Fully Automated Thrombus Segmentation on CT Images of Patients with Acute Ischemic Stroke
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
2.1.1. Ground Truth: Manual Annotations
2.1.2. Pre-Processing
2.2. Modelling and Analysis
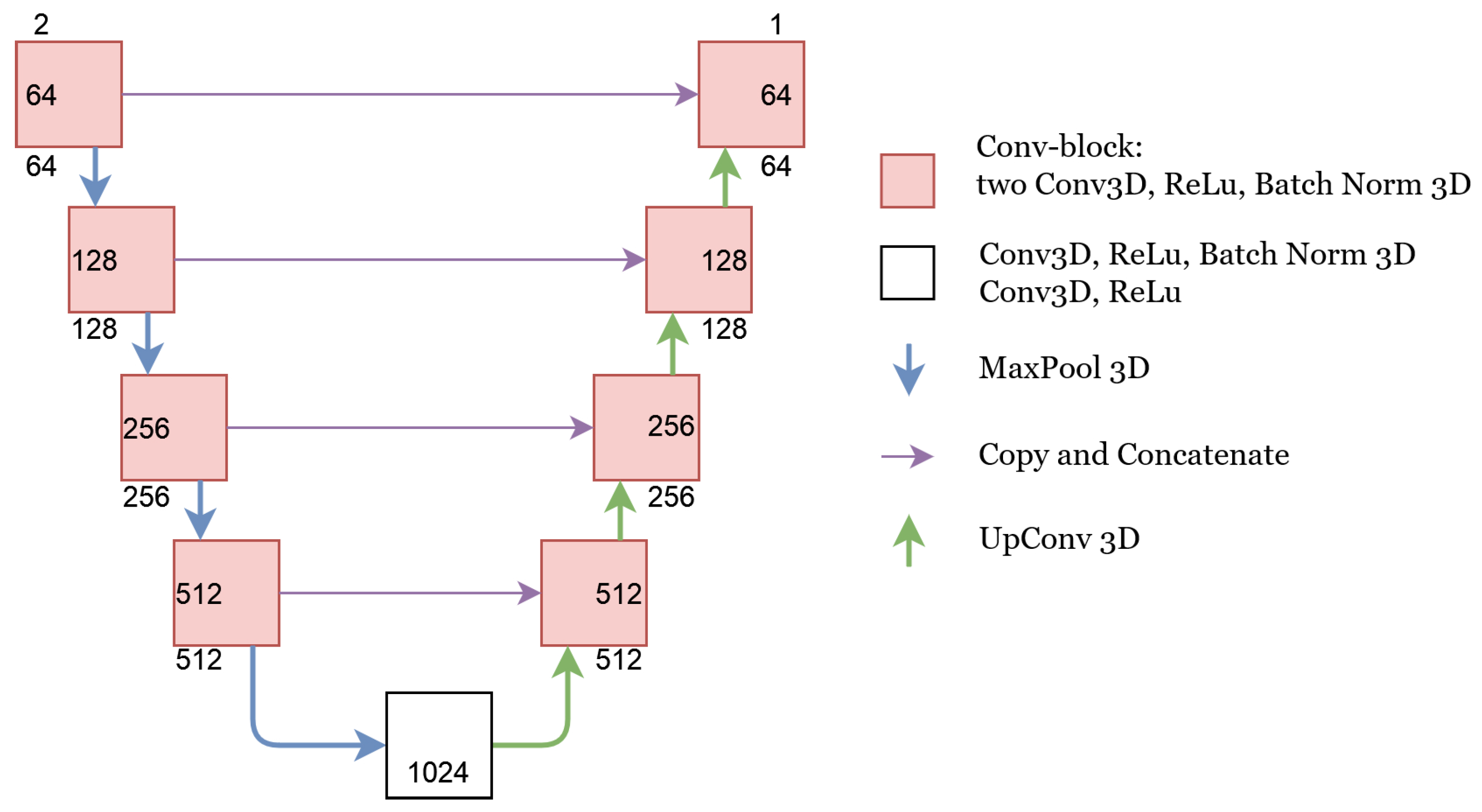
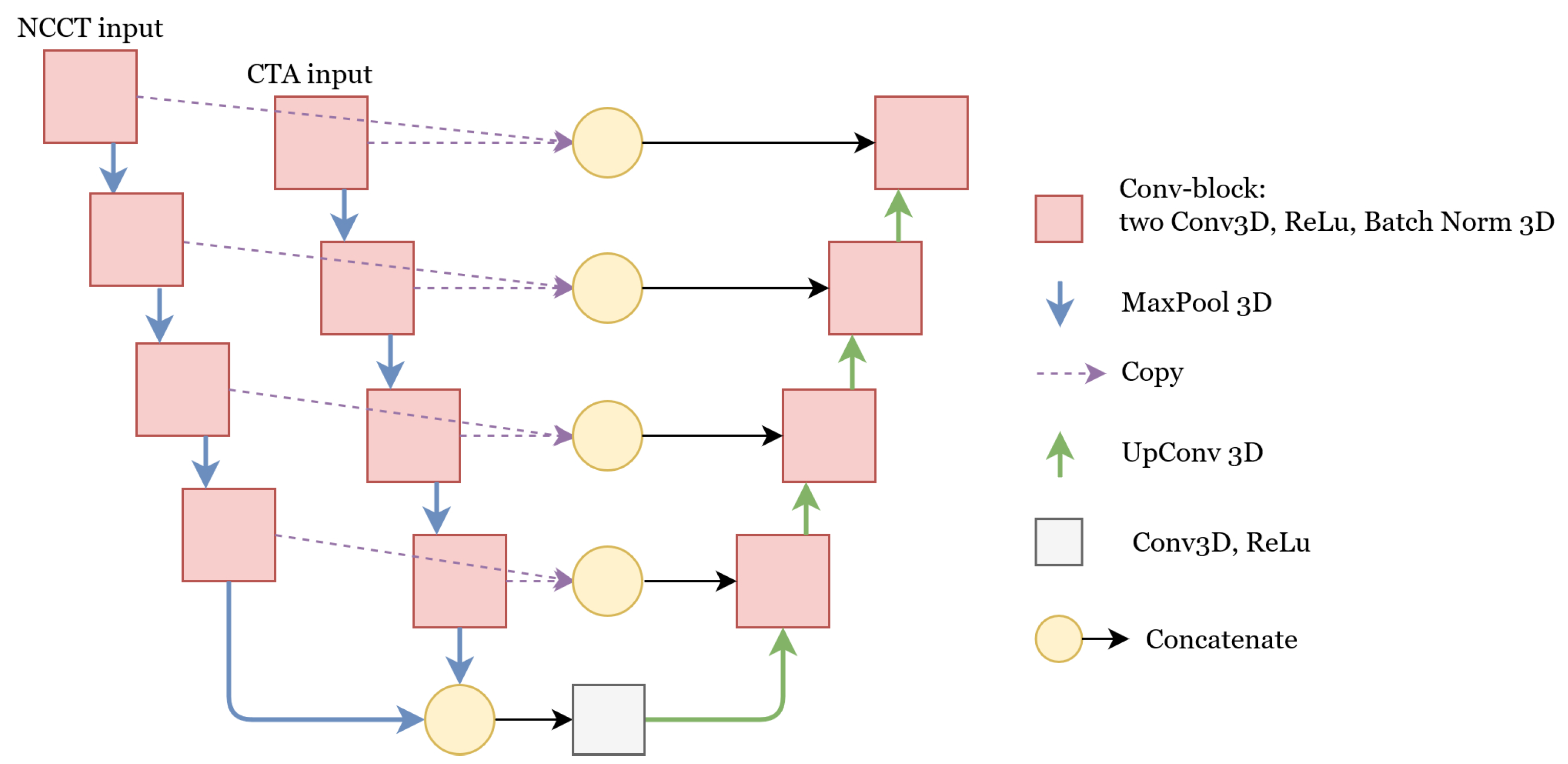
2.2.1. Segmentation Networks
U-Net
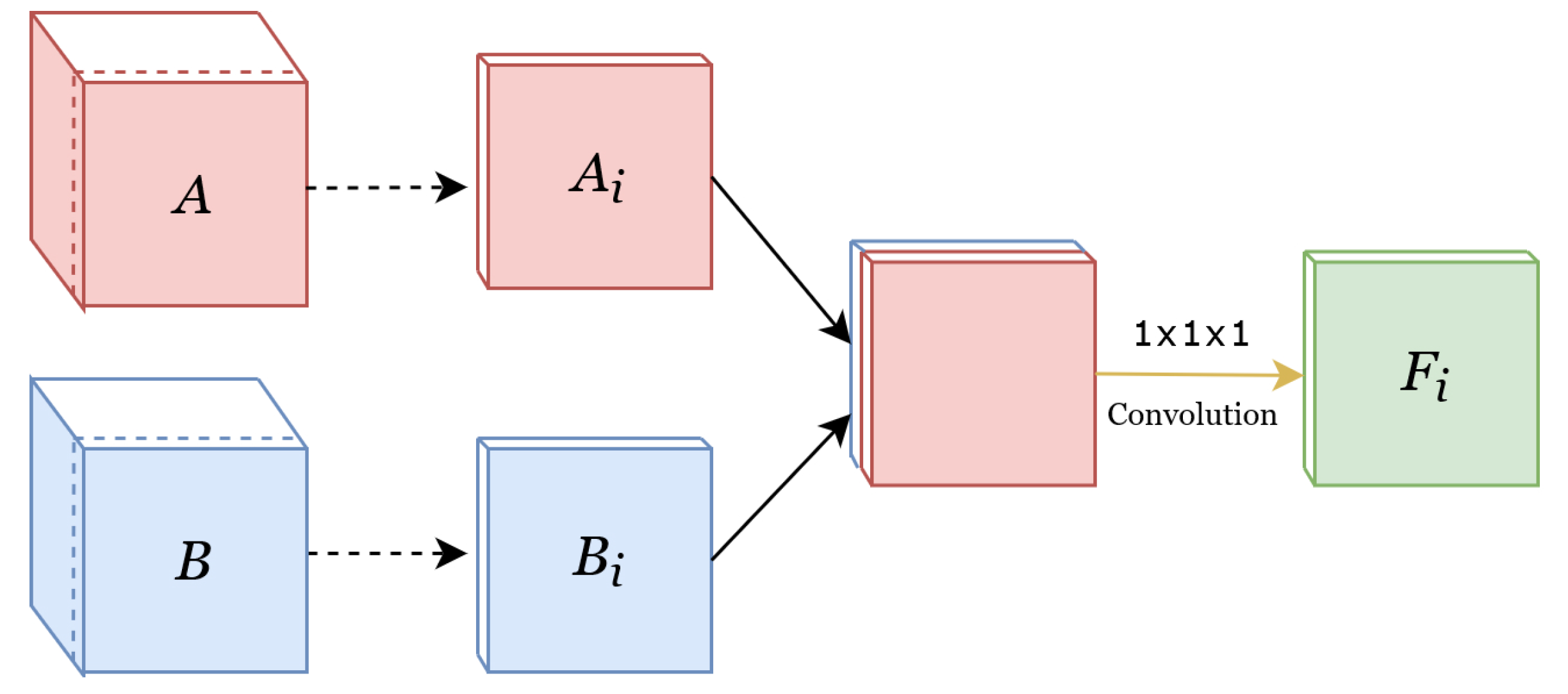
Concatenate
Add
Weighted-Sum
2.2.2. Network Training
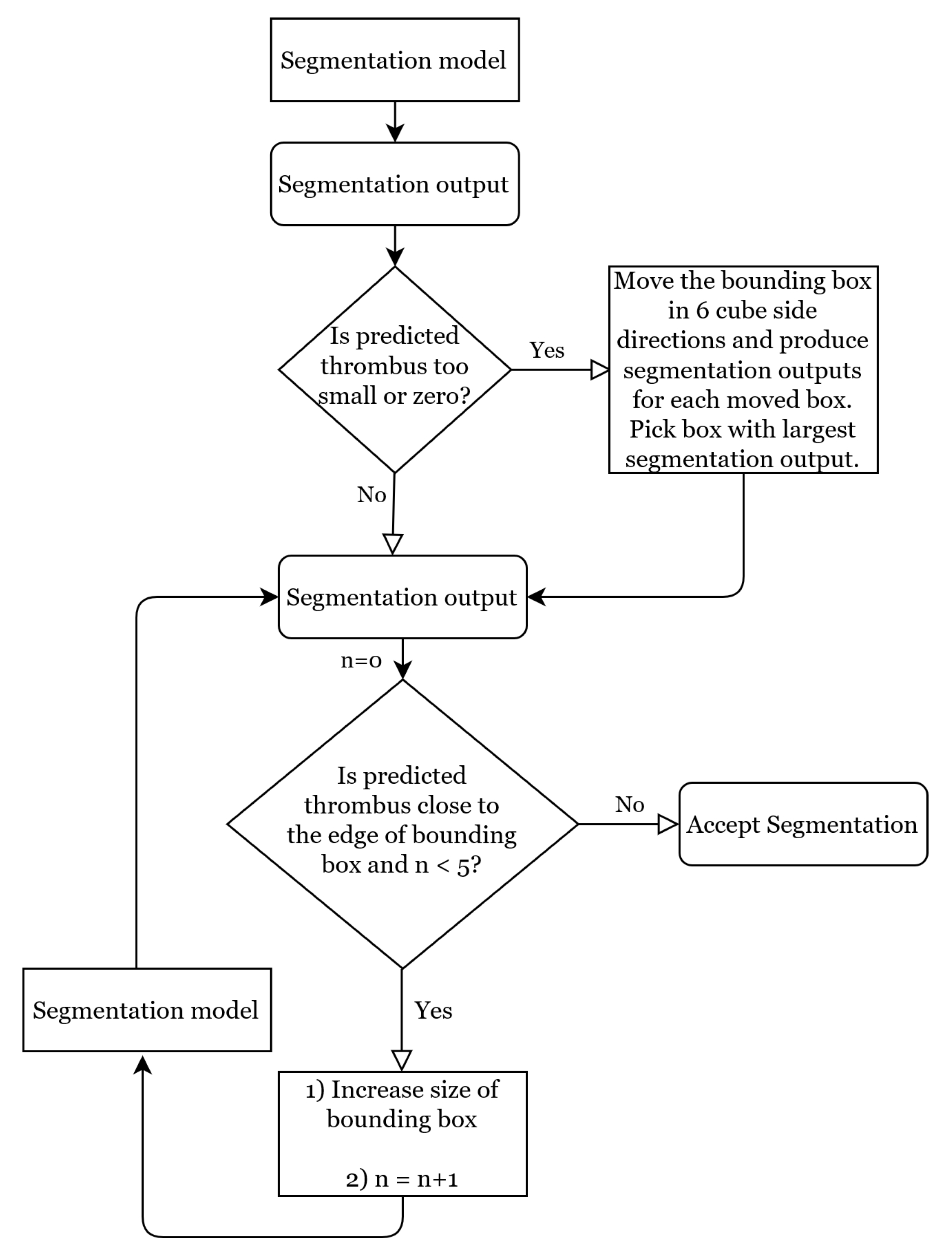
2.2.3. Dynamic Bounding Box Algorithm
Moving Bounding Box Method
Flexible Bounding Box Method
2.2.4. Post-Processing
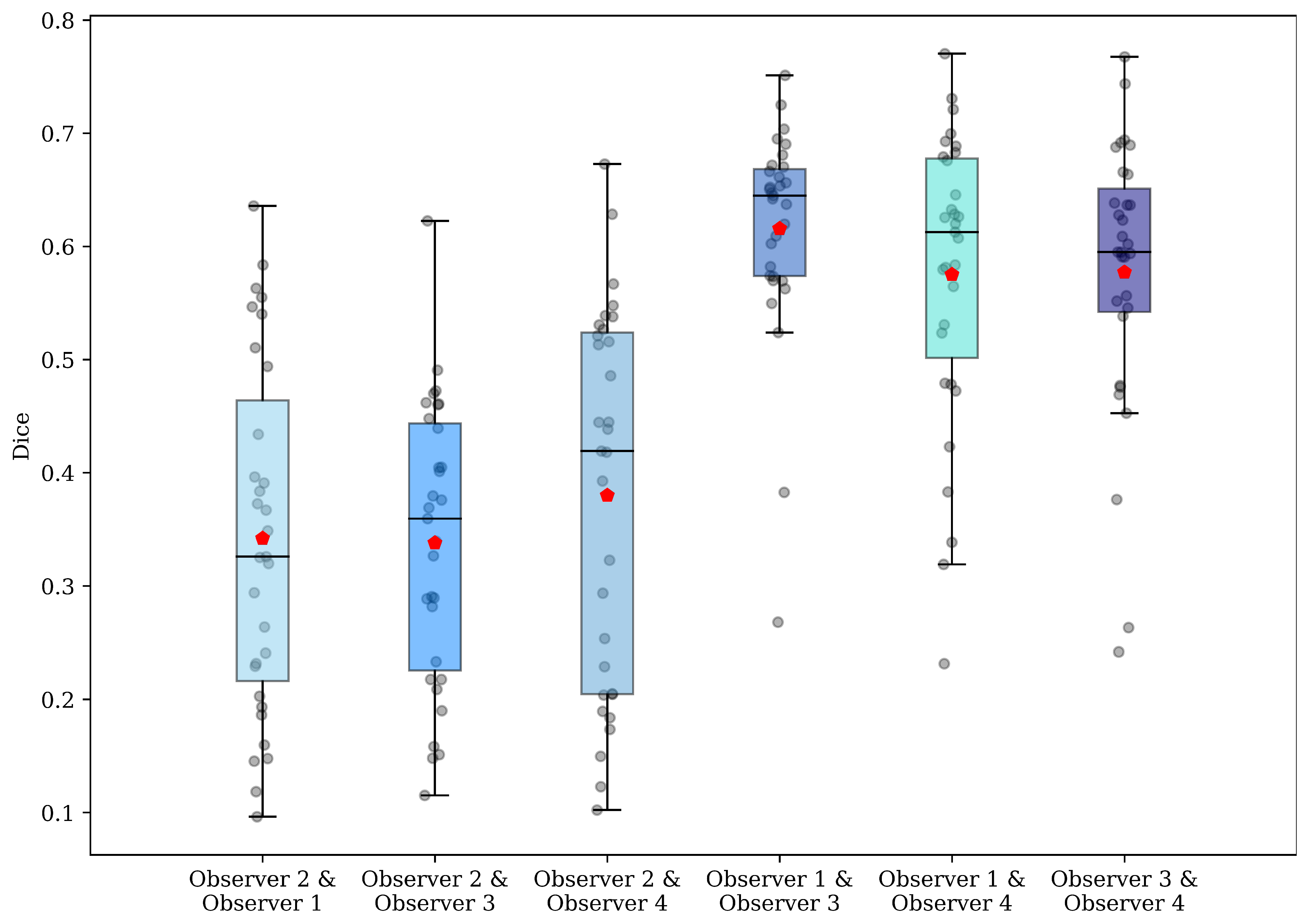
2.3. Evaluation Metrics
2.3.1. Dice
2.3.2. Surface Dice
2.3.3. 95th Percentile Hausdorff Distance (HD)
2.3.4. Intraclass Correlation Coefficient (ICC)
2.3.5. Number of Non-Overlapping Connected Components
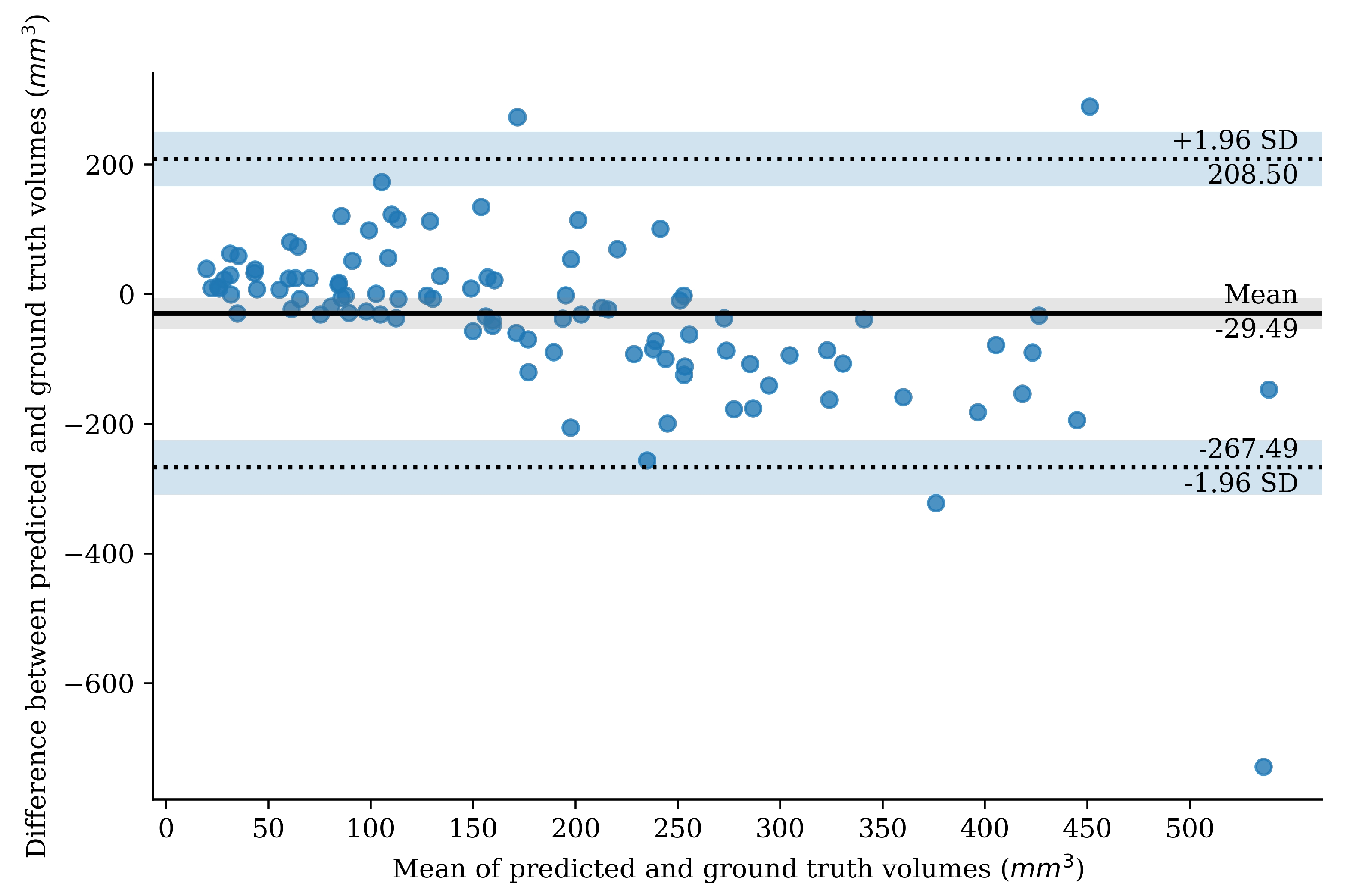
2.3.6. Bland–Altman Plot
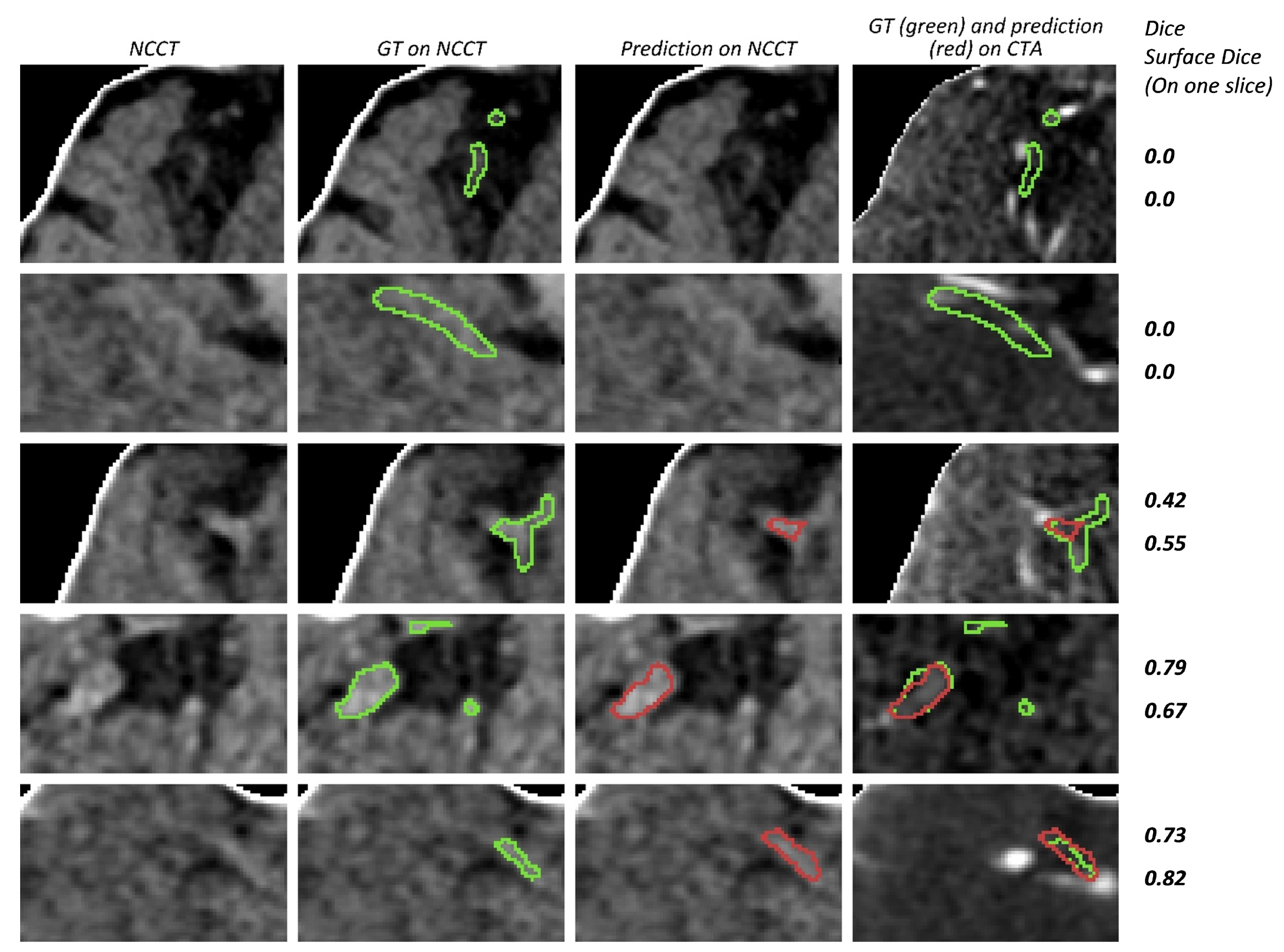
2.3.7. Missed Cases
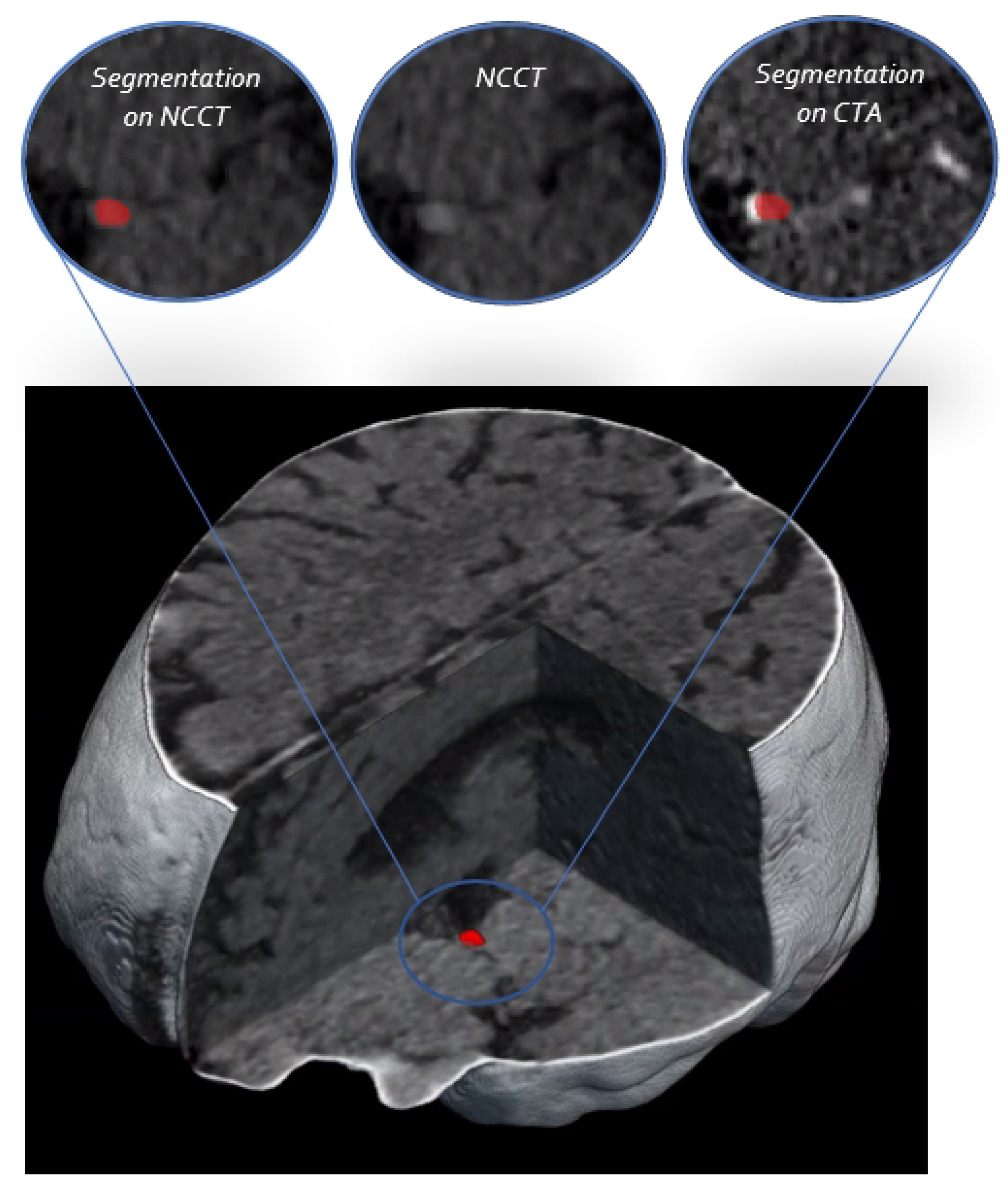
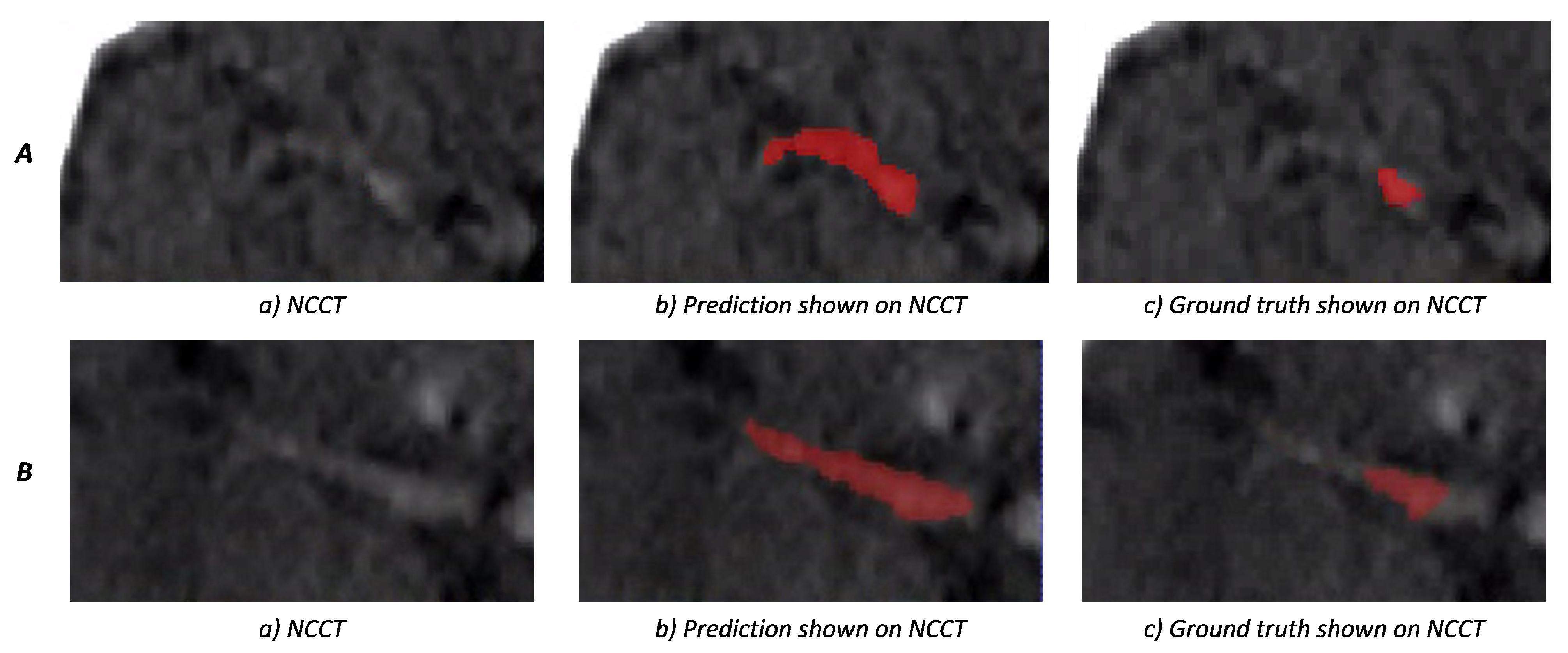
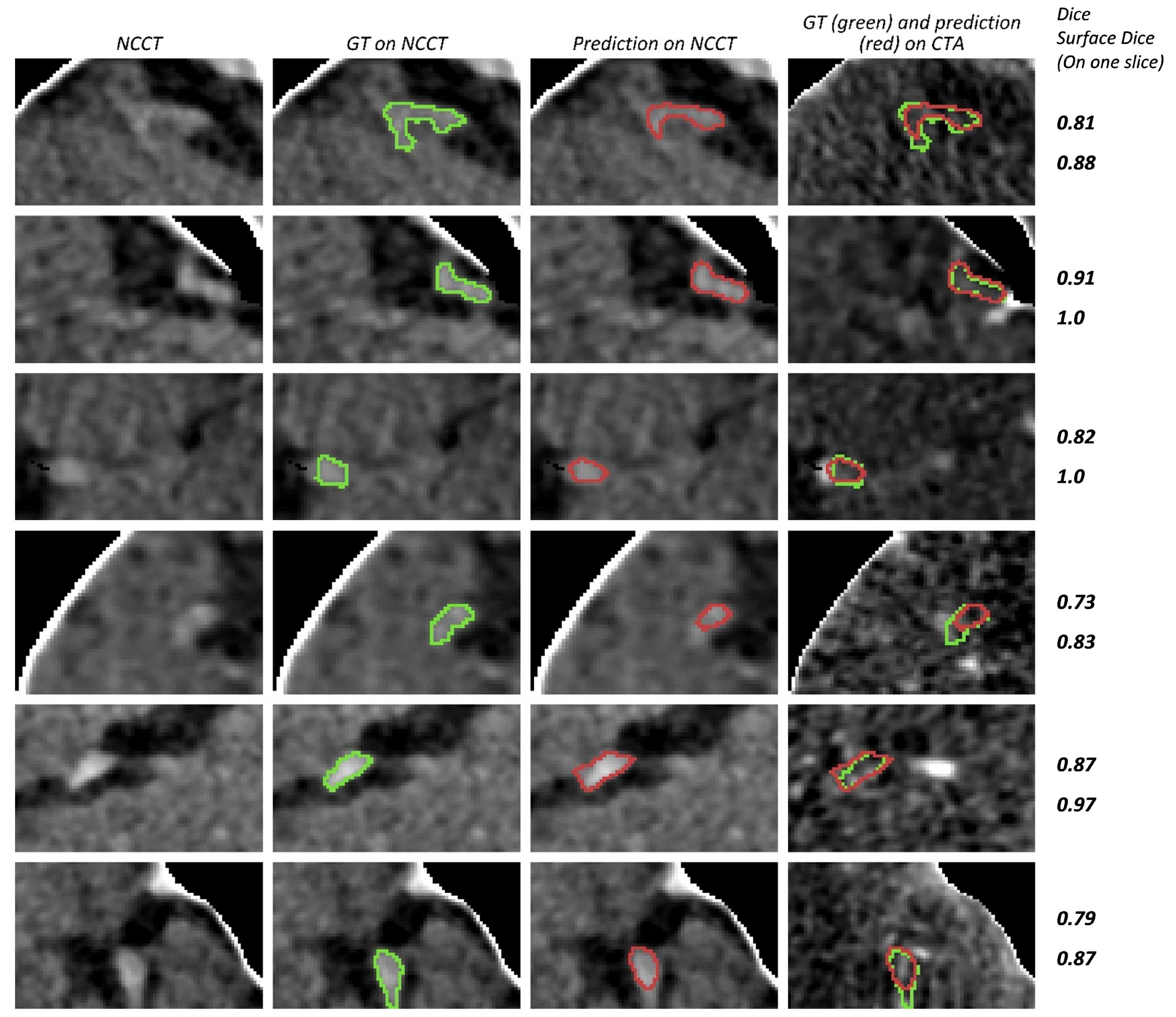
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
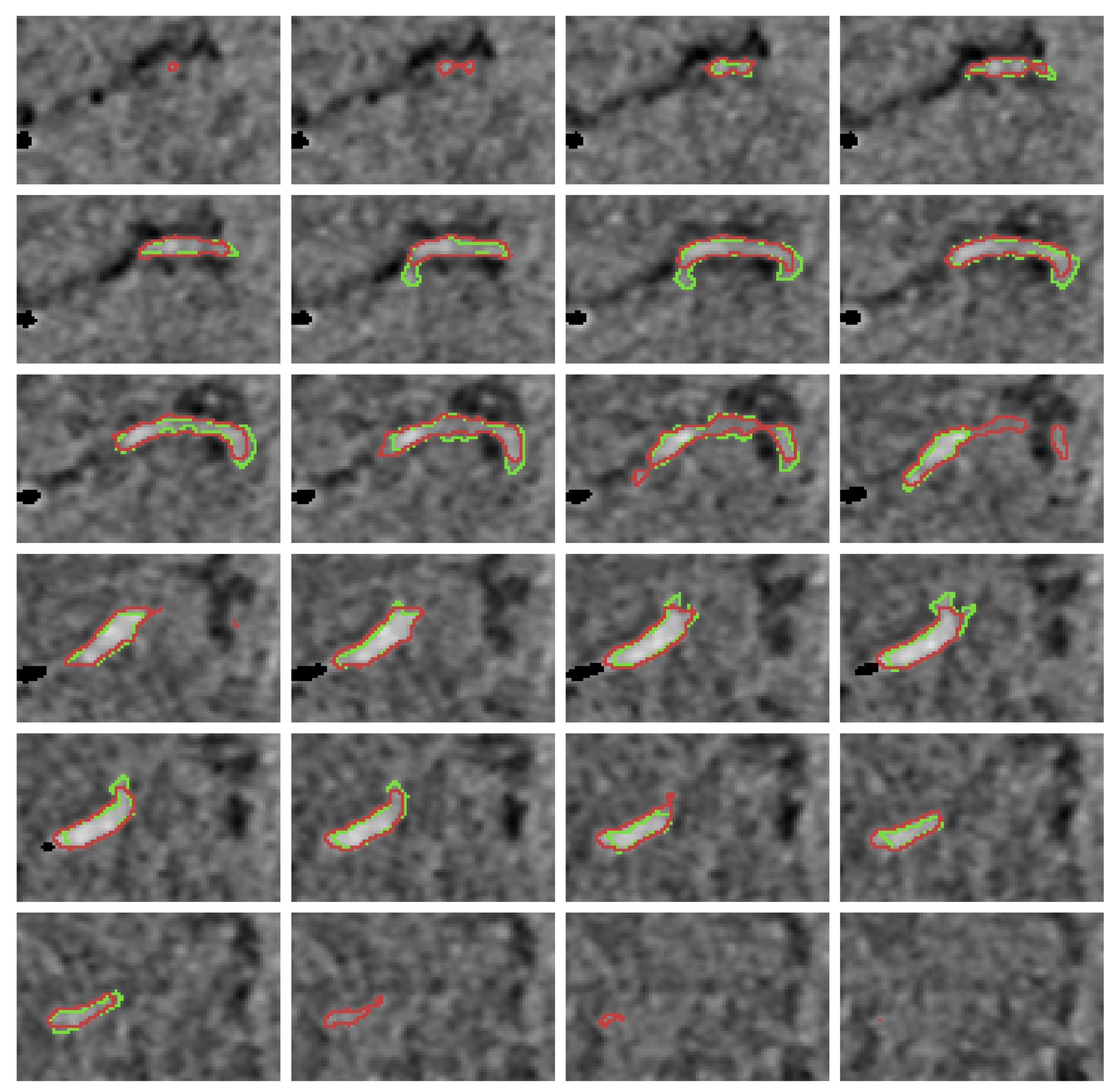
Appendix B
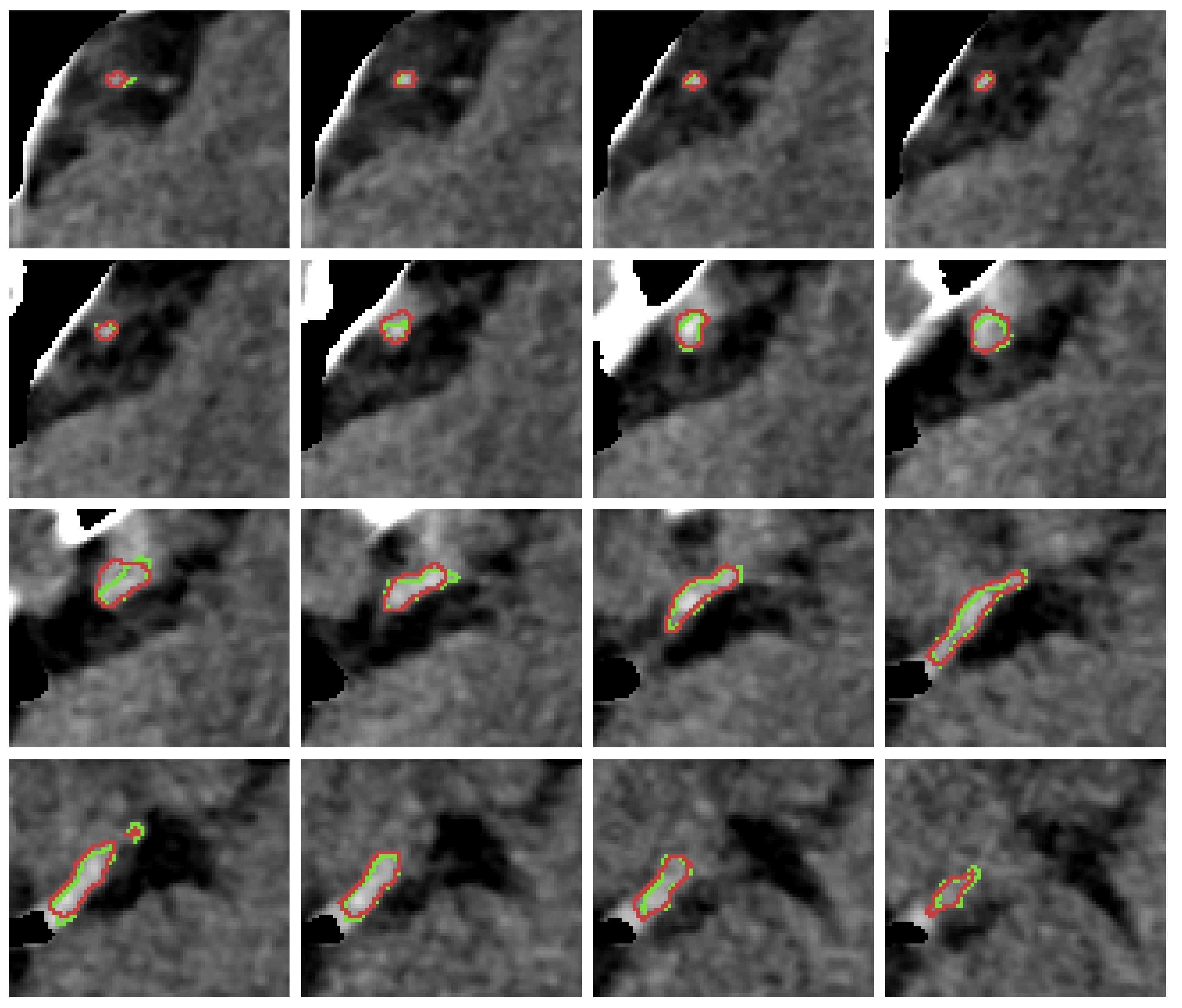
References
- Dutra, B.G.; Tolhuisen, M.L.; Alves, H.C.; Treurniet, K.M.; Kappelhof, M.; Yoo, A.J.; Jansen, I.G.; Dippel, D.W.; van Zwam, W.H.; van Oostenbrugge, R.J.; et al. Thrombus Imaging Characteristics and Outcomes in Acute Ischemic Stroke Patients Undergoing Endovascular Treatment. Stroke 2019, 50, 2057–2064. [Google Scholar] [CrossRef] [PubMed]
- Kappelhof, M.; Tolhuisen, M.L.; Treurniet, K.M.; Dutra, B.G.; Alves, H.; Zhang, G.; Brown, S.; Muir, K.W.; Dávalos, A.; Roos, Y.B.; et al. Endovascular Treatment Effect Diminishes With Increasing Thrombus Perviousness. Stroke 2021, 52, 3633–3641. [Google Scholar] [CrossRef] [PubMed]
- Santos, E.M.M.; Niessen, W.J.; Yoo, A.J.; Berkhemer, O.A.; Beenen, L.F.; Majoie, C.B.; Marquering, H.A. Automated Entire Thrombus Density Measurements for Robust and Comprehensive Thrombus Characterization in Patients with Acute Ischemic Stroke. PLoS ONE 2016, 11, e0145641. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qiu, W.; Kuang, H.; Nair, J.; Assis, Z.; Najm, M.; McDougall, C.; McDougall, B.; Chung, K.; Wilson, A.T.; Goyal, M.; et al. Radiomics-Based Intracranial Thrombus Features on CT and CTA Predict Recanalization with Intravenous Alteplase in Patients with Acute Ischemic Stroke. AJNR Am. J. Neuroradiol. 2019, 40, 39–44. [Google Scholar] [CrossRef]
- Hofmeister, J.; Bernava, G.; Rosi, A.; Vargas, M.I.; Carrera, E.; Montet, X.; Burgermeister, S.; Poletti, P.A.; Platon, A.; Lovblad, K.O.; et al. Clot-Based Radiomics Predict a Mechanical Thrombectomy Strategy for Successful Recanalization in Acute Ischemic Stroke. Stroke 2020, 51, 2488–2494. [Google Scholar] [CrossRef]
- Warner, J.J.; Harrington, R.A.; Sacco, R.L.; Elkind, M.S. Guidelines for the Early Management of Patients With Acute Ischemic Stroke: 2019 Update to the 2018 Guidelines for the Early Management of Acute Ischemic Stroke. Stroke 2019, 50, 3331–3332. [Google Scholar] [CrossRef] [PubMed]
- Koo, C.K.; Teasdale, E.; Muir, K.W. What constitutes a true hyperdense middle cerebral artery sign? Cerebrovasc. Dis. 2000, 10, 419–423. [Google Scholar] [CrossRef]
- Boodt, N.; Compagne, K.C.; Dutra, B.G.; Samuels, N.; Tolhuisen, M.L.; Alves, H.C.; Kappelhof, M.; Lycklama Nijeholt, G.J.; Marquering, H.A.; Majoie, C.B.; et al. Stroke Etiology and Thrombus Computed Tomography Characteristics in Patients with Acute Ischemic Stroke: A MR CLEAN Registry Substudy. Stroke 2020, 51, 1727–1735. [Google Scholar] [CrossRef]
- Chieng, J.; Singh, D.; Chawla, A.; Peh, W. The hyperdense vessel sign in cerebral computed tomography: Pearls and pitfalls. Singap. Med. J. 2020, 61, 230–237. [Google Scholar] [CrossRef]
- Tolhuisen, M.L.; Enthoven, J.; Santos, E.M.M.; Niessen, W.J.; Beenen, L.F.M.; Dippel, D.W.J.; van der Lugt, A.; van Zwam, W.H.; Roos, Y.B.W.E.M.; van Oostenbrugge, R.J.; et al. The Effect of Non-contrast CT Slice Thickness on Thrombus Density and Perviousness Assessment. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2017; Volume 10555 LNCS, pp. 168–175. [Google Scholar] [CrossRef]
- Santos, E.M.M.; Marquering, H.A.; Berkhemer, O.A.; van Zwam, W.H.; van der Lugt, A.; Majoie, C.B.; Niessen, W.J. Development and Validation of Intracranial Thrombus Segmentation on CT Angiography in Patients with Acute Ischemic Stroke. PLoS ONE 2014, 9, e101985. [Google Scholar] [CrossRef]
- Tolhuisen, M.L.; Ponomareva, E.; Boers, A.M.M.; Jansen, I.G.H.; Koopman, M.S.; Sales Barros, R.; Berkhemer, O.A.; van Zwam, W.H.; van der Lugt, A.; Majoie, C.B.L.M.; et al. A Convolutional Neural Network for Anterior Intra-Arterial Thrombus Detection and Segmentation on Non-Contrast Computed Tomography of Patients with Acute Ischemic Stroke. Appl. Sci. 2020, 10, 4861. [Google Scholar] [CrossRef]
- Amukotuwa, S.A.; Straka, M.; Dehkharghani, S.; Bammer, R. Fast automatic detection of large vessel occlusions on CT angiography. Stroke 2019, 50, 3431–3438. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Somayaji, N.R.; Kabakis, I.M. Abstract WMP16: Artificial Intelligence Detection of Cerebrovascular Large Vessel Occlusion—Nine Month, 650 Patient Evaluation of the Diagnostic Accuracy and Performance of the Viz.ai LVO Algorithm. Stroke 2019, 50. [Google Scholar] [CrossRef]
- Beesomboon, D.; Kaothanthong, N.; Songsaeng, D.; Chansumpao, T.; Sarampakhul, S. Thrombus localization in middle cerebral artery of patient with acute ischemic stroke on ncCT image. In Proceedings of the 2019 1st International Conference on Smart Technology and Urban Development (STUD), Chiang Mai, Thailand, 13–14 December 2019; pp. 48–54. [Google Scholar] [CrossRef]
- Lucas, C.; Schöttler, J.J.; Kemmling, A.; Aulmann, L.F.; Heinrich, M.P. Automatic Detection and Segmentation of the Acute Vessel Thrombus in Cerebral CT. In Informatik Aktuell; Springer: Berlin/Heidelberg, Germany, 2019; pp. 74–79. [Google Scholar] [CrossRef]
- Berkhemer, O.A.; Fransen, P.S.; Beumer, D.; van den Berg, L.A.; Lingsma, H.F.; Yoo, A.J.; Schonewille, W.J.; Vos, J.A.; Nederkoorn, P.J.; Wermer, M.J.; et al. A Randomized Trial of Intraarterial Treatment for Acute Ischemic Stroke. N. Engl. J. Med. 2015, 372, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Treurniet, K.M.; LeCouffe, N.E.; Kappelhof, M.; Emmer, B.J.; van Es, A.C.G.M.; Boiten, J.; Lycklama, G.J.; Keizer, K.; Yo, L.S.F.; Lingsma, H.F.; et al. MR CLEAN-NO IV: Intravenous treatment followed by endovascular treatment versus direct endovascular treatment for acute ischemic stroke caused by a proximal intracranial occlusion—Study protocol for a randomized clinical trial. Trials 2021, 22, 141. [Google Scholar] [CrossRef] [PubMed]
- Collins, D.L.; Neelin, P.; Peters, T.M.; Evans, A.C. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J. Comput. Assist. Tomogr. 1994, 18, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Marstal, K.; Berendsen, F.; Staring, M.; Klein, S. SimpleElastix: A User-Friendly, Multi-lingual Library for Medical Image Registration. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 574–582. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury Google, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. Adv. Neural Inf. Process. Syst. 2019, 32, 13435. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; 2017. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1–9. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 26 July 2017. [Google Scholar]
- Dice, L.R. Measures of the Amount of Ecologic Association between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging 2015, 15, 68. [Google Scholar] [CrossRef]
- Nikolov, S.; Blackwell, S.; Zverovitch, A.; Mendes, R.; Livne, M.; de Fauw, J.; Patel, Y.; Meyer, C.; Askham, H.; Romera-Paredes, B.; et al. Clinically applicable segmentation of head and neck anatomy for radiotherapy: Deep learning algorithm development and validation study. J. Med. Internet Res. 2021, 23, e26151. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Bland, J.M.; Altman, D.G. Measuring agreement in method comparison studies. Stat. Methods Med. Res. 1999, 8, 135–160. [Google Scholar] [CrossRef] [PubMed]
- Luijten, S.P.R.; Wolff, L.; Duvekot, M.H.C.; van Doormaal, P.J.; Moudrous, W.; Kerkhoff, H.; Lycklama a Nijeholt, G.J.; Bokkers, R.P.H.; Yo, L.S.F.; Hofmeijer, J.; et al. Diagnostic performance of an algorithm for automated large vessel occlusion detection on CT angiography. J. Neurointerventional Surg. 2021, 2021, 017842. [Google Scholar] [CrossRef] [PubMed]
- Zou, K.H.; Warfield, S.K.; Bharatha, A.; Tempany, C.M.; Kaus, M.R.; Haker, S.J.; Wells, W.M.; Jolesz, F.A.; Kikinis, R. Statistical validation of image segmentation quality based on a spatial overlap index1. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef]
- Riedel, C.H.; Zimmermann, P.; Jensen-Kondering, U.; Stingele, R.; Deuschl, G.; Jansen, O. The importance of size: Successful recanalization by intravenous thrombolysis in acute anterior stroke depends on thrombus length. Stroke 2011, 42, 1775–1777. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.M.; Dykeman, J.; Sajobi, T.T.; Trivedi, A.; Almekhlafi, M.; Sohn, S.I.; Bal, S.; Qazi, E.; Calleja, A.; Eesa, M.; et al. Early reperfusion rates with IV tPA are determined by CTA clot characteristics. AJNR Am. J. Neuroradiol. 2014, 35, 2265–2272. [Google Scholar] [CrossRef] [PubMed]
- Menon, B.K.; Al-Ajlan, F.S.; Najm, M.; Puig, J.; Castellanos, M.; Dowlatshahi, D.; Calleja, A.; Sohn, S.I.; Ahn, S.H.; Poppe, A.; et al. Association of Clinical, Imaging, and Thrombus Characteristics With Recanalization of Visible Intracranial Occlusion in Patients With Acute Ischemic Stroke. JAMA 2018, 320, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Santos, E.M.; Arrarte Terreros, N.; Kappelhof, M.; Borst, J.; Boers, A.M.; Lingsma, H.F.; Berkhemer, O.A.; Dippel, D.W.; Majoie, C.B.; Marquering, H.A.; et al. Associations of thrombus perviousness derived from entire thrombus segmentation with functional outcome in patients with acute ischemic stroke. J. Biomech. 2021, 128, 110700. [Google Scholar] [CrossRef]
- Liu, Y.; Brinjikji, W.; Abbasi, M.; Dai, D.; Arturo Larco, J.L.; Irfan Madhani, S.; Shahid, A.H.; Madalina Mereuta, O.; Nogueira, R.G.; Kvamme, P.; et al. Quantification of clot spatial heterogeneity and its impact on thrombectomy. J. Neurointerv. Surg. 2021; Ahead of print. [Google Scholar] [CrossRef]
- Joundi, R.A.; Menon, B.K. Thrombus Composition, Imaging, and Outcome Prediction in Acute Ischemic Stroke. Neurology 2021, 97, S68–S78. [Google Scholar] [CrossRef]
- Luthman, A.S.; Bouchez, L.; Botta, D.; Vargas, M.I.; Machi, P.; Lövblad, K.O. Imaging Clot Characteristics in Stroke and its Possible Implication on Treatment. Clin. Neuroradiol. 2020, 30, 27–35. [Google Scholar] [CrossRef]

| Dataset | % Right Hemisphere Affected | Average Thrombus Volume (mm) | Average Thrombus Length (mm) | Average Hounsfield Value of Thrombus on NCCT |
|---|---|---|---|---|
| Training set | 55 | 221 (196, 246) | 31 (28, 34) | 47 (45, 49) |
| Validation set | 55 | 187 (111, 263) | 23 (16, 29) | 45 (42, 48) |
| Test set | 48 | 168 (147, 188) | 32 (29, 36) | 50 (49, 51) |
| Model | Number of Trainable Parameters | Dice | Surface Dice | Number of Non-Overlapping Connected Components | Volume ICC | 95th Percentile HD | Missed Cases |
|---|---|---|---|---|---|---|---|
| U-Net | 82M | 0.59 (0.55, 0.64) | 0.75 (0.69, 0.80) | 2.5 (1.5, 3.5) | 0.70 | 10.46 (7.33, 13.58) | 6% |
| Concatenate | 115M | 0.60 (0.55, 0.65) | 0.76 (0.71, 0.82) | 0.6 (0.3, 0.9) | 0.68 | 5.89 (4.36, 7.42) | 8% |
| Weighted-sum | 97M | 0.62 (0.58, 0.66) | 0.78 (0.73, 0.83) | 0.7 (0.4, 0.9) | 0.60 | 5.88 (4.54, 7.22) | 4% |
| Add | 96M | 0.53 (0.48, 0.58) | 0.69 (0.63, 0.76) | 0.3 (0.1, 0.4) | 0.49 | 5.67 (4.30, 7.04) | 11% |
| U-Net with no CTA input | 82M | 0.38 (0.32, 0.45) | 0.51 (0.43, 0.58) | 0.2 (0.1, 0.3) | 0.41 | 7.75 (5.25, 10.26) | 26% |
| Metric | ICA-T | M1 | M2 |
|---|---|---|---|
| Dice | 0.64 (0.57, 0.72) | 0.63 (0.58, 0.68) | 0.51 (0.29, 0.74) |
| Surface Dice | 0.78 (0.69, 0.87) | 0.79 (0.74, 0.85) | 0.66 (0.37, 0.95) |
| Experiment | Dice | Surface Dice | Number of Non-Overlapping Connected Components | Volume ICC | 95th Percentile HD | Missed Cases |
|---|---|---|---|---|---|---|
| No moving bounding box | 0.60 (0.55, 0.64) | 0.75 (0.69, 0.80) | 0.6 (0.4, 0.9) | 0.67 | 5.58 (4.30, 6.85) | 9% |
| No flexible bounding box | 0.61 (0.57, 0.66) | 0.77 (0.73, 0.82) | 0.33 (0.2, 0.4) | 0.63 | 6.39 (4.30, 8.48) | 4% |
| No post-processing | 0.62 (0.58, 0.66) | 0.78 (0.73, 0.83) | 0.9 (0.6, 1.3) | 0.60 | 5.85 (4.51, 7.19) | 4% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mojtahedi, M.; Kappelhof, M.; Ponomareva, E.; Tolhuisen, M.; Jansen, I.; Bruggeman, A.A.E.; Dutra, B.G.; Yo, L.; LeCouffe, N.; Hoving, J.W.; et al. Fully Automated Thrombus Segmentation on CT Images of Patients with Acute Ischemic Stroke. Diagnostics 2022, 12, 698. https://doi.org/10.3390/diagnostics12030698
Mojtahedi M, Kappelhof M, Ponomareva E, Tolhuisen M, Jansen I, Bruggeman AAE, Dutra BG, Yo L, LeCouffe N, Hoving JW, et al. Fully Automated Thrombus Segmentation on CT Images of Patients with Acute Ischemic Stroke. Diagnostics. 2022; 12(3):698. https://doi.org/10.3390/diagnostics12030698
Chicago/Turabian StyleMojtahedi, Mahsa, Manon Kappelhof, Elena Ponomareva, Manon Tolhuisen, Ivo Jansen, Agnetha A. E. Bruggeman, Bruna G. Dutra, Lonneke Yo, Natalie LeCouffe, Jan W. Hoving, and et al. 2022. "Fully Automated Thrombus Segmentation on CT Images of Patients with Acute Ischemic Stroke" Diagnostics 12, no. 3: 698. https://doi.org/10.3390/diagnostics12030698
APA StyleMojtahedi, M., Kappelhof, M., Ponomareva, E., Tolhuisen, M., Jansen, I., Bruggeman, A. A. E., Dutra, B. G., Yo, L., LeCouffe, N., Hoving, J. W., van Voorst, H., Brouwer, J., Terreros, N. A., Konduri, P., Meijer, F. J. A., Appelman, A., Treurniet, K. M., Coutinho, J. M., Roos, Y., ... Marquering, H. (2022). Fully Automated Thrombus Segmentation on CT Images of Patients with Acute Ischemic Stroke. Diagnostics, 12(3), 698. https://doi.org/10.3390/diagnostics12030698








