.; Wang, L.; Zhu, H.T.; Tang, Z.C.; Wang, S.; Li, X.T.; Tian, J.; Sun, Y.S. Radiomics Analysis for Evaluation of Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Clin. Cancer Res. 2017, 23, 7253–7262. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.; Zhi, X.; Sun, Y.; Li, H.R.; Wang, J.J.; Xu, J.X.; Guo, J. A Nomogram Based on a Multiparametric Ultrasound Radiomics Model for Discrimination Between Malignant and Benign Prostate Lesions. Front. Oncol. 2021, 11, 610785. [Google Scholar] [CrossRef]
- Jin, Z.C.; Chen, L.; Zhong, B.Y.; Zhou, H.F.; Zhu, H.D.; Zhou, H.; Song, J.J.; Guo, J.H.; Zhu, X.L.; Ji, J.S.; et al. Machine-learning analysis of contrast-enhanced computed tomography radiomics predicts patients with hepatocellular carcinoma who are unsuitable for initial transarterial chemoembolization monotherapy: A multicenter study. Transl. Oncol. 2021, 14, 101034. [Google Scholar] [CrossRef]


| Characteristics | Training Cohort (n = 72) | Validation Cohort (n = 30) | p | ||
|---|---|---|---|---|---|
| HCC (n = 22) | Non-HCC (n = 50) | HCC (n = 9) | Non-HCC (n = 21) | ||
| Age | 0.269 | ||||
| ≤50 years | 10 (13.8%) | 20 (27.8%) | 5 (16.7%) | 4 (13.3%) | |
| >50 years | 12 (16.7%) | 30 (41.7%) | 4 (13.3%) | 17 (56.7%) | |
| Sex | 0.204 | ||||
| Male | 17 (23.6%) | 31 (43.1%) | 6 (20.0%) | 10 (33.3%) | |
| Female | 5 (6.9%) | 19 (26.4%) | 3 (10.0%) | 11 (36.7%) | |
| Location | 0.523 | ||||
| Left | 12 (16.7%) | 17 (23.6%) | 6(20.0%) | 6 (20.0%) | |
| Right | 8 (11.1%) | 27 (37.5%) | 3 (10.0%) | 9 (30.0%) | |
| Junction | 2 (2.8%) | 6 (8.3%) | 0 (0.0%) | 6 (20.0%) | |
| Tumour size (cm) | 0.550 | ||||
| ≤5 cm | 14 (19.5%) | 27 (37.5%) | 6 (20.0%) | 13 (43.3%) | |
| >5 cm | 8 (11.1%) | 23 (31.9%) | 3 (10.0%) | 8 (26.7%) | |
| Tumour Number | 0.867 | ||||
| One | 22 (30.6%) | 37 (51.4%) | 9 (30.0%) | 16 (53.3%) | |
| Multiple | 0 (0%) | 13 (18.0%) | 0 (0.0%) | 5 (16.7%) | |
| Pathological diagnosis | 1.000 | ||||
| HCC | 22 (30.6%) | - | 9 (30.0%) | - | |
| ICCA | - | 38 (52.7%) | - | 15 (50.0%) | |
| CHC | - | 1 (1.4%) | - | 0 (0%) | |
| metastases | - | 8 (11.1%) | - | 6 (20.0%) | |
| carcinosarcomas | - | 3 (4.2%) | - | 0 (0%) | |
| ALT | 0.178 | ||||
| ≤50 U/L | 17 (23.6%) | 38 (52.8%) | 5 (16.7%) | 14 (46.7%) | |
| >50 U/L | 5 (6.9%) | 12 (16.7%) | 4 (13.3%) | 7(23.3%) | |
| AST | 0.115 | ||||
| ≤40 U/L | 17 (23.6%) | 31 (43.1%) | 4 (13.3%) | 11 (36.7%) | |
| >40 U/L | 5 (6.9%) | 19 (26.4%) | 5 (16.7%) | 10 (33.3%) | |
| γ-GT | 0.209 | ||||
| ≤60 U/L | 12 (16.7%) | 14 (19.5%) | 4 (13.3%) | 3 (10.0%) | |
| >60 U/L | 10 (13.8%) | 36 (50.0%) | 5 (16.7%) | 18 (60.0%) | |
| ALP | 0.138 | ||||
| ≤125 U/L | 17 (23.6%) | 21 (29.2%) | 6 (20.0%) | 5 (16.7%) | |
| >125 U/L | 5 (6.9%) | 29 (40.3%) | 3 (10.0%) | 16 (53.3%) | |
| TBiL | 0.095 | ||||
| ≤20.4 µmol/L | 18 (25.0%) | 39 (54.2%) | 7 (23.3%) | 12 (40.0%) | |
| >20.4 µmol/L | 4 (5.5%) | 11 (15.3%) | 2 (6.7%) | 9 (30.0%) | |
| DBiL | 0.246 | ||||
| ≤11 µmol/L | 19 (26.4%) | 41 (56.9%) | 7 (23.3%) | 15 (50.0%) | |
| >11 µmol/L | 3 (4.2%) | 9 (12.5%) | 2 (6.7%) | 6 (20.0%) | |
| IBiL | 0.181 | ||||
| ≤10.2 µmol/L | 18 (25.0%) | 39 (54.2%) | 7 (23.3%) | 13 (43.3%) | |
| >10.2 µmol/L | 4 (5.5%) | 11 (15.3%) | 2 (6.7%) | 8 (26.7%) | |
| TP | 0.504 | ||||
| ≤85 g/L | 22 (30.5%) | 49 (68.1%) | 9 (30.0%) | 20 (66.7%) | |
| >85 g/L | 0 (0.0%) | 1 (1.4%) | 0 (0.0%) | 1 (3.3%) | |
| Albumin | 0.504 | ||||
| ≤55 g/L | 22 (30.5%) | 48 (66.7%) | 8 (26.7%) | 21 (70.0%) | |
| >55 g/L | 0 (0.0%) | 2 (2.8%) | 1 (3.3%) | 0 (0.0%) | |
| AFP | 0.411 | ||||
| ≤13.2 µg/L | 12 (16.7%) | 40 (55.7%) | 5 (16.7%) | 19 (63.3%) | |
| >13.2 µg/L | 10 (13.8%) | 10 (13.8%) | 4 (13.3%) | 2 (6.7%) | |
| CEA | 0.117 | ||||
| ≤5.5 ng/mL | 19 (26.4%) | 33 (45.8%) | 8 (26.7%) | 18 (60.0%) | |
| >5.5 ng/mL | 3 (4.2%) | 17 (23.6%) | 1 (3.3%) | 3 (10.0%) | |
| CA19-9 | 0.613 | ||||
| ≤37 U/mL | 16 (22.2%) | 31 (43.1%) | 8 (26.7%) | 10 (33.3%) | |
| >37 U/mL | 6 (8.3%) | 19 (26.4%) | 1 (3.3%) | 11 (36.7%) | |
| Model | Valuable Features | ||||||
|---|---|---|---|---|---|---|---|
| Original | Wavelet | ||||||
| Firstorder | Shape | GLCM | Firstorder | GLCM | NGTDM | GLSZM | |
| M1 | LLL_Skewness | HHL_Imc1 | |||||
| LLH_10Percentile | |||||||
| M2 | Minimum | Idmn | LHL_Mean | LLH_DependenceVariance | HLL_LargeAreaLowGrayLevelEmphasis | ||
| HHL_Kurtosis | LHL_Idn | ||||||
| M3 | Minimum | LLH_Skewness | LHL_Idmn | HLH_Strength | |||
| M4 | Minimum | Idmn | LHL_Mean | HHL_MCC | |||
| Skewness | |||||||
| M5 | Minimum | LLH_Skewness | |||||
| LLH_10Percentile | |||||||
| LLL_Skewness | |||||||
| M6 | Minimum | LHL_Mean | |||||
| HHL_Kurtosis | |||||||
| M7 | Minimum | Flatness | LHL_Mean | LHL_Idn | |||
| Skewness | LLH_RobustMeanAbsoluteDeviation | LHL_Idmn | |||||
| Model | Training Cohort (n = 72) | Validation Cohort (n = 30) | Delong | ||||
|---|---|---|---|---|---|---|---|
| AUC (95% CI) | Sensitivity | Specificity | AUC (95% CI) | Sensitivity | Specificity | ||
| M1 | 0.768 (0.647–0.889) | 0.56 | 0.864 | 0.759 (0.474–0.944) | 0.905 | 0.667 | 0.663 |
| M2 | 0.838 (0.739–0.938) | 0.9 | 0.636 | 0.836 (0.674–0.998) | 0.81 | 0.778 | 0.7 |
| M3 | 0.778 (0.657–0.900) | 0.7 | 0.864 | 0.762 (0.590–0.934) | 0.619 | 0.889 | 0.88 |
| M4 | 0.880 (0.802–0.958) | 0.84 | 0.773 | 0.836 (0.676–0.996) | 0.857 | 0.667 | 0.631 |
| M5 | 0.818 (0.707–0.929) | 0.74 | 0.818 | 0.783 (0.611–0.955) | 0.714 | 0.889 | 0.738 |
| M6 | 0.832 (0.735–0.929) | 0.74 | 0.818 | 0.815 (0.624–1.000) | 1 | 0.556 | 0.877 |
| M7 | 0.884 (0.804–0.963) | 0.82 | 0.864 | 0.873 (0.728–1.000) | 0.952 | 0.667 | 0.9 |
| Variable | AUC | 95% CI | Sensitivity | Specificity |
|---|---|---|---|---|
| Junior radiologist | 0.64 | 0.539–0.733 | 0.548 | 0.732 |
| Senior radiologist | 0.799 | 0.708–0.871 | 0.71 | 0.887 |
| Rim APHE | 0.723 | 0.625–0.807 | 0.516 | 0.93 |
| Peripheral washout | 0.685 | 0.585–0.773 | 0.581 | 0.789 |
| Delay central enhancement | 0.72 | 0.623–0.805 | 0.806 | 0.634 |
| Two targetoid masses | 0.621 | 0.519–0.715 | 0.903 | 0.338 |
| All targetoid masses | 0.754 | 0.658–0.833 | 1 | 0.507 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Guo, D.; Liu, H.; He, X.; Qiao, X.; Liu, X.; Liu, Y.; Zhou, J.; Zhou, Z.; Liu, X.; et al. MRI-Based Radiomics Models to Discriminate Hepatocellular Carcinoma and Non-Hepatocellular Carcinoma in LR-M According to LI-RADS Version 2018. Diagnostics 2022, 12, 1043. https://doi.org/10.3390/diagnostics12051043
Zhang H, Guo D, Liu H, He X, Qiao X, Liu X, Liu Y, Zhou J, Zhou Z, Liu X, et al. MRI-Based Radiomics Models to Discriminate Hepatocellular Carcinoma and Non-Hepatocellular Carcinoma in LR-M According to LI-RADS Version 2018. Diagnostics. 2022; 12(5):1043. https://doi.org/10.3390/diagnostics12051043
Chicago/Turabian StyleZhang, Haiping, Dajing Guo, Huan Liu, Xiaojing He, Xiaofeng Qiao, Xinjie Liu, Yangyang Liu, Jun Zhou, Zhiming Zhou, Xi Liu, and et al. 2022. "MRI-Based Radiomics Models to Discriminate Hepatocellular Carcinoma and Non-Hepatocellular Carcinoma in LR-M According to LI-RADS Version 2018" Diagnostics 12, no. 5: 1043. https://doi.org/10.3390/diagnostics12051043
APA StyleZhang, H., Guo, D., Liu, H., He, X., Qiao, X., Liu, X., Liu, Y., Zhou, J., Zhou, Z., Liu, X., & Fang, Z. (2022). MRI-Based Radiomics Models to Discriminate Hepatocellular Carcinoma and Non-Hepatocellular Carcinoma in LR-M According to LI-RADS Version 2018. Diagnostics, 12(5), 1043. https://doi.org/10.3390/diagnostics12051043






