Predictive Value of Heat-Shock Protein Gene Expression on Severe Neonatal Hypoxic-Ischemic Encephalopathy
Abstract
1. Introduction
2. Materials and Methods
2.1. Subject Selection
2.2. Exclusion Criteria
2.3. HIE Management for TH
2.3.1. RRNA Extraction, Alignment, and Next-Generation Sequencing
2.3.2. Gene Expression Analysis by Real-Time Quantitative RT-PCR (RT-qPCR)
2.4. Imaging Methods
2.5. Classification of Brain MRIs
2.6. Neurodevelopmental (ND) Assessment
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Volpe, J.J. Neurology of the Newborn, 4th ed.; WB Saunders Company: Philadelphia, PA, USA, 2001. [Google Scholar]
- Kurinczuk, J.J.; White-Koning, M.; Badawi, N. Epidemiology of neonatal encephalopathy and hypoxic–ischaemic encephalopathy. Early Hum. Dev. 2010, 86, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Finer, N.; Robertson, C.; Richards, R.; Pinnell, L.; Peters, K. Hypoxic-ischemic encephalopathy in term neonates: Perinatal factors and outcome. J. Pediatr. 1981, 98, 112–117. [Google Scholar] [CrossRef]
- Fernández-López, D.; Natarajan, N.; Ashwal, S.; Vexler, Z.S. Mechanisms of Perinatal Arterial Ischemic Stroke. J. Cereb. Blood Flow Metab. 2014, 34, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Gluckman, P.D.; Wyatt, J.S.; Azzopardi, D.; Ballard, R.; Edwards, A.D.; Ferriero, D.M.; Polin, R.A.; Robertson, C.M.; Thoresen, M.; Whitelaw, A.; et al. Selective head cooling with mild systemic hypothermia after neonatal encephalopathy: Multicentre randomised trial. Lancet 2005, 365, 663–670. [Google Scholar] [CrossRef]
- Shankaran, S.; Laptook, A.R.; Ehrenkranz, R.A.; Tyson, J.E.; McDonald, S.A.; Donovan, E.F.; Fanaroff, A.A.; Poole, W.K.; Wright, L.L.; Higgins, R.D.; et al. Whole-Body Hypothermia for Neonates with Hypoxic–Ischemic Encephalopathy. N. Engl. J. Med. 2005, 353, 1574–1584. [Google Scholar] [CrossRef]
- Locci, E.; Bazzano, G.; Demontis, R.; Chighine, A.; Fanos, V.; D’Aloja, E. Exploring Perinatal Asphyxia by Metabolomics. Metabolites 2020, 10, 141. [Google Scholar] [CrossRef]
- Samaiya, P.K.; Krishnamurthy, S.; Kumar, A. Mitochondrial dysfunction in perinatal asphyxia: Role in pathogenesis and potential therapeutic interventions. Mol. Cell. Biochem. 2021, 476, 4421–4434. [Google Scholar] [CrossRef]
- Lafemina, M.J.; Sheldon, R.A.; Ferriero, D.M. Acute Hypoxia-Ischemia Results in Hydrogen Peroxide Accumulation in Neonatal But Not Adult Mouse Brain. Pediatr. Res. 2006, 59, 680–683. [Google Scholar] [CrossRef]
- Matara, D.-I.; Pouliakis, A.; Xanthos, T.; Sokou, R.; Kafalidis, G.; Iliodromiti, Z.; Boutsikou, T.; Iacovidou, N.; Salakos, C. Microbial Translocation and Perinatal Asphyxia/Hypoxia: A Systematic Review. Diagnostics 2022, 12, 214. [Google Scholar] [CrossRef]
- Debuf, M.J.; Carkeek, K.; Piersigilli, F. A Metabolomic Approach in Search of Neurobiomarkers of Perinatal Asphyxia: A Review of the Current Literature. Front. Pediatr. 2021, 9, 674585. [Google Scholar] [CrossRef]
- Bersani, I.; Pluchinotta, F.; Dotta, A.; Savarese, I.; Campi, F.; Auriti, C.; Chuklantseva, N.; Piersigilli, F.; Gazzolo, F.; Varrica, A.; et al. Early predictors of perinatal brain damage: The role of neurobiomarkers. Clin. Chem. Lab. Med. (CCLM) 2019, 58, 471–486. [Google Scholar] [CrossRef] [PubMed]
- Tirapelli, D.P.D.C.; Junior, C.C.; Leite, J.; Tirapelli, L.F.; Colli, B.O. Expression of HSP70 in cerebral ischemia and neuroprotetive action of hypothermia and ketoprofen. Arq. Neuropsiquiatr. 2010, 68, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Bustelo, M.; Barkhuizen, M.; Hove, D.L.A.V.D.; Steinbusch, H.W.M.; Bruno, M.A.; Loidl, C.F.; Gavilanes, A.W. Clinical Implications of Epigenetic Dysregulation in Perinatal Hypoxic-Ischemic Brain Damage. Front. Neurol. 2020, 11, 483. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Barua, S.; Huang, M.Y.; Park, J.; Yenari, M.A.; Lee, J.E. Heat Shock Protein 70 (HSP70) Induction: Chaperonotherapy for Neuroprotection after Brain Injury. Cells 2020, 9, 2020. [Google Scholar] [CrossRef]
- Boskabadi, H.; Maamouri, G.; Ghayour-Mobarhan, M.; Bagheri, F.; Zakerihamidi, M.; Mollaey, M.K.; Abbasi, E.; Zareh, A.; Tamannanlo, A. Comparison of the predictive value of prooxidant-antioxidant balance and heat shock proteins in the diagnosis of neonatal asphyxia. Biomed. Res. Ther. 2017, 4, 1327. [Google Scholar] [CrossRef][Green Version]
- Ahmadshah, F.; Mojtaba, S.; Ashraf, M.; Reza, S.; Rana, A.; Majid, G.-M. Association of Neonatal Asphyxia with Serum Levels of Heat Shock Protein 27 in a Small Sample of Newborns. Acta Med. Iran. 2019, 57, 303–307. [Google Scholar]
- Dukay, B.; Csoboz, B.; Tóth, M.E. Heat-Shock Proteins in Neuroinflammation. Front. Pharmacol. 2019, 10, 920. [Google Scholar] [CrossRef]
- Turturici, G.; Sconzo, G.; Geraci, F. Hsp70 and Its Molecular Role in Nervous System Diseases. Biochem. Res. Int. 2011, 2011, 1–18. [Google Scholar] [CrossRef]
- Boskabadi, H.; Omidian, M.; Tavallai, S.; Mohammadi, S.; Parizadeh, M.; Mobarhan, M.G.; Ferns, G.A. Serum Hsp70 Antigen: Early Diagnosis Marker in Perinatal Asphyxia. Iran. J. Pediatr. 2015, 25, e381. [Google Scholar] [CrossRef]
- Sharp, F.R.; Massa, S.M.; Swanson, R.A. Heat-shock protein protection. Trends Neurosci. 1999, 22, 97–99. [Google Scholar] [CrossRef]
- Simbruner, G.; Mittal, R.A.; Rohlmann, F.; Muche, R.; Participants, N.T. Systemic Hypothermia After Neonatal Encephalopathy: Outcomes of neo.nEURO.network RCT. Pediatrics 2010, 126, e771–e778. [Google Scholar] [CrossRef] [PubMed]
- Azzopardi, D.V.; Strohm, B.; Edwards, A.D.; Dyet, L.; Halliday, H.; Juszczak, E.; Kapellou, O.; Levene, M.; Marlow, N.; Porter, E.; et al. Moderate Hypothermia to Treat Perinatal Asphyxial Encephalopathy. N. Engl. J. Med. 2009, 361, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Sarnat, H.B.; Sarnat, M.S. Neonatal encephalopathy following fetal distress. A clinical and electroencephalographic study. Obstet. Gynecol. Surv. 1977, 32, 295. [Google Scholar] [CrossRef]
- Shankaran, S.; McDonald, S.; Laptook, A.R.; Hintz, S.R.; Barnes, P.D.; Das, A.; Pappas, A.; Higgins, R.D.; Ehrenkranz, R.A.; Goldberg, R.N.; et al. Neonatal Magnetic Resonance Imaging Pattern of Brain Injury as a Biomarker of Childhood Outcomes following a Trial of Hypothermia for Neonatal Hypoxic-Ischemic Encephalopathy. J. Pediatr. 2015, 167, 987–993.e3. [Google Scholar] [CrossRef]
- Gunn, A.J.; Wyatt, J.S.; Whitelaw, A.; Barks, J.; Azzopardi, D.; Ballard, R.; Edwards, A.D.; Ferriero, D.M.; Gluckman, P.D.; Polin, R.A.; et al. Therapeutic Hypothermia Changes the Prognostic Value of Clinical Evaluation of Neonatal Encephalopathy. J. Pediatr. 2008, 152, 55–58.e1. [Google Scholar] [CrossRef] [PubMed]
- Thoresen, M.; Hellström-Westas, L.; Liu, X.; de Vries, L.S. Effect of hypothermia on amplitude-integrated electroencephalogram in infants with asphyxia. Pediatrics 2010, 126, e131–e139. [Google Scholar] [CrossRef]
- Massaro, A.N.; Tsuchida, T.; Kadom, N.; El-Dib, M.; Glass, P.; Baumgart, S.; Chang, T. aEEG Evolution during Therapeutic Hypothermia and Prediction of NICU Outcome in Encephalopathic Neonates. Neonatology 2012, 102, 197–202. [Google Scholar] [CrossRef]
- Nowak, T.S. Synthesis of a Stress Protein Following Transient Ischemia in the Gerbil. J. Neurochem. 1985, 45, 1635–1641. [Google Scholar] [CrossRef]
- Dienel, G.A.; Kiessling, M.; Jacewicz, M.; Pulsinelli, W.A. Synthesis of Heat Shock Proteins in Rat Brain Cortex after Transient Ischemia. J. Cereb. Blood Flow Metab. 1986, 6, 505–510. [Google Scholar] [CrossRef]
- Welsh, F.A.; Moyer, D.J.; Harris, V.A. Regional Expression of Heat Shock Protein-70 mRNA and c-Fos mRNA following Focal Ischemia in Rat Brain. J. Cereb. Blood Flow Metab. 1992, 12, 204–212. [Google Scholar] [CrossRef]
- Fernández-Fernández, M.R.; Gragera, M.; Ochoa-Ibarrola, L.; Quintana-Gallardo, L.; Valpuesta, J. Hsp70—A master regulator in protein degradation. FEBS Lett. 2017, 591, 2648–2660. [Google Scholar] [CrossRef] [PubMed]
- Kiang, J.G.; Tsokos, G.C. Heat shock protein 70 kDa: Molecular biology, biochemistry, and physiology. Pharmacol. Ther. 1998, 80, 183–201. [Google Scholar] [CrossRef]
- Yu, W.-W.; Cao, S.-N.; Zang, C.-X.; Wang, L.; Yang, H.-Y.; Bao, X.-Q.; Zhang, D. Heat shock protein 70 suppresses neuroinflammation induced by α-synuclein in astrocytes. Mol. Cell. Neurosci. 2018, 86, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Ma, X.; Liu, Z.; Gu, J.; Zhang, X.; Li, D.; Zhang, S. Different Heat Shock Proteins Bind α-Synuclein With Distinct Mechanisms and Synergistically Prevent Its Amyloid Aggregation. Front. Neurosci. 2019, 13, 1124. [Google Scholar] [CrossRef]
- Kasioumi, P.; Vrazeli, P.; Vezyraki, P.; Zerikiotis, S.; Katsouras, C.; Damalas, A.; Angelidis, C. Hsp70 (HSP70A1A) downregulation enhances the metastatic ability of cancer cells. Int. J. Oncol. 2018, 54, 821–832. [Google Scholar] [CrossRef]
- Schoel, B.; Zügel, U.; Ruppert, T.; Kaufmann, S.H.E. Elongated peptides, not the predicted nonapeptide stimulate a major histocompatibility complex class I-restricted cytotoxic T lymphocyte clone with specificity for a bacterial heat shock protein. Eur. J. Immunol. 1994, 24, 3161–3169. [Google Scholar] [CrossRef]
- Yamamoto, N.; Kinoshita, T.; Nohata, N.; Yoshino, H.; Itesako, T.; Fujimura, L.; Mitsuhashi, A.; Usui, H.; Enokida, H.; Nakagawa, M.; et al. Tumor-suppressive microRNA-29a inhibits cancer cell migration and invasion via targeting HSP47 in cervical squamous cell carcinoma. Int. J. Oncol. 2013, 43, 1855–1863. [Google Scholar] [CrossRef]
- Duarte, B.D.P.; Bonatto, D. The heat shock protein 47 as a potential biomarker and a therapeutic agent in cancer research. J. Cancer Res. Clin. Oncol. 2018, 144, 2319–2328. [Google Scholar] [CrossRef]
- Kelly, K.K.; MacPherson, A.M.; Grewal, H.; Strnad, F.; Jones, J.W.; Yu, J.; Pierzchalski, K.; Kane, M.A.; Herson, P.S.; Siegenthaler, J.A. Col1a1+ perivascular cells in the brain are a source of retinoic acid following stroke. BMC Neurosci. 2016, 17, 1–14. [Google Scholar] [CrossRef]
- Miyamura, T.; Sakamoto, N.; Kakugawa, T.; Taniguchi, H.; Akiyama, Y.; Okuno, D.; Moriyama, S.; Hara, A.; Kido, T.; Ishimoto, H.; et al. Small molecule inhibitor of HSP47 prevents pro-fibrotic mechanisms of fibroblasts in vitro. Biochem. Biophys. Res. Commun. 2020, 530, 561–565. [Google Scholar] [CrossRef]
- Blumenfeld, K.S.; Welsh, F.A.; Harris, V.A.; Pesenson, M.A. Regional Expression of c-Fos and Heat Shock Protein-70 mRNA following Hypoxia-Ischemia in Immature Rat Brain. J. Cereb. Blood Flow Metab. 1992, 12, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Kraio, R.; Nicholson, C. Extracellular ionic variations during spreading depression. Neuroscience 1978, 3, 1045–1059. [Google Scholar] [CrossRef]
- Morgan, J.I.; Curran, T. Role of ion flux in the control of c-fos expression. Nature 1986, 322, 552–555. [Google Scholar] [CrossRef]
- Graham, E.M.; Ruis, K.A.; Hartman, A.; Northington, F.J.; Fox, H.E. A systematic review of the role of intrapartum hypoxia-ischemia in the causation of neonatal encephalopathy. Am. J. Obstet. Gynecol. 2008, 199, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Youn, Y.-A.; Kim, J.H.; Yum, S.-K.; Moon, C.-J.; Lee, I.-G.; Sung, I.K. The hospital outcomes compared between the early and late hypothermia-treated groups in neonates. J. Matern. Neonatal Med. 2015, 29, 2288–2292. [Google Scholar] [CrossRef] [PubMed]
- Awaysheh, F.; Bsharat, A.; Alhmaiedeen, N. Early Biochemical Markers of Hypoxic Ischemic Encephalopathy in Neonates. J. R. Med. Serv. 2016, 23, 6–10. [Google Scholar] [CrossRef][Green Version]
- Massaro, A.N.; Wu, Y.W.; Bammler, T.K.; Comstock, B.; Mathur, A.; McKinstry, R.C.; Chang, T.; Mayock, D.E.; Mulkey, S.B.; Van Meurs, K.; et al. Plasma Biomarkers of Brain Injury in Neonatal Hypoxic-Ischemic Encephalopathy. J. Pediatr. 2018, 194, 67–75. [Google Scholar] [CrossRef]
- Agut, T.; León, M.; Rebollo, M.; Muchart, J.; Arca, G.; Garcia-Alix, A. Early identification of brain injury in infants with hypoxic ischemic encephalopathy at high risk for severe impairments: Accuracy of MRI performed in the first days of life. BMC Pediatr. 2014, 8, 177. [Google Scholar] [CrossRef]
- Semenza, G.L.; Agani, F.; Feldser, D.; Iyer, N.; Kotch, L.; Laughner, E.; Yu, A. Hypoxia, HIF-1, and the pathophysiology of common human diseases. Adv. Exp. Med. Biol. 2000, 475, 123–130. [Google Scholar]
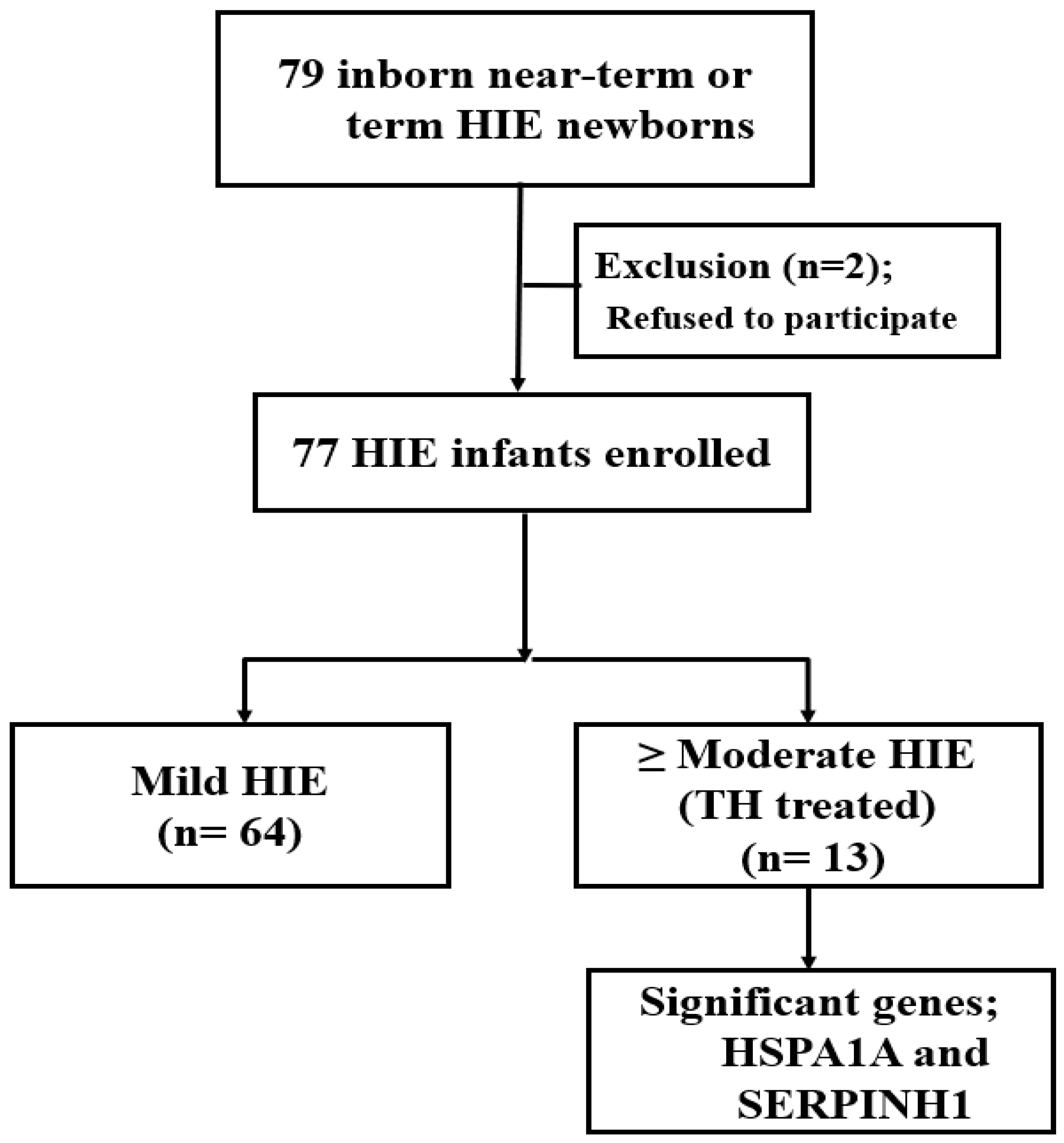
| Mild HIE (n = 64) | ≥Moderate HIE (TH, n = 13) | p-Value | |
|---|---|---|---|
| Gestational age, week | 37.882 ± 2.228 | 37.956 ± 1.679 | 0.713 |
| Birth weight, kg | 3.026 ± 0.533 | 2.915 ± 0.723 | 0.678 |
| Emergent delivery | 25 (39.1) | 9 (69.2) | 0.046 |
| Apgar score at 1 min | 7.063 ± 2.137 | 4.385 ± 2.785 | 0.002 |
| Apgar score at 5 min | 8.391 ± 1.857 | 5.692 ± 3.225 | 0.002 |
| Outborn, n (%) | |||
| Male, n (%) | 40 (62.5) | 7 (53.8) | 0.560 |
| MAS, n (%) | 4 (6.3) | 5 (38.5) | 0.006 |
| SGA, n (%) | 7 (10.9) | 3 (23.1) | 0.359 |
| Fetal heart deceleration | 7 (10.9) | 4 (30.8) | 0.083 |
| Initial PH < 7.0 | 2 (3.1) | 1 (7.7) | 0.430 |
| LDH (initial) | 1115.125 ± 1115.292 | 2192.615 ± 1588.452 | <0.001 |
| CPK (initial) | 649.922 ± 549.473 | 1357.154 ± 1444.528 | 0.022 |
| Sarnat Stage on Day 1 | N/A | ||
| Stage 2 | 1 (1.6) | 3 (23.1) | |
| Stage 3 | 2 (3.1) | 7 (53.8) | |
| Sarnat Stage on Day 4 | N/A | ||
| Stage 2 | 0 (0) | 0 (0) | |
| Stage 3 | 1 (1.6) | 4 (30.8) | |
| Clinical seizure | 6 (9.4) | 9 (69.1) | <0.001 |
| AED, phenobarbital | 7 (10.9) | 10 (76.9) | <0.001 |
| AED, keppra | 3 (4.7) | 4 (30.8) | 0.014 |
| AED, phenytoin | 0 | 0 | N/A |
| Ventilator care, days | 3.09 ± 4.166 | 28.08 ± 48.683 | <0.001 |
| Abnormal MRI | 12 (18.8) | 10 (76.9) | 0.001 |
| Developmental delay | 16 (25) | 5 (38.5) | 0.325 |
| Hospital days | 15.25 ± 12.80 | 42.46 ± 44.70 | <0.001 |
| Death | 3 (4.7) | 3 (23.1) | 0.057 |
| Genes | |||
| GAPDH | 23.509 ± 1.678 | 22.680 ± 1.089 | 0.142 |
| DEFA3 | 26.397 ± 46.884 | 17.684 ± 12.209 | 0.174 |
| FOS | 0.399 ± 0.639 | 0.377 ± 0.375 | 0.751 |
| HSPA1A | 0.303 ± 0.852 | 2.093 ± 5.608 | <0.001 |
| IL-1b | 0.049 ± 0.050 | 0.031 ± 0.026 | 0.479 |
| ORM1 | 0.172 ± 0.191 | 0.215 ± 0.248 | 0.860 |
| OSM | 0.005 ± 0.006 | 0.003 ± 0.003 | 0.146 |
| SERPINH1 | 0.012 ± 0.040 | 0.065 ± 0.163 | 0.006 |
| TF | 0.008 ± 0.023 | 0.002 ± 0.003 | 0.812 |
| ZDHHC19 | 0.001 ± 0.001 | 0.001 ± 0.001 | 0.548 |
| Normal (n = 62) | Seizure (n = 15) | p-Value | |
|---|---|---|---|
| Gestational age, week | 38.009 ± 2.214 | 37.419 ± 2.192 | 0.207 |
| Birth weight, kg | 3.054 ± 0.541 | 2.814 ± 0.642 | 0.179 |
| Emergent delivery | 24 (38.7) | 10 (66.7) | 0.050 |
| Apgar score at 1 min | 7.06 ± 2.137 | 4.38 ± 2.785 | 0.040 |
| Apgar score at 5 min | 8.39 ± 1.857 | 5.69 ± 3.225 | 0.010 |
| Outborn, n (%) | |||
| Male, n (%) | 39 (62.9) | 8 (53.3) | 0.495 |
| MAS, n (%) | 6 (9.7) | 3 (20.0) | 0.366 |
| SGA, n (%) | 7 (11.3) | 3 (20.0) | 0.399 |
| Fetal heart deceleration | 7 (11.3) | 4 (26.7) | 0.210 |
| Emergent call | 13 (21.0) | 6 (40.0) | 0.180 |
| Initial PH < 7.0 | 2 (3.2) | 1 (6.7) | 0.483 |
| Initial BE | −6.589 ± −7.104 | −10.433 ± −8.827 | 0.059 |
| LDH (initial) | 1005.145 ± 426.325 | 2503.533 ± 2510.961 | <0.001 |
| CPK (initial) | 635.952 ± 552.600 | 1320.600 ± 1341.018 | 0.008 |
| Sarnat Stage on Day 1 | N/A | ||
| Stage 2 | 1 (1.6) | 3 (20.0) | |
| Stage 3 | 3 (4.8) | 6 (40.0) | |
| Sarnat Stage on Day 4 | N/A | ||
| Stage 2 | 1 (1.6) | 3 (20.0) | |
| Stage 3 | 2 (3.2) | 3 (20.0) | |
| AED, phenobarbital | 2 (3.2) | 15 (100) | <0.001 |
| AED, keppra | 1 (1.6) | 6 (40.0) | <0.001 |
| AED, phenytoin | 0 | 0 | N/A |
| Ventilator care, days | 5.129 ± 16.423 | 16.333 ± 36.164 | <0.001 |
| Developmental delay | 14 (22.7) | 7 (46.7) | 0.102 |
| Hospital days | 17.419 ± 19.065 | 29.867 ± 36.109 | 0.124 |
| Mortality | 2 (3.2) | 4 (26.7) | 0.012 |
| Genes | |||
| GAPDH | 23.465 ± 1.758 | 22.974 ± 0.735 | 0.643 |
| DEFA3 | 26.141 ± 47.230 | 19.905 ± 18.027 | 0.598 |
| FOS | 0.407 ± 0.648 | 0.349 ± 0.356 | 0.471 |
| HSPA1A | 0.251 ± 0.727 | 2.068 ± 5.248 | 0.001 |
| IL-1b | 0.049 ± 0.051 | 0.031 ± 0.027 | 0.258 |
| ORM1 | 0.161 ± 0.176 | 0.254 ± 0.275 | 0.375 |
| OSM | 0.005 ± 0.006 | 0.003 ± 0.003 | 0.361 |
| SERPINH1 | 0.011 ± 0.036 | 0.062 ± 0.155 | 0.025 |
| TF | 0.008 ± 0.023 | 0.006 ± 0.010 | 0.450 |
| ZDHHC19 | 0.001 ± 0.001 | 0.001 ± 0.001 | 0.225 |
| Normal (n = 56) | Abnormal MRI (n = 21) | p-Value | |
|---|---|---|---|
| Gestational age, week | 37.88 ± 2.23 | 37.96 ± 1.68 | 0.877 |
| Birth weight, kg | 3.03 ± 0.53 | 2.92 ± 0.72 | 0.991 |
| Emergent delivery | 22 (39.3) | 12 (57.1) | 0.160 |
| Apgar score at 1 min | 7.06 ± 2.137 | 4.38 ± 2.785 | 0.012 |
| Apgar score at 5 min | 8.39 ± 1.857 | 5.69 ± 3.225 | 0.056 |
| Outborn, n (%) | |||
| Male, n (%) | 37 (66.1) | 10 (47.6) | 0.139 |
| MAS, n (%) | 7(12.5) | 2 (9.5) | 1.000 |
| SGA, n (%) | 8 (14.3) | 2 (9.5) | 0.719 |
| Fetal heart deceleration | 7 (12.5) | 4 (19.0) | 0.479 |
| Emergent call | 11 (19.6) | 8 (38.1) | 0.094 |
| Initial PH < 7.0 | 3 (5.4) | 0 (0) | 0.558 |
| Initial BE | 6.122 ± 6.304 | 13.323 ± 10.345 | 0.740 |
| LDH (initial) | 1115.125 ± 1115.292 | 2192.615 ± 1588.452 | 0.063 |
| CPK (initial) | 649.922 ± 549.473 | 1357.154 ± 1444.528 | 0.277 |
| Sarnat Stage on Day 1 | N/A | ||
| Stage 2 | 2 (3.6) | 2 (9.5) | |
| Stage 3 | 4 (7.1) | 5 (23.8) | |
| Clinical seizure | 4 (7.1) | 11 (52.4) | <0.001 |
| AED, phenobarbital | 5 (8.9) | 12 (57.1) | <0.001 |
| AED, keppra | 2 (3.6) | 5 (23.8) | 0.014 |
| AED, phenytoin | 0 | 0 | N/A |
| Ventilator care, days | 4.875 ± 17.024 | 13.810 ± 30.919 | 0.001 |
| Developmental delay | 11 (19.6) | 10 (47.6) | 0.014 |
| Hospital days | 15.25 ± 12.80 | 42.46 ± 44.70 | 0.001 |
| Death | 5 (8.9) | 1 (4.8) | 1.000 |
| Genes | |||
| GAPDH | 23.435 ± 1.803 | 23.194 ± 0.986 | 0.855 |
| DEFA3 | 22.989 ± 42.572 | 30.091 ± 45.078 | 0.507 |
| FOS | 0.424 ± 0.670 | 0.320 ± 0.359 | 0.042 |
| HSPA1A | 0.361 ± 0.914 | 1.257 ± 4.470 | 0.058 |
| IL-1b | 0.048 ± 0.051 | 0.040 ± 0.036 | 0.714 |
| ORM1 | 0.174 ± 0.201 | 0.193 ± 0.201 | 0.656 |
| OSM | 0.005 ± 0.006 | 0.003 ± 0.003 | 0.123 |
| SERPINH1 | 0.015 ± 0.043 | 0.038 ± 0.130 | 0.020 |
| TF | 0.009 ± 0.024 | 0.003 ± 0.006 | 0.850 |
| ZDHHC19 | 0.001 ± 0.001 | 0.001 ± 0.001 | 0.210 |
| Mild HIE (n = 60) | Moderate-to-Severe HIE (TH-Treated) (n = 10) | p-Value | |
|---|---|---|---|
| Score, mean ±SD | |||
| Cognitive | 101.47 ± 13.04 | 102.7 ± 16.09 | 0.789 |
| Language | 100.3 ± 11.14 | 83.6 ± 13.30 | 0.041 |
| Motor | 98.52 ± 16.19 | 95.3 ± 14.97 | 0.559 |
| At risk, n (%) | |||
| Cognitive delay | 1 (1.7%) | 0 (0.0%) | 1.000 a |
| Language delay | 0 (0.0%) | 1 (10.0%) | 0.304 a |
| Motor delay | 2 (3.3%) | 1 (10.0%) | 0.904 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, Y.-M.; Hwang-Bo, S.; Im, S.-A.; Kim, M.; Youn, Y.-A. Predictive Value of Heat-Shock Protein Gene Expression on Severe Neonatal Hypoxic-Ischemic Encephalopathy. Diagnostics 2022, 12, 981. https://doi.org/10.3390/diagnostics12040981
Seo Y-M, Hwang-Bo S, Im S-A, Kim M, Youn Y-A. Predictive Value of Heat-Shock Protein Gene Expression on Severe Neonatal Hypoxic-Ischemic Encephalopathy. Diagnostics. 2022; 12(4):981. https://doi.org/10.3390/diagnostics12040981
Chicago/Turabian StyleSeo, Yu-Mi, Seok Hwang-Bo, Soo-Ah Im, Myungshin Kim, and Young-Ah Youn. 2022. "Predictive Value of Heat-Shock Protein Gene Expression on Severe Neonatal Hypoxic-Ischemic Encephalopathy" Diagnostics 12, no. 4: 981. https://doi.org/10.3390/diagnostics12040981
APA StyleSeo, Y.-M., Hwang-Bo, S., Im, S.-A., Kim, M., & Youn, Y.-A. (2022). Predictive Value of Heat-Shock Protein Gene Expression on Severe Neonatal Hypoxic-Ischemic Encephalopathy. Diagnostics, 12(4), 981. https://doi.org/10.3390/diagnostics12040981






