Promising Genomic Testing for Biliary Tract Cancer Using Endoscopic Ultrasound-Guided Fine-Needle Aspiration/Biopsy Specimens
Abstract
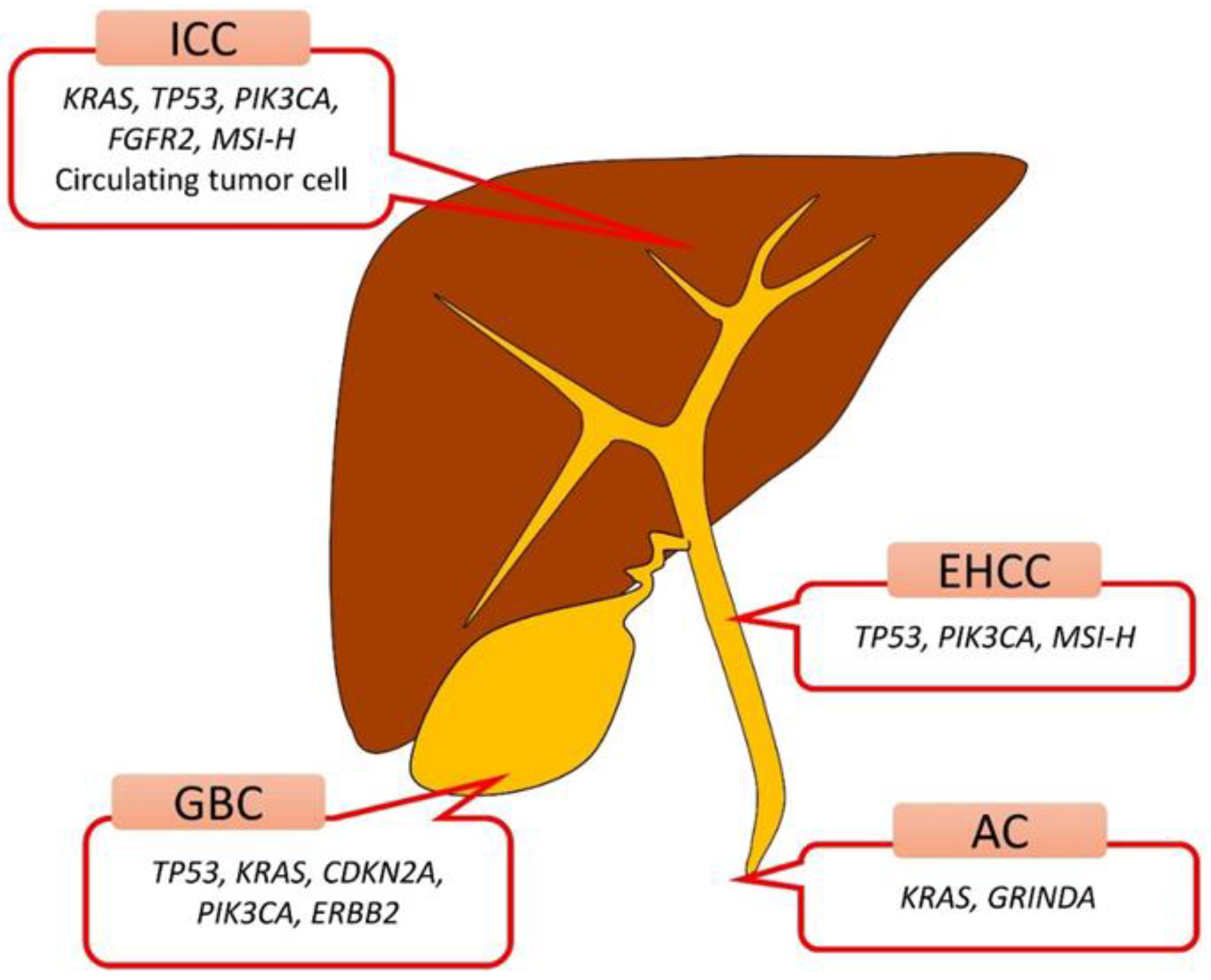
1. Introduction
2. Bile Duct
3. Gallbladder
4. Needle Size and Sample Volume
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- National Cancer Center Japan. Cancer Registry and Statistics. Cancer Information Service. Available online: https://ganjoho.jp/public/qa_links/report/statistics/2021_en.html (accessed on 1 January 2022).
- Chandrasekhara, V.; Khashab, M.A.; Muthusamy, V.R.; Acosta, R.D.; Agrawal, D.; Bruining, D.H.; Eloubeidi, M.A.; Fanelli, R.D.; Faulx, A.L.; Gurudu, S.R.; et al. Adverse events associated with ERCP. Gastrointest. Endosc. 2017, 85, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Dumonceau, J.-M.; Kapral, C.; Aabakken, L.; Papanikolaou, I.S.; Tringali, A.; Vanbiervliet, G.; Beyna, T.; Dinis-Ribeiro, M.; Hritz, I.; Mariani, A.; et al. ERCP-related adverse events: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy 2020, 52, 127–149. [Google Scholar] [CrossRef] [PubMed]
- Ponchon, T.; Gagnon, P.; Berger, F.; Labadie, M.; Liaras, A.; Chavaillon, A.; Bory, R. Value of endobiliary brush cytology and biopsies for the diagnosis of malignant bile duct stenosis: Results of a prospective study. Gastrointest. Endosc. 1995, 42, 565–572. [Google Scholar] [CrossRef]
- Pugliese, V.; Conio, M.; Nicolò, G.; Saccomanno, S.; Gatteschi, B. Endoscopic retrograde forceps biopsy and brush cytology of biliary strictures: A prospective study. Gastrointest. Endosc. 1995, 42, 520–526. [Google Scholar] [CrossRef]
- Nanda, A.; Brown, J.M.; Berger, S.H.; Lewis, M.M.; Fritcher, E.G.B.; Gores, G.J.; Keilin, S.A.; Woods, K.E.; Cai, Q.; Willingham, F.F. Triple modality testing by endoscopic retrograde cholangiopancreatography for the diagnosis of cholangiocarcinoma. Ther. Adv. Gastroenterol. 2015, 8, 56–65. [Google Scholar] [CrossRef]
- Naitoh, I.; Nakazawa, T.; Kato, A.; Hayashi, K.; Miyabe, K.; Shimizu, S.; Kondo, H.; Nishi, Y.; Yoshida, M.C.; Umemura, S.; et al. Predictive factors for positive diagnosis of malignant biliary strictures by transpapillary brush cytology and forceps biopsy. J. Dig. Dis. 2016, 17, 44–51. [Google Scholar] [CrossRef]
- Navaneethan, U.; Njei, B.; Lourdusamy, V.; Konjeti, R.; Vargo, J.J.; Parsi, M.A. Comparative effectiveness of biliary brush cytology and intraductal biopsy for detection of malignant biliary strictures: A systematic review and meta-analysis. Gastrointest. Endosc. 2015, 81, 168–176. [Google Scholar] [CrossRef]
- Weber, A.; von Weyhern, C.; Fend, F.; Schneider, J.; Neu, B.; Meining, A.; Weidenbach, H.; Schmid, R.M.; Prinz, C. Endoscopic transpapillary brush cytology and forceps biopsy in patients with hilar cholangiocarcinoma. World J. Gastroenterol. 2008, 14, 1097–1101. [Google Scholar] [CrossRef]
- Aabakken, L.; Karlsen, T.H.T.; Albert, J.G.J.; Arvanitakis, M.; Chazouilleres, O.O.; Dumonceau, J.-M.; Färkkilä, M.; Fickert, P.P.; Hirschfield, G.; Laghi, A.; et al. Role of endoscopy in primary sclerosing cholangitis: European Society of Gastrointestinal Endoscopy (ESGE) and European Association for the Study of the Liver (EASL) Clinical Guideline. Endoscopy 2017, 49, 588–608. [Google Scholar]
- Lee, S.J.; Lee, Y.S.; Lee, M.G.; Lee, S.H.; Shin, E.; Hwang, J.-H. Triple-Tissue Sampling during Endoscopic Retrograde Cholangiopancreatography Increases the Overall Diagnostic Sensitivity for Cholangiocarcinoma. Gut Liver 2014, 8, 669–673. [Google Scholar] [CrossRef]
- Valle, J.; Wasan, H.; Palmer, D.H.; Cunningham, D.; Anthoney, A.; Maraveyas, A.; Madhusudan, S.; Iveson, T.; Hughes, S.; Pereira, S.P.; et al. Cisplatin plus Gemcitabine versus Gemcitabine for Biliary Tract Cancer. N. Engl. J. Med. 2010, 362, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Sakai, D.; Kanai, M.; Kobayashi, S.; Eguchi, H.; Baba, H.; Seo, S.; Taketomi, A.; Takayama, T.; Yamaue, H.; Ishioka, C.; et al. Randomized phase III study of gemcitabine, cisplatin plus S-1 (GCS) versus gemcitabine, cisplatin (GC) for advanced biliary tract cancer (KHBO1401-MITSUBA). Ann. Oncol. 2018, 29 (Suppl. 8), viii205–viii270. [Google Scholar] [CrossRef]
- Itano, O.; Takemura, Y.; Kishida, N.; Tamagawa, E.; Shinozaki, H.; Ikeda, K.; Urakami, H.; Ei, S.; Hayatsu, S.; Suzuki, K.; et al. A prospective feasibility study of one-year administration of adjuvant S-1 therapy for resected biliary tract cancer in a multi-institutional trial (Tokyo Study Group for Biliary Cancer: TOSBIC01). BMC Cancer 2020, 20, 688. [Google Scholar] [CrossRef] [PubMed]
- Ebata, T.; Hirano, S.; Konishi, M.; Uesaka, K.; Tsuchiya, Y.; Ohtsuka, M.; Kaneoka, Y.; Yamamoto, M.; Ambo, Y.; Shimizu, Y.; et al. Faculty Opinions recommendation of Randomized clinical trial of adjuvant gemcitabine chemotherapy versus observation in resected bile duct cancer. Br. J. Surg. 2018, 105, 192–202. [Google Scholar] [CrossRef]
- Edeline, J.; Benabdelghani, M.; Bertaut, A.; Watelet, J.; Hammel, P.; Joly, J.-P.; Boudjema, K.; Fartoux, L.; Bouhier-Leporrier, K.; Jouve, J.L.; et al. Gemcitabine and oxaliplatin chemotherapy or sur-veillance in resected biliary tract cancer (PRODIGE 12-ACCORD 18-UNICANCER GI): A randomized phase III study. J. Clin. Oncol. 2019, 37, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Primrose, J.N.; Fox, R.P.; Palmer, D.H.; Malik, H.Z.; Prasad, R.; Mirza, D.; Anthony, A.; Corrie, P.; Falk, S.; Finch-Jones, M.; et al. Capecitabine compared with observation in resected biliary tract cancer (BILCAP): A randomised, controlled, multicentre, phase 3 study. Lancet Oncol. 2019, 20, 663–673. [Google Scholar] [CrossRef]
- Byrne, M.F.; Gerke, H.; Mitchell, R.M.; Stiffler, H.L.; McGrath, K.; Branch, M.S.; Baillie, J.; Jowell, P.S. Yield of Endoscopic Ultrasound-Guided Fine-Needle Aspiration of Bile Duct Lesions. Endoscopy 2004, 36, 715–719. [Google Scholar] [CrossRef]
- Eloubeidi, M.A.; Chen, V.K.; Jhala, N.C.; Eltoum, I.E.; Jhala, D.; Chhieng, D.C.; Syed, S.A.; Vickers, S.M.; Wilcox, C.M. Endoscopic ultrasound-guided fine needle aspiration biopsy of suspected cholangiocarcinoma. Clin. Gastroenterol. Hepatol. 2004, 2, 209–213. [Google Scholar] [CrossRef]
- Fritscher-Ravens, A.; Broering, D.C.; Knoefel, W.T.; Rogiers, X.; Swain, P.; Thonke, F.; Bobrowski, C.; Topalidis, T.; Soehendra, N. EUS-Guided Fine-Needle Aspiration of Suspected Hilar Cholangiocarcinoma in Potentially Operable Patients with Negative Brush Cytology. Am. J. Gastroenterol. 2004, 99, 45–51. [Google Scholar] [CrossRef]
- Lee, J.H.; Salem, R.; Aslanian, H.; Chacho, M.; Topazian, M. Endoscopic Ultrasound and Fine-Needle Aspiration of Unexplained Bile Duct Strictures. Am. J. Gastroenterol. 2004, 99, 1069–1073. [Google Scholar] [CrossRef]
- DeWitt, J.; Misra, V.L.; Leblanc, J.K.; McHenry, L.; Sherman, S. EUS-guided FNA of proximal biliary strictures after negative ERCP brush cytology results. Gastrointest. Endosc. 2006, 64, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Meara, R.S.; Jhala, D.; Eloubeidi, M.A.; Eltoum, I.; Chhieng, D.C.; Crowe, D.R.; Varadarajulu, S.; Jhala, N. Endoscopic ultrasound-guided FNA biopsy of bile duct and gallbladder: Analysis of 53 cases. Cytopathology 2006, 17, 42–49. [Google Scholar] [CrossRef]
- Agarwal, B.; Labundy, J.L.; Krishna, N.B.; Collins, B.T. Endoscopic Ultrasound Guided Fine Needle Aspiration Biopsy for Diagnostic Evaluation of Proximal Biliary Strictures. Gastrointest. Endosc. 2007, 67, AB304. [Google Scholar] [CrossRef]
- Ascunce, G.; Ribeiro, A.; Rocha-Lima, C.; Larsen, M.; Sleeman, D.; Merchan, J.; Szabo, D.; Levi, J.U. Single-session endoscopic ultrasonography and endoscopic retrograde cholangiopancreatography for evaluation of pancreaticobiliary disorders. Surg. Endosc. 2010, 24, 1447–1450. [Google Scholar] [CrossRef] [PubMed]
- Fargahi, S.; Lee, J.; Lin, F.; Gu, M. Endoscopic retrograde cholangiopancreatography (ERCP) and endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) in the cytologic diagnosis of cholangiocarcinoma-a comparative and morphologic study. Cancer Cytopathol. 2010, 118 (Suppl. 5), 323. [Google Scholar]
- Novis, M.; Ardengh, J.C.; Della Libera, E.; Nakao, F.S.; Ornellas, L.C.; Santo, G.C.; Venco, F.; Ferrari, A.P. Prospective comparative study of ERCP brush cytology and EUS-FNA for the differential diagnosis of biliary strictures. Rev. Col. Bras. Cir. 2010, 37, 190–198. [Google Scholar] [CrossRef]
- Oppong, K.; Raine, D.; Nayar, M.; Wadehra, V.; Ramakrishnan, S.; Charnley, R.M. EUS-FNA versus biliary brushings and assessment of simultaneous performance in jaundiced patients with suspected malignant obstruction. JOP 2010, 11, 560–567. [Google Scholar]
- Mohamadnejad, M.; DeWitt, J.M.; Sherman, S.; LeBlanc, J.K.; Pitt, H.A.; House, M.G.; Jones, K.J.; Fogel, E.L.; McHenry, L.; Watkins, J.L.; et al. Role of EUS for preoperative evaluation of cholangiocarcinoma: A large single-center experience. Gastrointest. Endosc. 2011, 73, 71–78. [Google Scholar] [CrossRef]
- Nayar, M.K.; Manu, N.K.; Manas, D.M.; Derek, M.M.; Wadehra, V.; Viney, W.; Oppong, K.E.; Kofi, O.E. Role of EUS/EUS-guided FNA in the management of proximal biliary strictures. Hepatogastroenterology 2011, 58, 1862–1865. [Google Scholar]
- Ohshima, Y.; Yasuda, I.; Kawakami, H.; Kuwatani, M.; Mukai, T.; Iwashita, T.; Doi, S.; Nakashima, M.; Hirose, Y.; Asaka, M.; et al. EUS-FNA for suspected malignant biliary strictures after negative endoscopic transpapillary brush cytology and forceps biopsy. J. Gastroenterol. 2011, 46, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Krishna, N.B.; Tummala, P.; Labundy, J.L.; Agarwal, B. EUS guided fine needle aspiration is useful in diagnostic evaluation of in-determinate proximal biliary strictures. Open J. Gastroenterol. 2012, 2, 33–39. [Google Scholar] [CrossRef]
- Putta, S.; Croagh, D.; Boulton, R.; Brahmall, S.; Forde, C.; Mahon, B. Endoscopic ultrasound in the evaluation of liver hilar pathology [abstract]. Gut 2012, 61, A355. [Google Scholar] [CrossRef][Green Version]
- de la Mora Levy, J.G.; Alonso-Larraga, J.; Solis, M.R.; Guerrero, A.H. EUS-FNA for proximal bile duct strictures, brushings or cholangioscopy? J. Gastroenterol. Hepatol. 2013, 28 (Suppl. 3), 651. [Google Scholar]
- Nguyen, N.Q.; Schoeman, M.N.; Ruszkiewicz, A. Clinical utility of EUS before cholangioscopy in the evaluation of difficult biliary strictures. Gastrointest. Endosc. 2013, 78, 868–874. [Google Scholar] [CrossRef] [PubMed]
- Tummala, P.; Munigala, S.; Eloubeidi, M.A.; Agarwal, B. Patients with obstructive jaundice and biliary stricture ± mass lesion on imaging: Prevalence of malignancy and potential role of EUS-FNA. J. Clin. Gastroenterol. 2013, 47, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Weilert, F.; Bhat, Y.M.; Binmoeller, K.F.; Kane, S.; Jaffee, I.M.; Shaw, R.E.; Cameron, R.; Hashimoto, Y.; Shah, J.N. EUS-FNA is superior to ERCP-based tissue sampling in suspected malignant biliary obstruction: Results of a prospective, single-blind, comparative study. Gastrointest. Endosc. 2014, 80, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, A.; Mohamadnejad, M.; Islami, F.; Keshtkar, A.; Biglari, M.; Malekzadeh, R.; Eloubeidi, M.A. Diagnostic yield of EUS-guided FNA for malignant biliary stricture: A systematic review and meta-analysis. Gastrointest. Endosc. 2016, 83, 290–298. [Google Scholar] [CrossRef] [PubMed]
- de Moura, D.T.H.; Ryou, M.; de Moura, E.G.H.; Ribeiro, I.B.; Bernardo, W.M.; Thompson, C.C. Endoscopic ultrasound-guided fine needle aspiration and endoscopic retrograde cholangiopancreatography-based tissue sampling in suspected malignant biliary strictures: A meta-analysis of same-session procedures. Clin. Endosc. 2020, 53, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Arai, Y.; Totoki, Y.; Shirota, T.; El Zawahry, A.; Kato, M.; Hama, N.; Hosoda, F.; Urushidate, T.; Ohashi, S.; et al. Genomic spectra of biliary tract cancer. Nat. Genet. 2015, 47, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Lowery, M.; Ptashkin, R.N.; Jordan, E.J.; Berger, M.F.; Zehir, A.; Capanu, M.; Kemeny, N.E.; O’Reilly, E.M.; El-Dika, I.; Jarnagin, W.R.; et al. Comprehensive Molecular Profiling of Intrahepatic and Extrahepatic Cholangiocarcinomas: Potential Targets for Intervention. Clin. Cancer Res. 2018, 24, 4154–4161. [Google Scholar] [CrossRef]
- Lowery, M.; Jordan, E.J.; Basturk, O.; Ptashkin, R.N.; Zehir, A.; Berger, M.F.; Leach, T.; Herbst, B.; Askan, G.; Maynard, H.; et al. Real-Time Genomic Profiling of Pancreatic Ductal Adenocarcinoma: Potential Actionability and Correlation with Clinical Phenotype. Clin. Cancer Res. 2017, 23, 6094–6100. [Google Scholar] [CrossRef] [PubMed]
- Rashid, A.; Ueki, T.; Gao, Y.-T.; Houlihan, P.S.; Wallace, C.; Wang, B.-S.; Shen, M.-C.; Deng, J.; Hsing, A.W. K-ras mutation, p53 overexpression, and microsatellite instability in biliary tract cancers: A population-based study in China. Clin. Cancer Res. 2002, 8, 3156–3163. [Google Scholar]
- Chan-On, W.; Nairismägi, M.L.; Ong, C.K.; Lim, W.K.; Dima, S.; Pairojkul, C.; Lim, K.H.; McPherson, J.R.; Cutcutache, I.; Heng, H.L.; et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat. Genet. 2013, 45, 1474–1478. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.K.; Subimerb, C.; Pairojkul, C.; Wongkham, S.; Cutcutache, I.; Yu, W.; McPherson, J.R.; Allen, G.E.; Ng, C.C.; Wong, B.H.; et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat. Genet. 2012, 44, 690–693. [Google Scholar] [CrossRef]
- Ross, J.S.; Wang, K.; Javle, M.M.; Catenacci, D.V.T.; Shroff, R.T.; Ali, S.M.; Elvin, J.A.; Chmielecki, J.; Yelensky, R.; Lipson, D.; et al. Comprehensive genomic profiling of biliary tract cancers to reveal tumor-specific differences and frequency of clinically relevant genomic alterations. J. Clin. Oncol. 2015, 33, 4009. [Google Scholar] [CrossRef]
- Jain, A.; Javle, M. Molecular profiling of biliary tract cancer: A target rich disease. J. Gastrointest. Oncol. 2016, 7, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Singhi, A.D.; Nikiforova, M.N.; Chennat, J.; Papachristou, G.I.; Khalid, A.; Rabinovitz, M.; Das, R.; Sarkaria, S.; Ayasso, M.S.; Wald, A.I.; et al. Integrating next-generation sequencing to endoscopic retrograde cholangiopancreatography (ERCP)-obtained biliary specimens improves the detection and management of patients with malignant bile duct strictures. Gut 2020, 69, 52–61. [Google Scholar] [CrossRef]
- Malhotra, N.; Jackson, S.A.; Freed, L.L.; Styn, M.A.; Sidawy, M.K.; Haddad, N.G.; Finkelstein, S.D. The added value of using mutational profiling in addition to cytology in diagnosing aggressive pancreaticobiliary disease: Review of clinical cases at a single center. BMC Gastroenterol. 2014, 14, 135. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, F.C.; Kerr, S.E.; Kipp, B.R.; Voss, J.S.; Minot, D.M.; Tu, Z.J.; Henry, M.R.; Graham, R.P.; Vasmatzis, G.; Cheville, J.C.; et al. Targeted next generation sequencing of endoscopic ultrasound acquired cytology from ampullary and pancreatic adeno-carcinoma has the potential to aid patient stratification for optimal therapy selection. Oncotarget 2016, 34, 54526–54536. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Moon, J.H.; Kim, H.K.; Lee, Y.N.; Lee, T.H.; Cha, S.W.; Cho, Y.D.; Park, S.H. KRAS mutation analysis by next-generation sequencing in endoscopic ultrasound-guided sampling for solid liver masses. J. Gastroenterol. Hepatol. 2017, 32, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Maruki, Y.; Morizane, C.; Arai, Y.; Ikeda, M.; Ueno, M.; Ioka, T.; Naganuma, A.; Furukawa, M.; Mizuno, N.; Uwagawa, T.; et al. Molecular detection and clinicopathological characteristics of advanced/recurrent biliary tract carcinomas harboring the FGFR2 rearrangements: A prospective observational study (PRELUDE Study). J. Gastroenterol. 2021, 56, 250–260. [Google Scholar] [CrossRef]
- Jacobson, B.C.; Pitman, M.B.; Brugge, W.R. EUS-guided FNA for the diagnosis of gallbladder masses. Gastrointest. Endosc. 2003, 57, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Varadarajulu, S.; Eloubeidi, M.A. Endoscopic Ultrasound-Guided Fine-Needle Aspiration in the Evaluation of Gallbladder Masses. Endoscopy 2005, 37, 751–754. [Google Scholar] [CrossRef] [PubMed]
- Hijioka, S.; Mekky, M.A.; Bhatia, V.; Sawaki, A.; Mizuno, N.; Hara, K.; Hosoda, W.; Shimizu, Y.; Tamada, K.; Niwa, Y.; et al. Can EUS-guided FNA distinguish between gallbladder cancer and xanthogranulomatous cholecystitis? Gastrointest. Endosc. 2010, 72, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Hijioka, S.; Hara, K.; Mizuno, N.; Imaoka, H.; Ogura, T.; Haba, S.; Mekky, M.A.; Bhatia, V.; Hosoda, W.; Yatabe, Y.; et al. Diagnostic yield of endoscopic retrograde cholangiography and of EUS-guided fine needle aspiration sampling in gallbladder carcinomas. J. Hepato-Biliary-Pancreat. Sci. 2011, 19, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Singla, V.; Agarwal, R.; Anikhindi, S.A.; Puri, P.; Kumar, M.; Ranjan, P.; Kumar, A.; Sharma, P.; Bansal, N.; Bakshi, P.; et al. Role of EUS-FNA for gallbladder mass lesions with biliary obstruction: A large single-center experience. Endosc. Int. Open 2019, 7, E1403–E1409. [Google Scholar] [CrossRef] [PubMed]
- Hijioka, S.; Nagashio, Y.; Ohba, A.; Maruki, Y.; Okusaka, T. The Role of EUS and EUS-FNA in Differentiating Benign and Malignant Gallbladder Lesions. Diagnostics 2021, 11, 1586. [Google Scholar] [CrossRef]
- Maruta, A.; Iwata, K.; Iwashita, T.; Mizoguchi, K.; Kimura, M.; Takeyama, H.; Joh, T. Factors affecting technical success of endoscopic transpapillary gallbladder drainage for acute cholecystitis. J. Hepato-Biliary-Pancreat. Sci. 2020, 27, 429–436. [Google Scholar] [CrossRef]
- Jiao, Y.; Pawlik, T.M.; Anders, R.A.; Selaru, F.M.; Streppel, M.M.; Lucas, D.J.; Niknafs, N.; Guthrie, V.B.; Maitra, A.; Argani, P.; et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat. Genet. 2013, 45, 1470–1473. [Google Scholar] [CrossRef]
- Kuipers, H.; de Bitter, T.J.J.; de Boer, M.T.; van der Post, R.S.; Nijkamp, M.W.; de Reuver, P.R.; Fehrmann, R.S.N.; Hoogwater, F.J.H. Gallbladder Cancer: Current Insights in Genetic Alterations and Their Possible Therapeutic Implications. Cancers 2021, 13, 5257. [Google Scholar] [CrossRef]
- Spizzo, G.; Puccini, A.; Xiu, J.; Goldberg, R.M.; Grothey, A.; Shields, A.F.; Arora, S.P.; Khushmann, M.; Salem, M.E.; Battaglin, F.; et al. Molecular profile of BRCA-mutated biliary tract cancers. ESMO Open 2020, 5, e000682. [Google Scholar] [CrossRef] [PubMed]
- Hirata, K.; Kuwatani, M.; Suda, G.; Ishikawa, M.; Sugiura, R.; Kato, S.; Kawakubo, K.; Sakamoto, N. A Novel Approach for the Genetic Analysis of Biliary Tract Cancer Specimens Obtained Through Endoscopic Ultrasound-Guided Fine Needle Aspiration Using Targeted Amplicon Sequencing. Clin. Transl. Gastroenterol. 2019, 10, e00022. [Google Scholar] [CrossRef] [PubMed]
- Kai, Y.; Ikezawa, K.; Takada, R.; Daiku, K.; Maeda, S.; Abe, Y.; Yamai, T.; Fukutake, N.; Nakabori, T.; Uehara, H.; et al. Success rate of microsatellite instability examination and complete response with pembrolizumab in biliary tract cancer. JGH Open 2021, 5, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Akagi, K.; Oki, E.; Taniguchi, H.; Nakatani, K.; Aoki, D.; Kuwata, T.; Yoshino, T. Nationwide large-scale investigation of microsatellite instability status in more than 18,000 patients with various advanced solid cancers. J. Clin. Oncol. 2020, 38 (Suppl. 4), 803. [Google Scholar] [CrossRef]
- Takahashi, K.; Yasuda, I.; Hanaoka, T.; Hayashi, Y.; Motoo, I.; Kajiura, S.; Ando, T.; Fujinami, H.; Tajiri, K.; Imura, J.; et al. Comparison of Histological Sample Volumes among Various Endoscopic Ultrasound-Guided Biopsy Needles. J. Clin. Med. 2021, 10, 3560. [Google Scholar] [CrossRef]
- Asokkumar, R.; Ka, C.Y.; Loh, T.; Ling, L.K.; San, T.G.; Ying, H.; Tan, D.; Khor, C.; Lim, T.; Soetikno, R. Comparison of tissue and molecular yield between fine-needle biopsy (FNB) and fine-needle aspiration (FNA): A randomized study. Endosc. Int. Open 2019, 7, E955–E963. [Google Scholar] [CrossRef] [PubMed]
- Mukai, S.; Itoi, T.; Yamaguchi, H.; Sofuni, A.; Tsuchiya, T.; Tanaka, R.; Tonozuka, R.; Honjo, M.; Fujita, M.; Yamamoto, K.; et al. A retrospective histological comparison of EUS-guided fine-needle biopsy using a novel franseen needle and a conventional end-cut type needle. Endosc. Ultrasound 2019, 8, 50–57. [Google Scholar]
- Bang, J.Y.; Hebert-Magee, S.; Navaneethan, U.; Hasan, M.K.; Hawes, R.; Varadarajulu, S. Randomized trial comparing the Franseen and Fork-tip needles for EUS-guided fine-needle biopsy sampling of solid pancreatic mass lesions. Gastrointest. Endosc. 2018, 87, 1432–1438. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.; Kong, J.; Ko, S.W.; Hong, S.-M.; So, H.; Hwang, J.S.; Song, T.J.; Lee, S.K.; Kim, M.-H.; Lee, S.S. A comparison between 25-gauge and 22-gauge Franseen needles for endoscopic ultrasound-guided sampling of pancreatic and peripancreatic masses: A randomized non-inferiority study. Endoscopy 2021, 53, 1122–1129. [Google Scholar] [CrossRef]
- Kuwatani, M.; Sakamoto, N. Evolution and a promising role of EUS-FNA in gene and future analyses. Endosc. Ultrasound 2020, 9, 151–153. [Google Scholar] [PubMed]
- Catenacci, D.V.; Chapman, C.G.; Xu, P.; Koons, A.; Konda, V.J.; Siddiqui, U.D.; Waxman, I. Acquisition of Portal Venous Circulating Tumor Cells from Patients with Pancreaticobiliary Cancers by Endoscopic Ultrasound. Gastroenterology 2015, 149, 1794–1803. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Zhang, D.; Yin, L.; Qiu, Y.; Liu, J.; Yu, W.; Fu, X.; Zhu, B.; Xu, X.; Duan, A.; et al. Bile cell-free DNA as a novel and powerful liquid biopsy for detecting somatic variants in biliary tract cancer. Oncol. Rep. 2019, 42, 549–560. [Google Scholar] [CrossRef]
- Arechederra, M.; Rullán, M.; Amat, I.; Oyon, D.; Zabalza, L.; Elizalde, M.; Latasa, M.U.; Mercado, M.R.; Ruiz-Clavijo, D.; Saldaña, C.; et al. Next-generation sequencing of bile cell-free DNA for the early detection of patients with malignant biliary strictures. Gut 2021. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Sereewattanawoot, S.; Suzuki, A. A new era of long-read sequencing for cancer genomics. J. Hum. Genet. 2020, 65, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Alcaide, M.; Cheung, M.; Bushell, K.; Arthur, S.E.; Wong, H.-L.; Karasinska, J.; Renouf, D.; Schaeffer, D.F.; McNamara, S.; du Tertre, M.C.; et al. A Novel Multiplex Droplet Digital PCR Assay to Identify and Quantify KRAS Mutations in Clinical Specimens. J. Mol. Diagn. 2019, 21, 214–227. [Google Scholar] [CrossRef] [PubMed]
| Year | Author | Sampling Site | Sampling Method | Patient ** Number | Lesion Size (mm) | Needle Size (Number) | Analysis Target | Analysis Method | |
|---|---|---|---|---|---|---|---|---|---|
| 2014 | Malhotra et al. | ND | FNA § | ND | ND | ND | LOH of 10 genes and KRAS point mutations | PCR and subsequent capillary gel electrophoresis | |
| 2016 | Gleeson et al. | Ampulla | FNA || | 4 | 30 (25–41) †† | ND | 160 cancer gene | NGS | |
| 2017 | Choi et al. | Liver left lobe | FNB || | 13 | 33 (23–39) †† | 22 G (25); 25 G (3) | KRAS | peptide nucleic acid-PCR vs. NGS | |
| 2019 | Hirata et al. | BT, LN | FNA/FNB || | 21 | ND | 22 G (19), 25 G (2) | 50 cancer gene | Targeted Amplicon Sequencing | |
| 2021 | Maruki et al. | ND | FNA ¶ | 423 | ND | ND | FGFR2 | FISH + targeted RNA sequencing | |
| 2021 | Kai et al. | ND | FNA ¶ | 60 | 29 (12–85) ‡‡ | 22 G (11), 20 G (1) | MSI (BAT-26, NR-21, BAT-25, MONO-27, NR-24) | MSI Kit | |
| Cancer Type (Number) * | Cancer Stage * | DNA Extraction | SN * | SP | ACC | PPV | NPV | ||
| Pancreaticobiliary mass (26) | ND | 26/26 | 8/17 | 9/9 | 17/26 | 8/8 | 9/18 | ||
| AC (4), PDAC (22), IPMC (3) | Ia 2 (7%) Ib 3 (10%) IIa 4 (14%) IIb 18 (62%) III 1 (4%) IV 1 (3%) | 29/47 | KRAS 93% TP53 72% SMAD4 31% GNAS 10% | NA | NA | NA | NA | ||
| HCC (4), ICC (7), NEC (1), m-PC (7), m-GBC (5), m-AC (1), m-Others (3) | ND | PCR: 27/28 NGS: 16/28 | PCR 14.3% NGS 25% | NA | Pathology + KRAS = 96.4% | NA | NA | ||
| p-GBC (10), p-ICC (1), p-EHCC (1); n-GBC (2), n-ICC (5), n-EHCC (2) | II/III/IV, 2/2/17 | 21/21 | GBC/ICC/EHCC: TP53, 38/20/40%; KRAS 21/30/0%; CDKN2A, 9/0/0%; PIK3CA, 4/10/20%; ERBB2 4/0/0% | NA | NA | NA | NA | ||
| ICC, PCC, DCC, GBC, AC | IV/recurrence | † | ICC 7.4% PCC 3.6% | NA | NA | NA | NA | ||
| ICC (24), PCC (12), DCC (4), GBC (16), AC (4) | ND | MSI 59/60 | MSI-H 3.3% ‡ | NA | NA | NA | NA | ||
| FNA/FNB Needle | 19G-FNB | 19G-FNA | 22G-FNB | 22G-FNA | 25G-FNB |
|---|---|---|---|---|---|
| Pathological tissue area (mm2) (IQR) | 15.20 (6.89–25.75) | 5.44 (3.19–25.75) | 4.49 (1.69–6.63) 2.2 2.13 0.9 (0.3–2.8)–1.0 (0.4–2.7) | 0.9 0.45 | NA (Optimal histologic core procurement: 87.1%, 25G-FNB vs. 97.1%, 22G-FNB) |
| DNA amount (ng) | 2185 (1478–3066) | 1477 (1151–2522) | |||
| DNA amount (ng) (predicted) | 15,000 | 5000 | 1000–4000 | 500–1000 | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuwatani, M.; Kawakubo, K.; Sakamoto, N. Promising Genomic Testing for Biliary Tract Cancer Using Endoscopic Ultrasound-Guided Fine-Needle Aspiration/Biopsy Specimens. Diagnostics 2022, 12, 900. https://doi.org/10.3390/diagnostics12040900
Kuwatani M, Kawakubo K, Sakamoto N. Promising Genomic Testing for Biliary Tract Cancer Using Endoscopic Ultrasound-Guided Fine-Needle Aspiration/Biopsy Specimens. Diagnostics. 2022; 12(4):900. https://doi.org/10.3390/diagnostics12040900
Chicago/Turabian StyleKuwatani, Masaki, Kazumichi Kawakubo, and Naoya Sakamoto. 2022. "Promising Genomic Testing for Biliary Tract Cancer Using Endoscopic Ultrasound-Guided Fine-Needle Aspiration/Biopsy Specimens" Diagnostics 12, no. 4: 900. https://doi.org/10.3390/diagnostics12040900
APA StyleKuwatani, M., Kawakubo, K., & Sakamoto, N. (2022). Promising Genomic Testing for Biliary Tract Cancer Using Endoscopic Ultrasound-Guided Fine-Needle Aspiration/Biopsy Specimens. Diagnostics, 12(4), 900. https://doi.org/10.3390/diagnostics12040900







