Evaluation of Expressed MicroRNAs as Prospective Biomarkers for Detection of Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Blood Sampling
2.2. RNA Extraction and cDNA Synthesis
2.3. Real-Time PCR Analysis of miRs
2.4. Statistical Analysis
3. Results
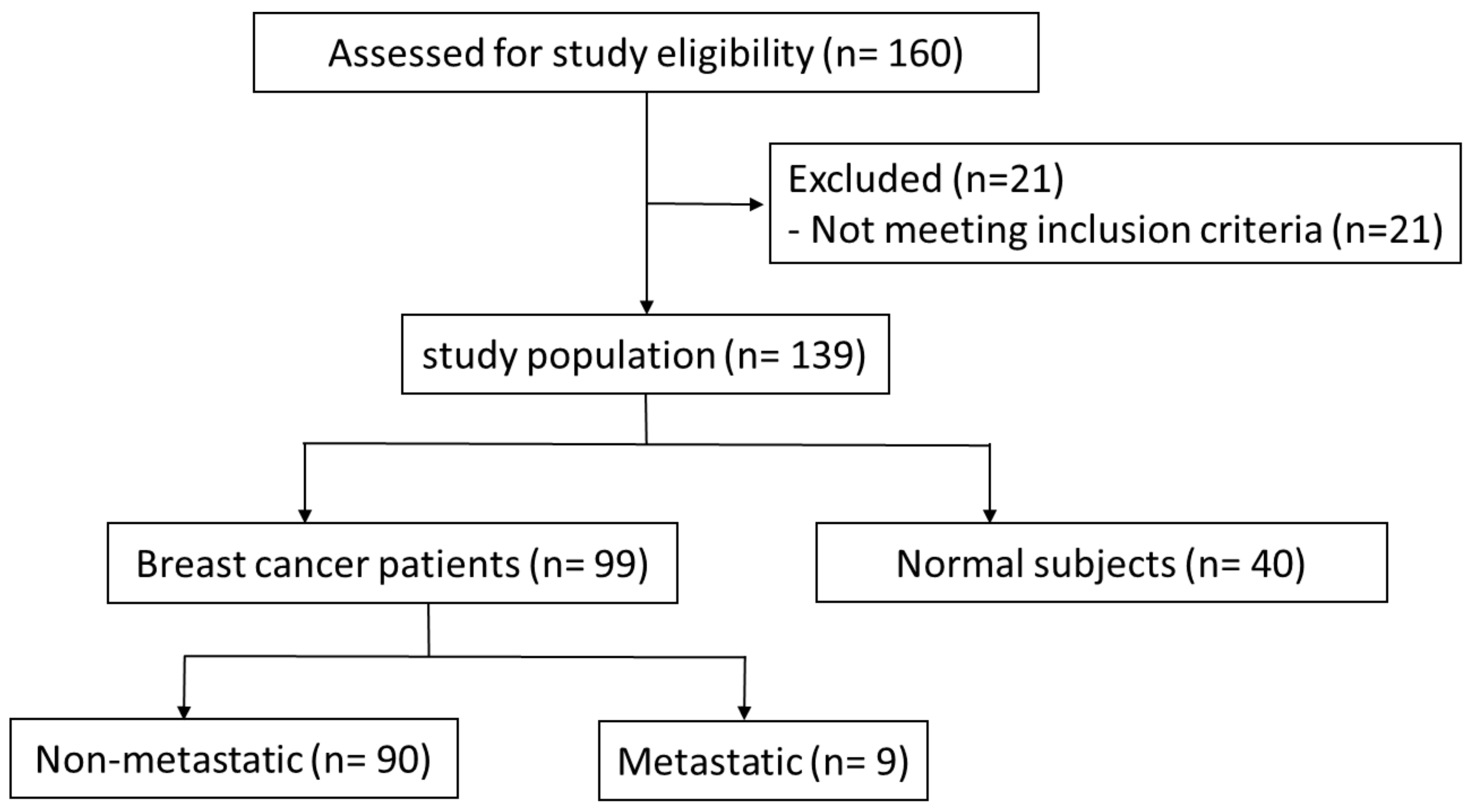
3.1. Demographic and Clinical Data
3.2. Correlation of miR Expression with Clinicopathological Data
3.3. Serum miR Expression in BC
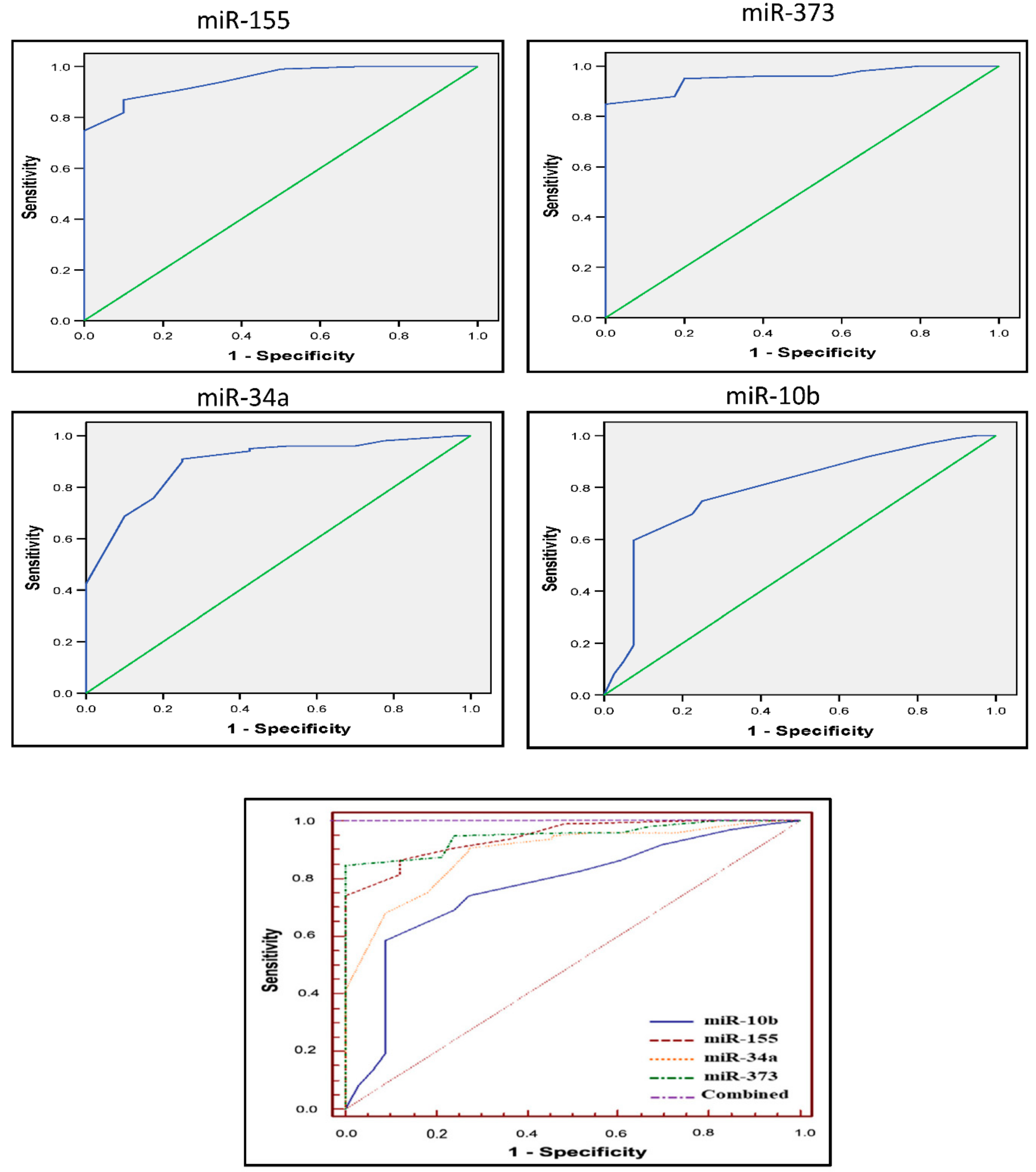
3.4. Diagnostic Accuracy of the Four miRs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Akoko, L.O.; Rutashobya, A.K.; Lutainulwa, E.W.; Mwanga, A.H.; Kivuyo, S.L. The effect of reproductive, hormonal, nutritional and lifestyle on breast cancer risk among black Tanzanian women: A case control study. PLoS ONE 2022, 17, e0263374. [Google Scholar] [CrossRef] [PubMed]
- Zeeneldin, A.A.; Ramadan, M.; Gaber, A.A.; Taha, F.M. Clinico-pathological features of breast carcinoma in elderly Egyptian patients: A comparison with the non-elderly using population-based data. J. Egypt. Natl. Cancer Inst. 2013, 25, 5–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Omar, S.; Khaled, H.; Gaafar, R.; Zekry, A.R.; Eissa, S.; El Khatib, O. Breast cancer in Egypt:a review of disease presentation and detection strategies. East. Mediterr. Heal. J. 2021, 9, 448–463. [Google Scholar] [CrossRef]
- Najjar, H.; Easson, A. Age at diagnosis of breast cancer in Arab nations. Int. J. Surg. 2010, 8, 448–452. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.S.; Khaled, H.M.; Mikhail, N.N.; Baraka, H.; Kamel, H. Cancer Incidence in Egypt: Results of the National Population-Based Cancer Registry Program. J. Cancer Epidemiol. 2014, 2014, 1–18. [Google Scholar] [CrossRef]
- Kerlikowske, K.; Zhu, W.; Hubbard, R.; Geller, B.; Dittus, K.; Braithwaite, D.; Wernli, K.J.; Miglioretti, D.L.; O’Meara, E.S. For the Breast Cancer Surveillance Consortium Outcomes of Screening Mammography by Frequency, Breast Density, and Postmenopausal Hormone Therapy. JAMA Intern. Med. 2013, 173, 807–816. [Google Scholar] [CrossRef] [Green Version]
- Bleyer, A.; Welch, H.G. Effect of Three Decades of Screening Mammography on Breast-Cancer Incidence. N. Engl. J. Med. 2012, 367, 1998–2005. [Google Scholar] [CrossRef] [Green Version]
- Afzal, S.; Hassan, M.; Ullah, S.; Abbas, H.; Tawakkal, F.; Khan, M.A. Breast Cancer; Discovery of Novel Diagnostic Biomarkers, Drug Resistance, and Therapeutic Implications. Front. Mol. Biosci. 2022, 9, 783450. [Google Scholar] [CrossRef]
- Haider, M.-T.; Smit, D.J.; Taipaleenmäki, H. MicroRNAs: Emerging Regulators of Metastatic Bone Disease in Breast Cancer. Cancers 2022, 14, 729. [Google Scholar] [CrossRef] [PubMed]
- Kandettu, A.; Radhakrishnan, R.; Chakrabarty, S.; Sriharikrishnaa, S.; Kabekkodu, S.P. The emerging role of miRNA clusters in breast cancer progression. Biochim. Biophys. Acta 2020, 1874, 188413. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Zhu, B.; Wang, M.; Jin, J. Knockdown of long non-coding RNA DDX11-AS1 inhibits the proliferation, migration and paclitaxel resistance of breast cancer cells by upregulating microRNA-497 expression. Mol. Med. Rep. 2022, 25, 1–14. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F. Oncomirs—microRNAs with a role in cancer. Nat. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Eichelser, C.; Flesch-Janys, D.; Chang-Claude, J.; Pantel, K.; Schwarzenbach, H. Deregulated Serum Concentrations of Circulating Cell–Free MicroRNAs miR-17, miR-34a, miR-155, and miR-373 in Human Breast Cancer Development and Progression. Clin. Chem. 2013, 59, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Nassar, F.; El Sabban, M.; Zgheib, N.K.; Tfayli, A.; Boulos, F.; Jabbour, M.; Saghir, N.S.E.L.; Talhouk, R.; Bazarbachi, A.; Calin, G.A.; et al. miRNA as Potential Biomarkers of Breast Cancer in the Lebanese Population and in Young Women: A Pilot Study. PLoS ONE 2014, 9, e107566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef]
- Iorio, M.; Casalini, P.; Tagliabue, E.; Ménard, S.; Croce, C.M. MicroRNA profiling as a tool to understand prognosis, therapy response and resistance in breast cancer. Eur. J. Cancer 2008, 44, 2753–2759. [Google Scholar] [CrossRef]
- Pourteimoor, V.; Paryan, M.; Mohammadi-Yeganeh, S. microRNA as a systemic intervention in the specific breast cancer subtypes with C-MYC impacts; introducing subtype-based appraisal tool. J. Cell. Physiol. 2018, 233, 5655–5669. [Google Scholar] [CrossRef]
- Eichelser, C.; Stückrath, I.; Müller, V.; Milde-Langosch, K.; Wikman, H.; Pantel, K.; Schwarzenbach, H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014, 5, 9650–9663. [Google Scholar] [CrossRef] [Green Version]
- Rice, J.; Roberts, H.; Rai, S.N.; Galandiuk, S. Housekeeping genes for studies of plasma microRNA: A need for more precise standardization. Surgery 2015, 158, 1345–1351. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Chang, S.; Sharan, S.K. BRCA1 and MicroRNAs: Emerging networks and potential therapeutic targets. Mol. Cells 2012, 34, 425–432. [Google Scholar] [CrossRef] [Green Version]
- Chang, S.; Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab); Wang, R.-H.; Akagi, K.; Kim, K.-A.; Martin, B.K.; Cavallone, L.; Haines, D.C.; Basik, M.; Mai, P.; et al. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat. Med. 2011, 17, 1275–1282. [Google Scholar] [CrossRef] [Green Version]
- Kong, W.; Yang, H.; He, L.; Zhao, J.-J.; Coppola, D.; Dalton, W.S.; Cheng, J.Q. MicroRNA-155 Is Regulated by the Transforming Growth Factor β/Smad Pathway and Contributes to Epithelial Cell Plasticity by Targeting RhoA. Mol. Cell. Biol. 2008, 28, 6773–6784. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Qin, W.; Atasoy, U.; Sauter, E.R. Circulating microRNAs in breast cancer and healthy subjects. BMC Res. Notes 2009, 2, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Shaker, O.; Maher, M.; Nassar, Y.; Morcos, G.; Gad, Z. Role of microRNAs -29b-2, −155, −197 and −205 as diagnostic biomarkers in serum of breast cancer females. Gene 2015, 560, 77–82. [Google Scholar] [CrossRef]
- Hafez, M.M.; Hassan, Z.K.; Zekri, A.-R.; Gaber, A.; Al Rejaie, S.S.; Sayed-Ahmed, M.M.; Al Shabanah, O. MicroRNAs and Metastasis-related Gene Expression in Egyptian Breast Cancer Patients. Asian Pac. J. Cancer Prev. 2012, 13, 591–598. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Zheng, Z.; Guo, J.; Ding, X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol. Oncol. 2010, 119, 586–593. [Google Scholar] [CrossRef]
- Hamam, R.; Ali, A.; Alsaleh, K.A.; Kassem, M.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci. Rep. 2016, 6, 25997. [Google Scholar] [CrossRef] [Green Version]
- Zonari, E.; Pucci, F.; Saini, M.; Mazzieri, R.; Politi, L.S.; Gentner, B.; Naldini, L. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood 2013, 122, 243–252. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Mm, B.-C.W.; Tang, J.-H. Clinical significance of MicoRNA-155 expression in human breast cancer. J. Surg. Oncol. 2012, 106, 260–266. [Google Scholar] [CrossRef]
- Yan, G.-R.; Xu, S.-H.; Tan, Z.-L.; Liu, L.; He, Q.-Y. Global identification of miR-373-regulated genes in breast cancer by quantitative proteomics. Proteomics 2011, 11, 912–920. [Google Scholar] [CrossRef]
- Van Roy, F. Beyond E-cadherin: Roles of other cadherin superfamily members in cancer. Nat. Cancer 2014, 14, 121–134. [Google Scholar] [CrossRef]
- Chen, W.; Cai, F.; Zhang, B.; Barekati, Z.; Zhong, X.Y. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: Potential biomarkers. Tumor Biol. 2013, 34, 455–462. [Google Scholar] [CrossRef]
- Li, X.J.; Ren, Z.J.; Tang, J.H. MicroRNA-34a: A potential therapeutic target in human cancer. Cell Death Dis. 2014, 5, e1327. [Google Scholar] [CrossRef]
- Chang, T.-C.; Wentzel, E.A.; Kent, O.; Ramachandran, K.; Mullendore, M.; Lee, K.H.; Feldmann, G.; Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J.; et al. Transactivation of miR-34a by p53 Broadly Influences Gene Expression and Promotes Apoptosis. Mol. Cell 2007, 26, 745–752. [Google Scholar] [CrossRef] [Green Version]
- He, L.; He, X.; Lim, L.P.; De Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodygin, D.; Tarasov, V.; Epanchintsev, A.; Berking, C.; Knyazeva, T.; Körner, H.; Knyazev, P.; Diebold, J.; Hermeking, H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 2008, 7, 2591–2600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Li, Y.; Gao, J.; Zhang, T.; Li, S.; Luo, A.; Chen, H.; Ding, F.; Wang, X.; Liu, Z. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene 2012, 32, 4294–4303. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Srivastava, A.K.; Suman, S.; Kumar, V.; Shukla, Y. Circulating miRNAs revealed as surrogate molecular signatures for the early detection of breast cancer. Cancer Lett. 2015, 369, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Hagrass, H.A.; Sharaf, S.; Pasha, H.F.; Tantawy, E.A.; Mohamed, R.; Kassem, R. Circulating microRNAs—A new horizon in molecular diagnosis of breast cancer. Genes Cancer 2015, 6, 281–287. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Reinhardt, F.; Pan, E.; Soutschek, J.; Bhat, B.; Marcusson, E.G.; Teruya-Feldstein, J.; Bell, G.W.; A Weinberg, R. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010, 28, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Ma, L. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 2010, 12, 210. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Jiang, Y.; Zhou, J.; Liu, S.; Gu, Y.; Jin, G.; Hu, Z.; Ma, H.; Shen, H.; Dai, J. Genetic Variants in the Promoter Region of miR-10b and the Risk of Breast Cancer. BioMed Res. Int. 2017, 2017, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, F.-L.; Hu, G.-D.; Wang, X.-F.; Zhang, X.-H.; Zhang, Y.-K.; Yu, Z.-S. Serum Overexpression of MicroRNA-10b in Patients with Bone Metastatic Primary Breast Cancer. J. Int. Med Res. 2012, 40, 859–866. [Google Scholar] [CrossRef]
- Iorio, M.; Ferracin, M.; Liu, C.-G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [Green Version]
- Heneghan, H.M.; Miller, N.; Lowery, A.J.; Sweeney, K.J.; Newell, J.; Kerin, M.J. Circulating microRNAs as Novel Minimally Invasive Biomarkers for Breast Cancer. Ann. Surg. 2010, 251, 499–505. [Google Scholar] [CrossRef]
 Control group
Control group  BC group.
BC group.
 Control group
Control group  BC group.
BC group.
| Patient Presentation | Relative Expression of miRs and p-Value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | miR-155 | p | miR-10b | p | miR-34a | p | miR-373 | p | ||
| Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | |||||||
| Age | <40 | 9 | 11.67 (4.15) | 0.699 | 12.33 (3.32) | 0.204 | 5.78 (3.03) | 0.275 | 13.44 (3.58) | 0.872 |
| ≥40 | 90 | 12.16 (3.55) | 10.53 (4.09) | 6.93 (3.01) | 13.26 (3.32) | |||||
| BMI | Obese | 35 | 11.77 (3.34) | 0.489 | 11.43 (3.99) | 0.184 | 7.11 (3.21) | 0.488 | 12.6 (3.33) | 0.137 |
| Normal/ overweight | 64 | 12.3 (3.73) | 10.3 (4.05) | 6.67 (2.92) | 13.64 (3.29) | |||||
| Grade | 1 | 18 | 6.89 (1.61) | <0.001 | 9.94 (4.4) | 0.43 | 6.44 (2.38) | 0.839 | 12.61 (3.58) | 0.049 |
| 2 | 51 | 12.53 (2.64) | 10.53 (4.35) | 6.92 (2.9) | 14.06 (2.89) | |||||
| 3 | 30 | 14.53 (2.58) | 11.43 (3.2) | 6.9 (3.58) | 12.33 (3.62) | |||||
| LN stage | 0 | 33 | 10.36 (3.55) | 0.001 | 6.39 (2.14) | <0.001 | 6.18 (2.11) | 0.516 | 13.79 (3.57) | 0.096 |
| 1 | 27 | 11.62 (2.81) | 11 (2.68) | 7.19 (3.81) | 13.33 (2.65) | |||||
| 2 | 33 | 14.00 (2.90) | 13.79 (2.26) | 7.09 (2.84) | 12.3 (3.6) | |||||
| 3 | 6 | 16.80 (1.07) | 16 (1.27) | 7.33 (4.23) | 15.5 (1.23) | |||||
| Tumor size | 1 | 25 | 8.04 (2.89) | <0.001 | 9.24 (3.99) | 0.001 | 6.96 (2.48) | 0.968 | 12.84 (3.85) | 0.749 |
| 2 | 58 | 13.17 (2.69) | 10.45 (4.01) | 6.78 (3.21) | 13.45 (3.2) | |||||
| 3 | 16 | 14.63 (2.13) | 13.88 (2.39) | 6.81 (3.23) | 13.31 (3.01) | |||||
| Metastasis | 0 | 90 | 12.04 (3.64) | 0.562 | 10.62 (4.03) | 0.563 | 6.73 (2.88) | 0.324 | 13.33 (3.28) | 0.569 |
| 1 | 9 | 12.78 (3.15) | 11.44 (4.3) | 7.78 (4.24) | 12.67 (3.91) | |||||
| Stage | I | 14 | 7.57 (3.28) | <0.001 | 6.86 (2.25) | <0.001 | 6.64 (1.87) | 0.855 | 13.14 (4.11) | 0.845 |
| II | 36 | 12.97 (3.07) | 8.35 (3.37) | 6.54 (3.14) | 13.65 (2.9) | |||||
| III | 40 | 12.63 (3.2) | 13.88 (2.29) | 7.1 (3.08) | 13.08 (3.32) | |||||
| IV | 9 | 6.89 (1.61) | 12.38 (3.5) | 7.13 (4.02) | 12.75 (4.17) | |||||
| Her2/neu receptor | Positive | 27 | 11.59 (4.02) | 0.382 | 10.63 (4.25) | 0.96 | 6.41 (2.06) | 0.365 | 10.78 (4.33) | <0.001 |
| Negative | 71 | 12.31 (3.45) | 10.68 (4) | 7.03 (3.3) | 14.23 (2.28) | |||||
| ER status | Positive | 48 | 12.25 (3.59) | 0.711 | 10.63 (4.04) | 0.865 | 6.6 (2.85) | 0.476 | 14.23 (3.1) | 0.005 |
| Negative | 51 | 11.98 (3.62) | 10.76 (4.09) | 7.04 (3.18) | 12.37 (3.3) | |||||
| PR status | Negative | 41 | 15.23 (2.31) | 0.001 | 10.78 (4.11) | 0.315 | 6.76 (3.42) | 0.863 | 12.78 (3.24) | 0.024 |
| Positive | 50 | 10.54 (2.83) | 10.96 (4) | 6.96 (2.87) | 14.04 (3.14) | |||||
| Pathology type | IDC | 80 | 12.08 (3.48) | 0.72 | 10.68 (4.06) | 0.96 | 6.69 (3.14) | 0.455 | 13.16 (3.35) | 0.066 |
| ILC | 11 | 12.82 (4.12) | 11 (4.03) | 7.91 (2.39) | 15.18 (1.72) | |||||
| Mixed | 8 | 11.5 (4.28) | 10.5 (4.44) | 6.75 (2.44) | 11.75 (3.88) | |||||
| Control vs. M1 cases | Control | 40 | 4.7 (2.38) | <0.001 | 6.33 (3.63) | 0.003 | 12.55 (3.37) | 0.004 | 5.55 (2.51) | <0.001 |
| M1 cases | 9 | 12.78 (3.15) | 11.44 (4.3) | 7.78 (4.24) | 12.67 (3.91 | |||||
| Test Result Variable(s) | AUC | Cut-Off Value | Sensitivity | Specificity | Std. Error a | Asymptotic Significance b | Asymptotic 95% CI | |
|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||
| miR-155 | 0.944 | 7.5 | 86.9 | 90 | 0.019 | 0.001 | 0.889 | 0.977 |
| miR-373 | 0.948 | 10 | 85 | 100 | 0.018 | 0.001 | 0.895 | 0.979 |
| miR-10b | 0.768 | 9.5 | 60 | 93 | 0.043 | 0.001 | 0.686 | 0.838 |
| miR-34a | 0.887 | 10.5 | 91 | 75 | 0.039 | 0.001 | 0.820 | 0.936 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, A.A.; Allam, A.E.; Aref, A.M.; Mahmoud, M.O.; Eldesoky, N.A.; Fawazy, N.; Sakr, Y.; Sobeih, M.E.; Albogami, S.; Fayad, E.; et al. Evaluation of Expressed MicroRNAs as Prospective Biomarkers for Detection of Breast Cancer. Diagnostics 2022, 12, 789. https://doi.org/10.3390/diagnostics12040789
Mohamed AA, Allam AE, Aref AM, Mahmoud MO, Eldesoky NA, Fawazy N, Sakr Y, Sobeih ME, Albogami S, Fayad E, et al. Evaluation of Expressed MicroRNAs as Prospective Biomarkers for Detection of Breast Cancer. Diagnostics. 2022; 12(4):789. https://doi.org/10.3390/diagnostics12040789
Chicago/Turabian StyleMohamed, Amal Ahmed, Ahmed E. Allam, Ahmed M. Aref, Maha Osama Mahmoud, Noha A. Eldesoky, Naglaa Fawazy, Yasser Sakr, Mohamed Emam Sobeih, Sarah Albogami, Eman Fayad, and et al. 2022. "Evaluation of Expressed MicroRNAs as Prospective Biomarkers for Detection of Breast Cancer" Diagnostics 12, no. 4: 789. https://doi.org/10.3390/diagnostics12040789
APA StyleMohamed, A. A., Allam, A. E., Aref, A. M., Mahmoud, M. O., Eldesoky, N. A., Fawazy, N., Sakr, Y., Sobeih, M. E., Albogami, S., Fayad, E., Althobaiti, F., Jafri, I., Alsharif, G., El-Sayed, M., Abdelgeliel, A. S., & Abdel Aziz, R. S. (2022). Evaluation of Expressed MicroRNAs as Prospective Biomarkers for Detection of Breast Cancer. Diagnostics, 12(4), 789. https://doi.org/10.3390/diagnostics12040789






