Four Types of Multiclass Frameworks for Pneumonia Classification and Its Validation in X-ray Scans Using Seven Types of Deep Learning Artificial Intelligence Models
Abstract
:1. Introduction
2. Related Work
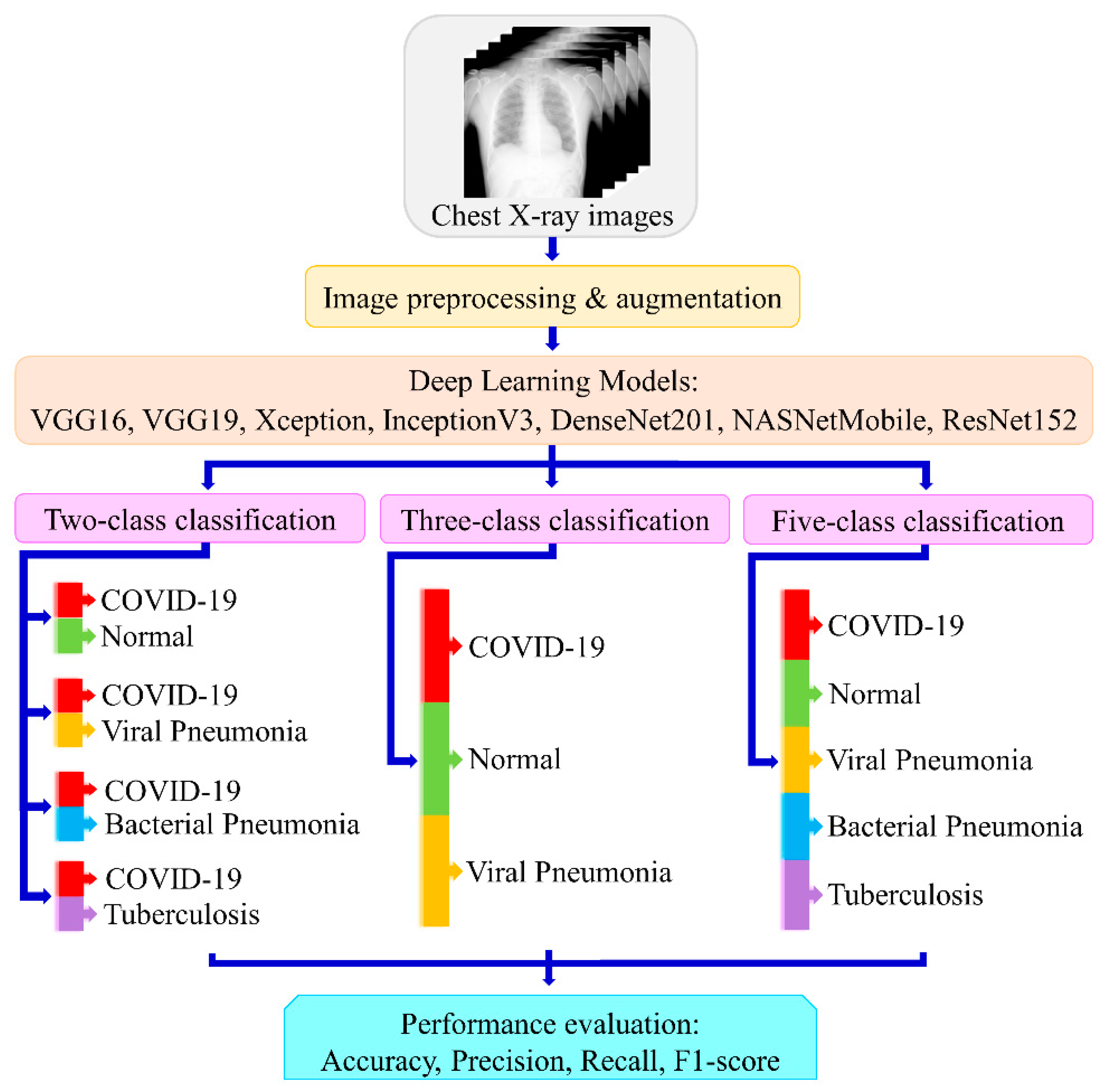
3. Methodology
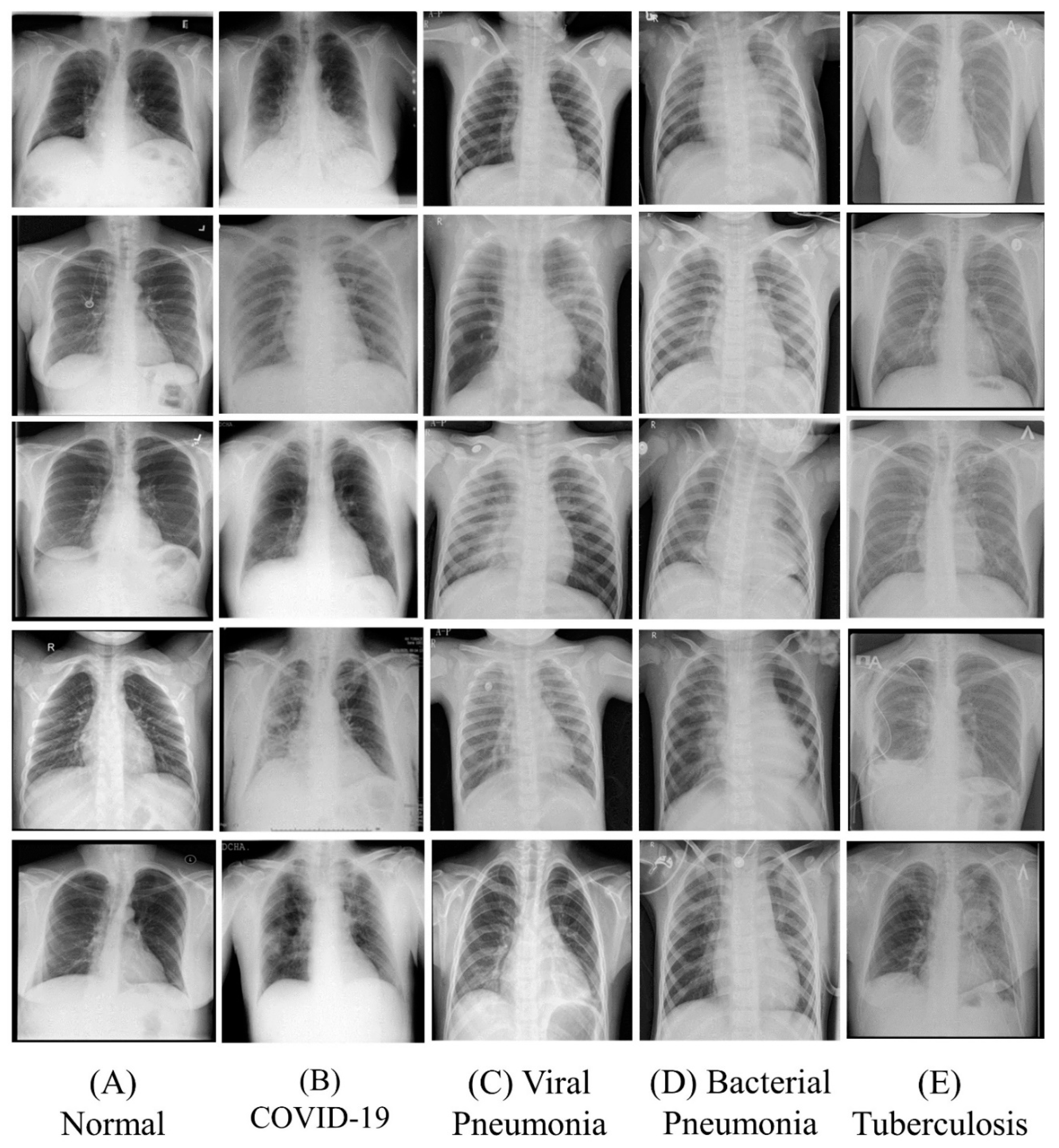
3.1. Dataset
3.1.1. COVID-19 Radiography Database
3.1.2. Chest X-ray Pneumonia Images
3.1.3. Tuberculosis Chest X-ray Database
3.2. Image Processing
3.3. Experimental Setup
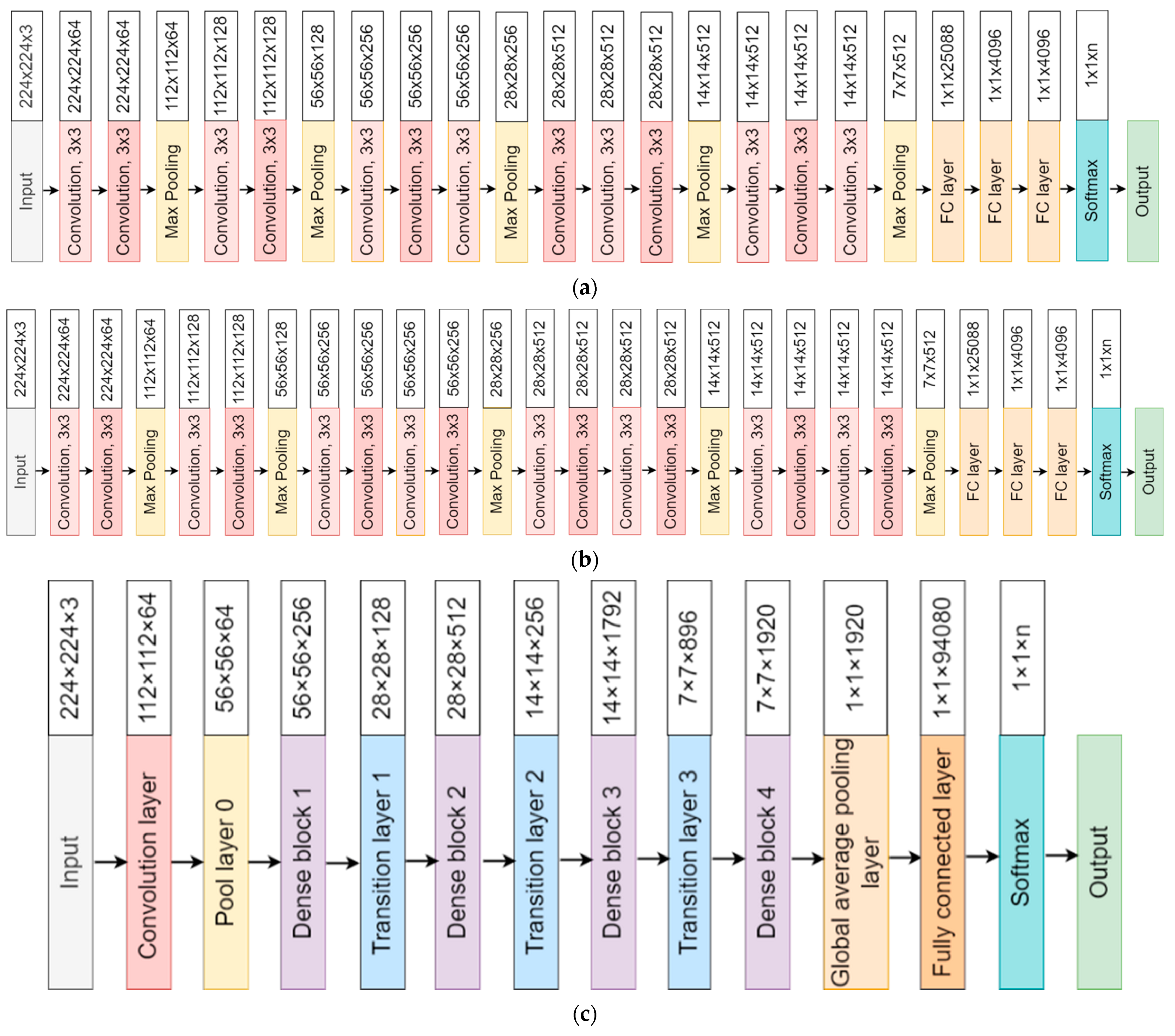
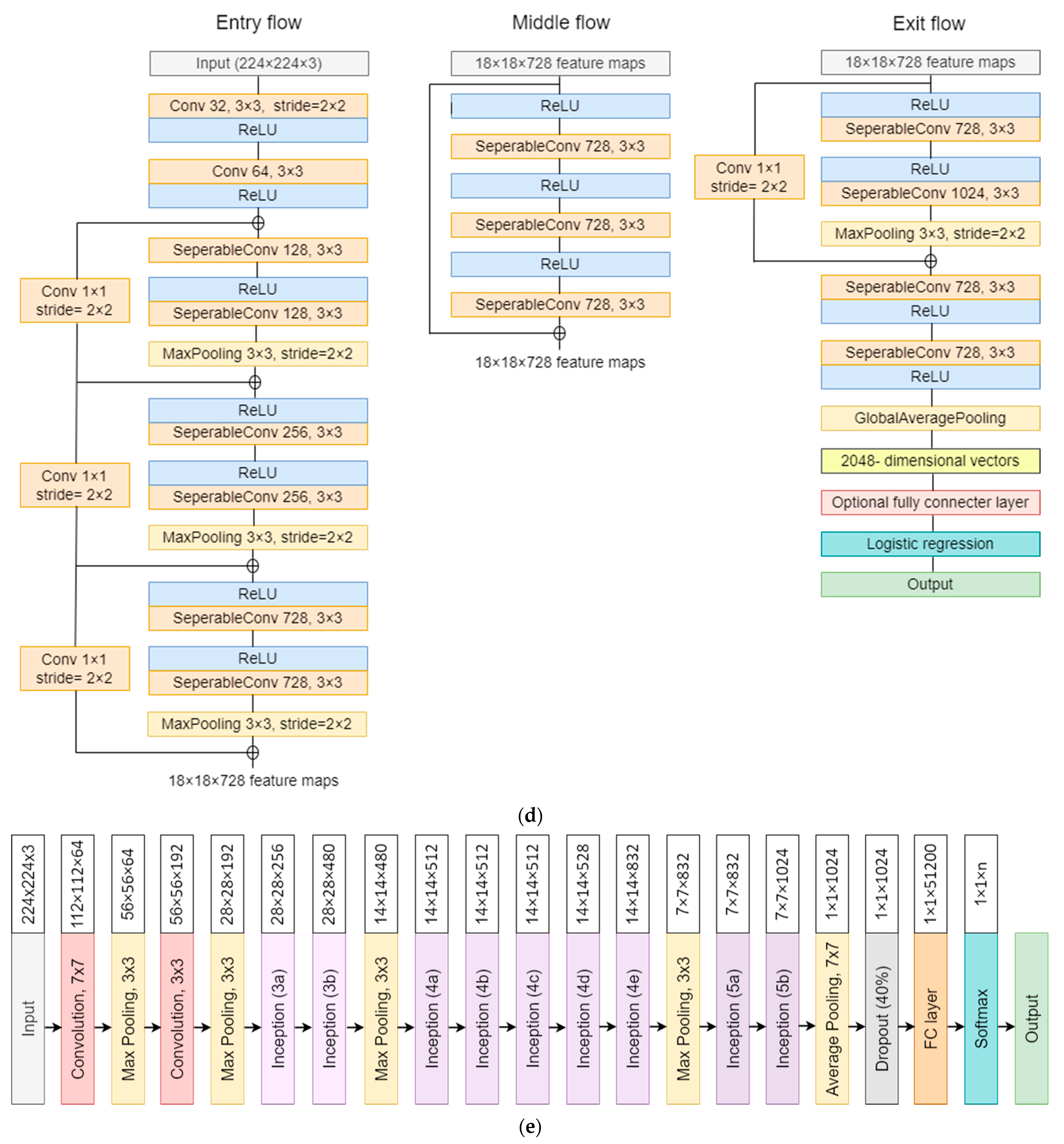
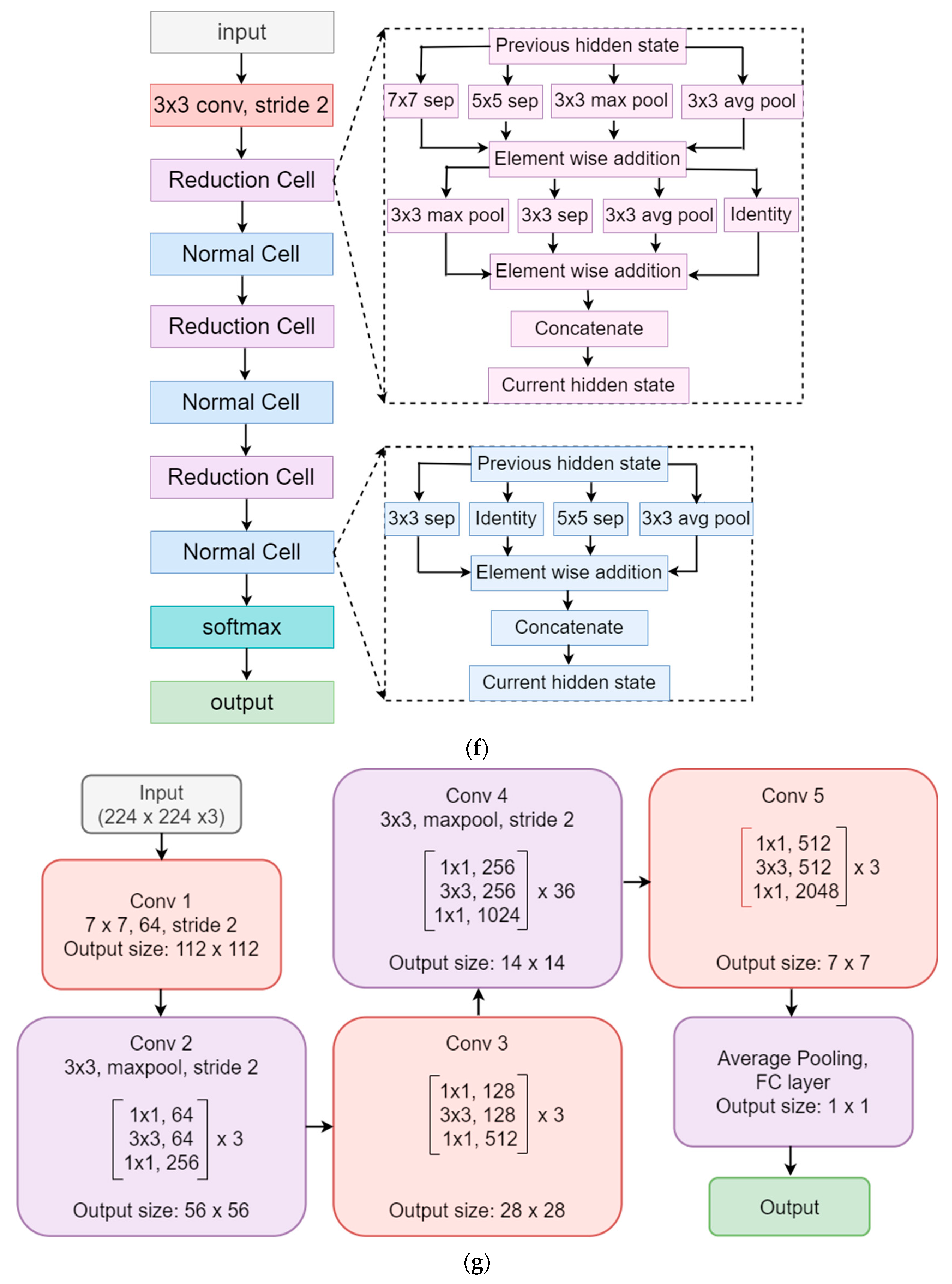
3.4. Model Architectures
3.5. Cross-Entropy Loss Function for Models
3.6. Performance Metrics Used for Classification Evaluation
- (a)
- Accuracy: Accuracy is the most significant criterion for the analysis of the convolutional neural network’s performance. Accuracy is the sum of true positive and true negative values divided by the entire component of the confusion matrix. It is represented as given in Equation (3) [88].
- (b)
- Precision: Precision is an important measure of the results of the CNN models. It counts how many correct positive predictions have been made. Precision is evaluated as the ratio between true positive predicted components and the sum of positive predicted components. It is represented as given in Equation (4) [88].
- (c)
- Recall (Sensitivity): Recall is another important metric for the analysis of the classifier’s performance. It is defined as the ratio between the true positive predicted components and the sum of true positive and false negative predicted components. It is represented as given in Equation (5) [91].
- (d)
- F1-score: The F1-score is an important measure for assessing the test’s accuracy. It is the harmonic mean between precision and recall. It is defined as twice the ratio between multiplication of precision and recall to the sum of precision and recall. It is represented as given in Equation (6) [91].
4. Results
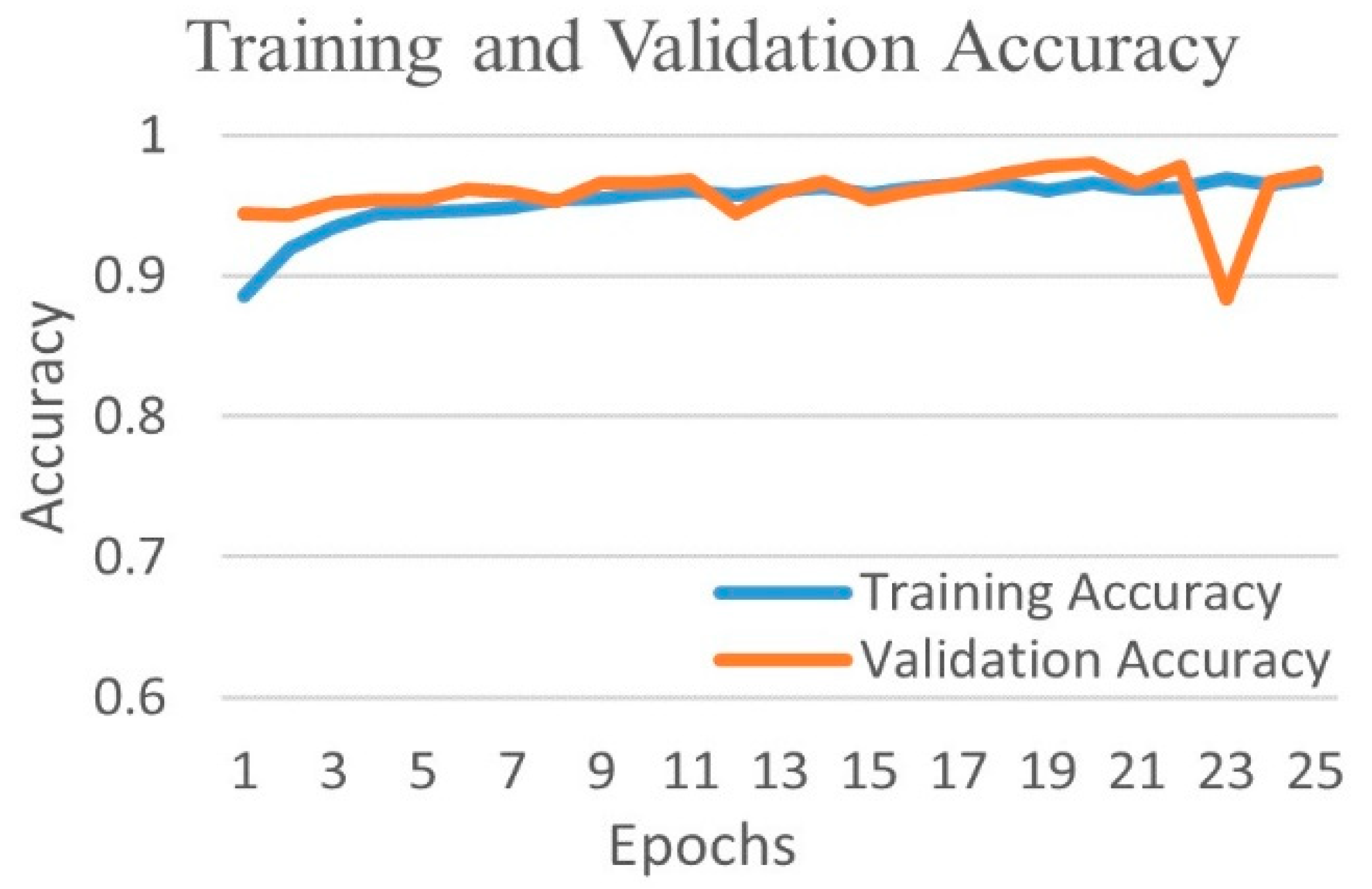
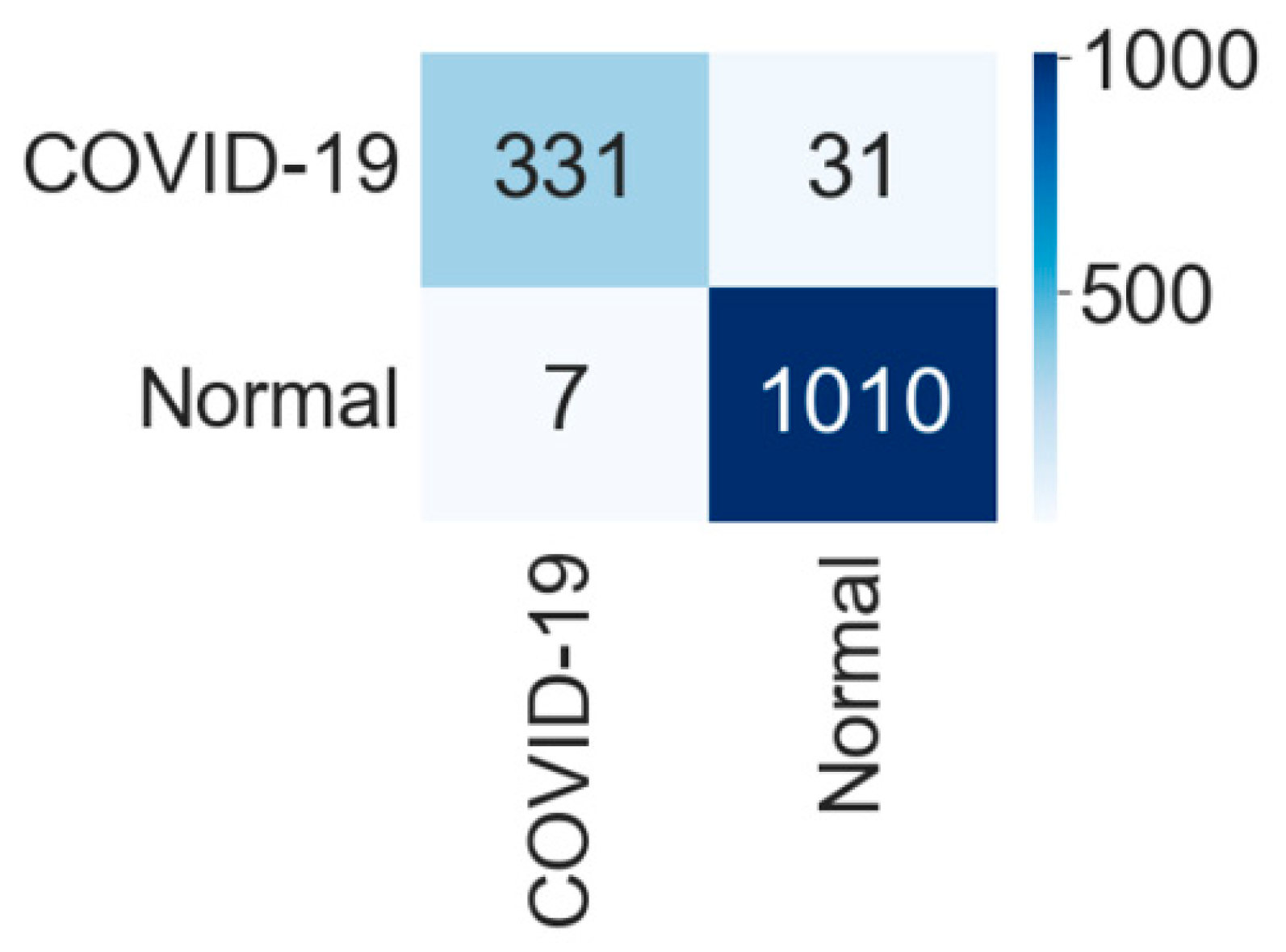
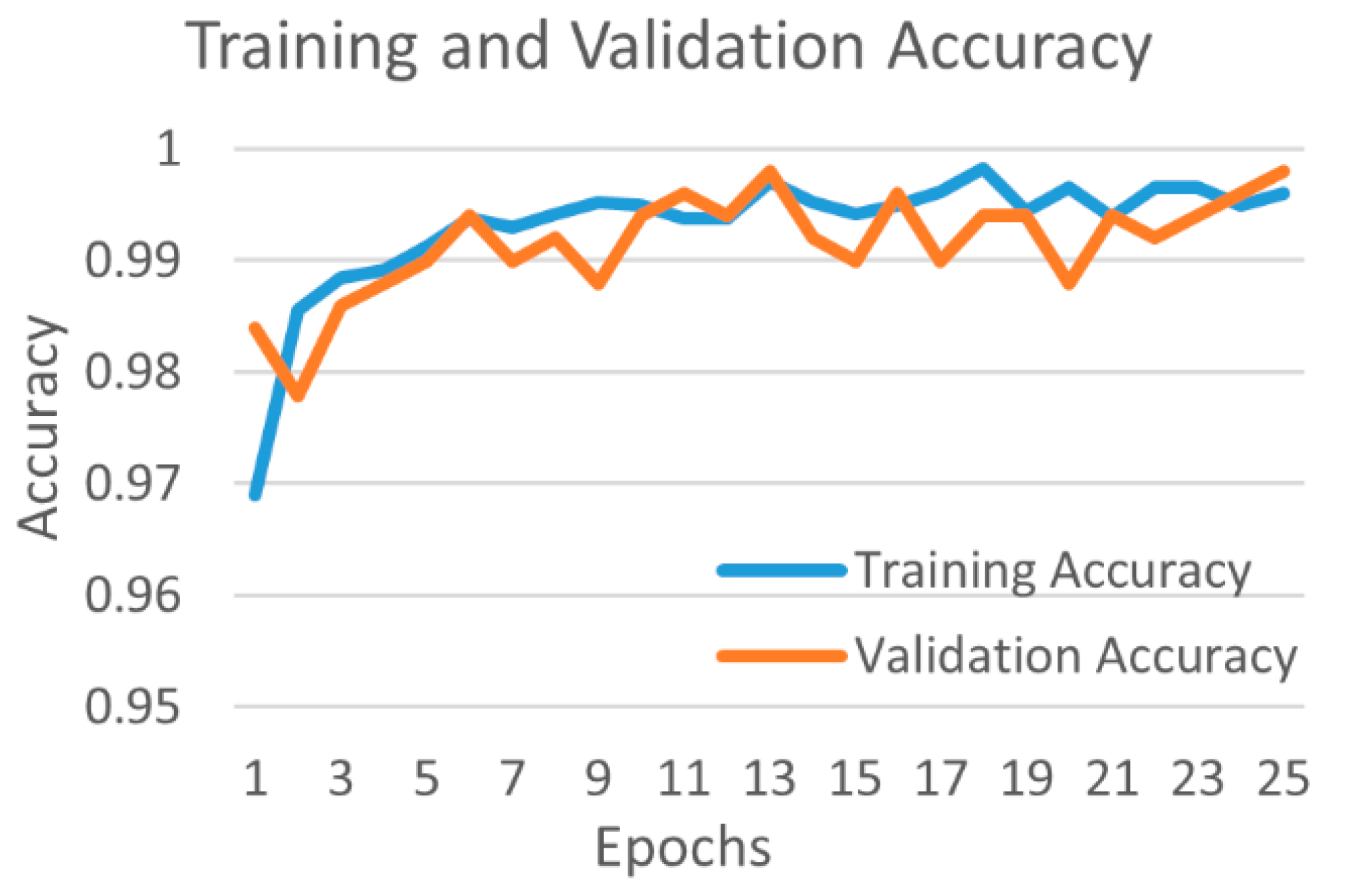
4.1. Binary Classification
4.1.1. Binary Class Case 1: COVID-19 vs. Normal
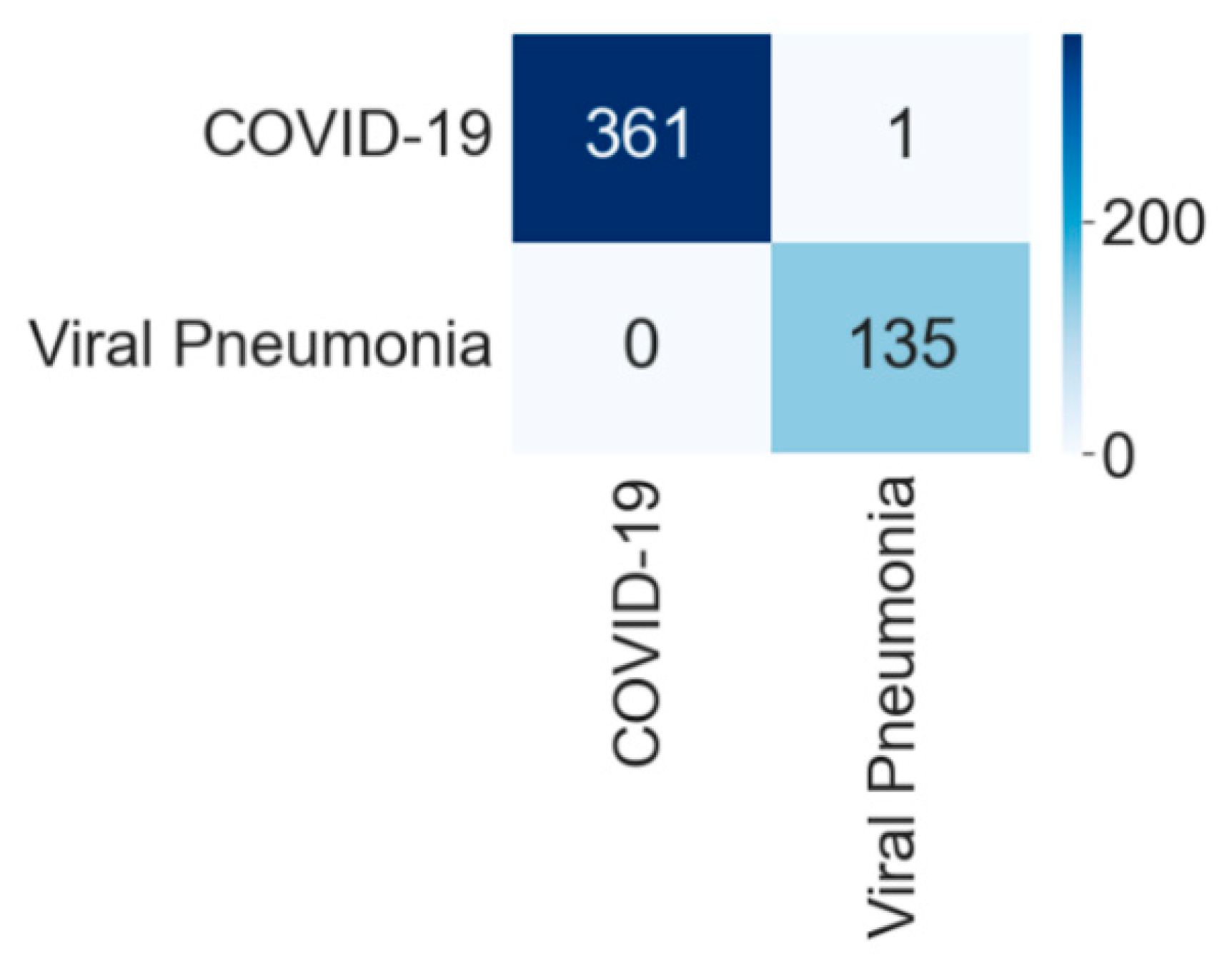
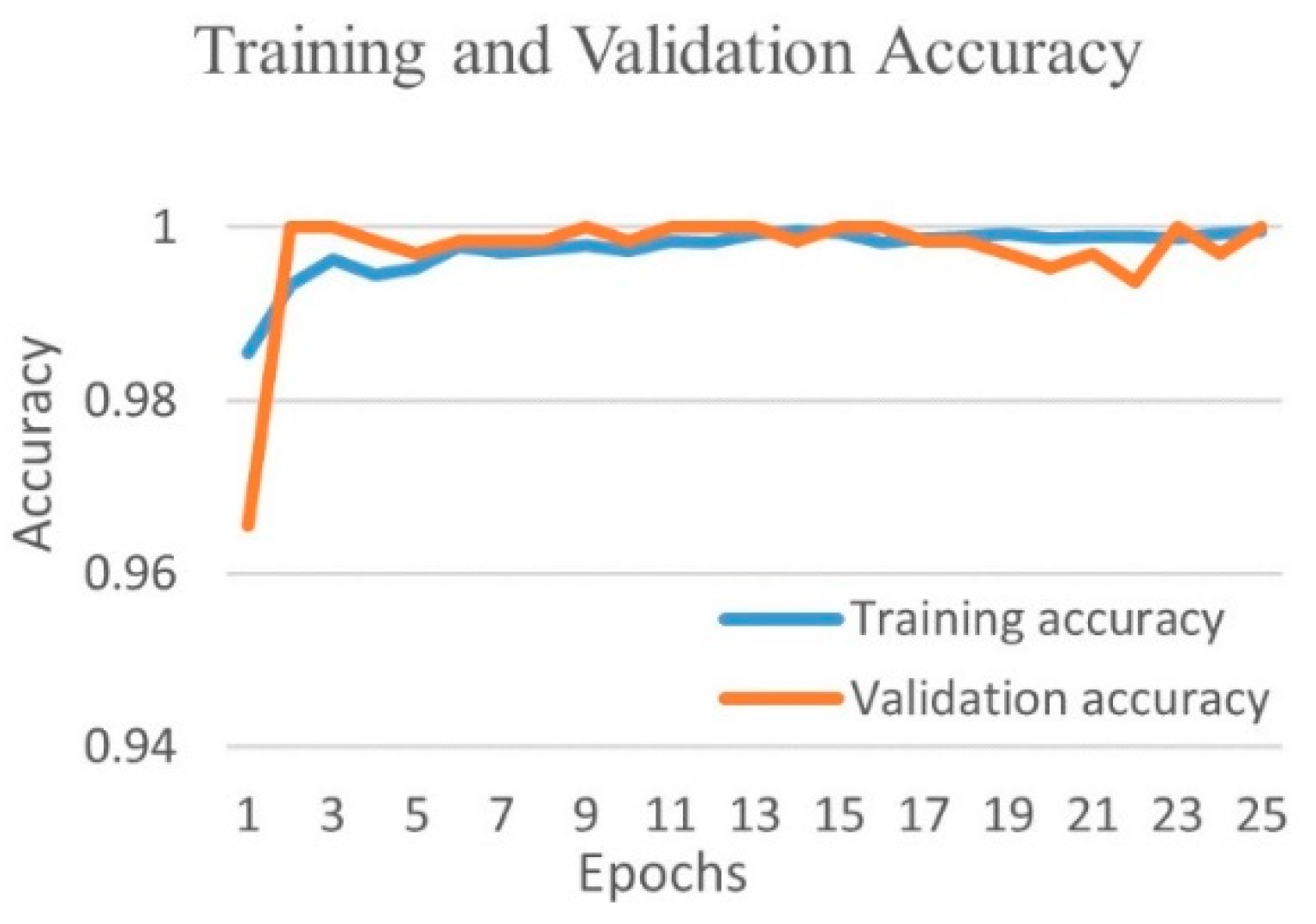
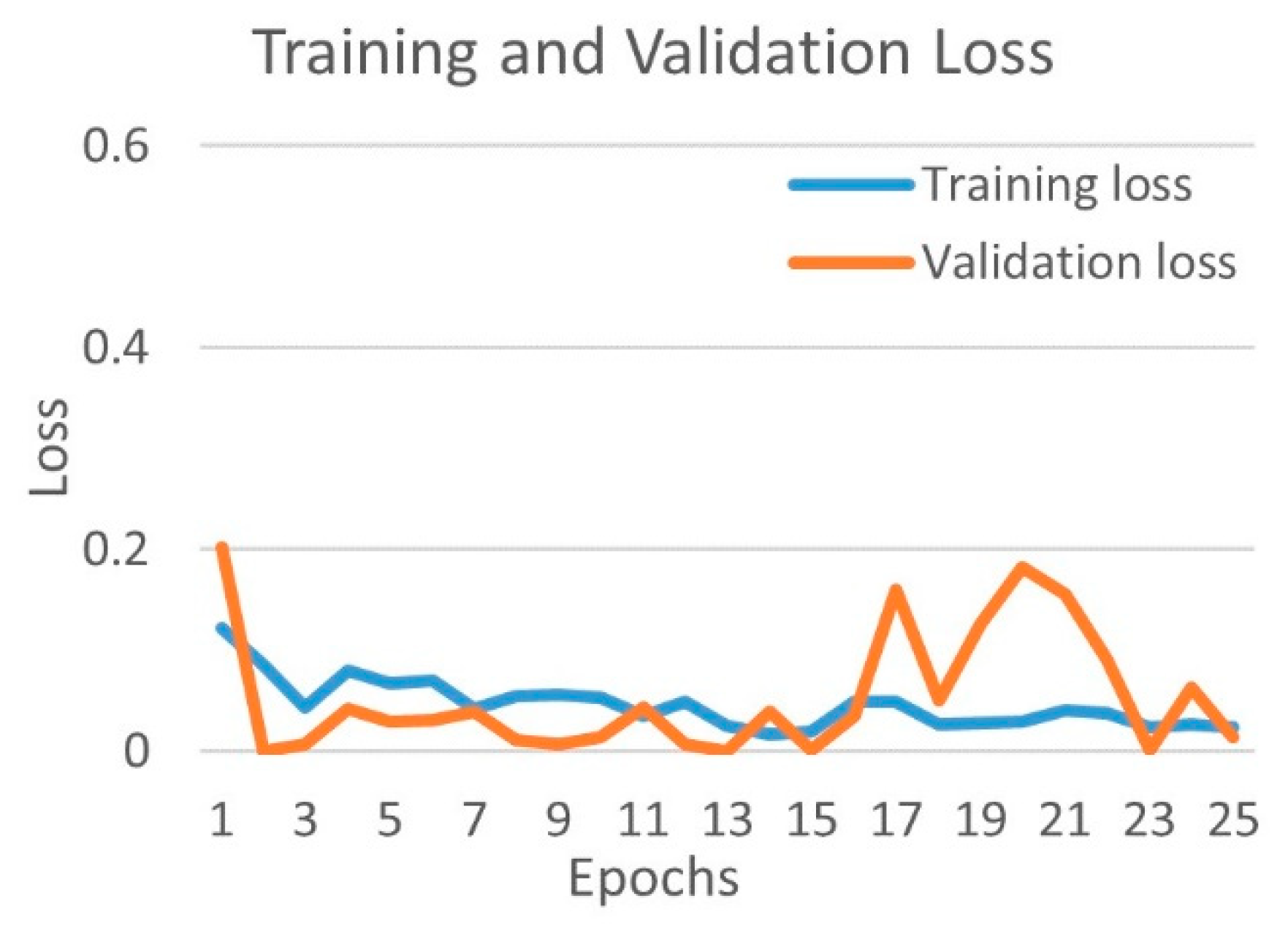
4.1.2. Binary Class Case 2: COVID-19 vs. Viral Pneumonia
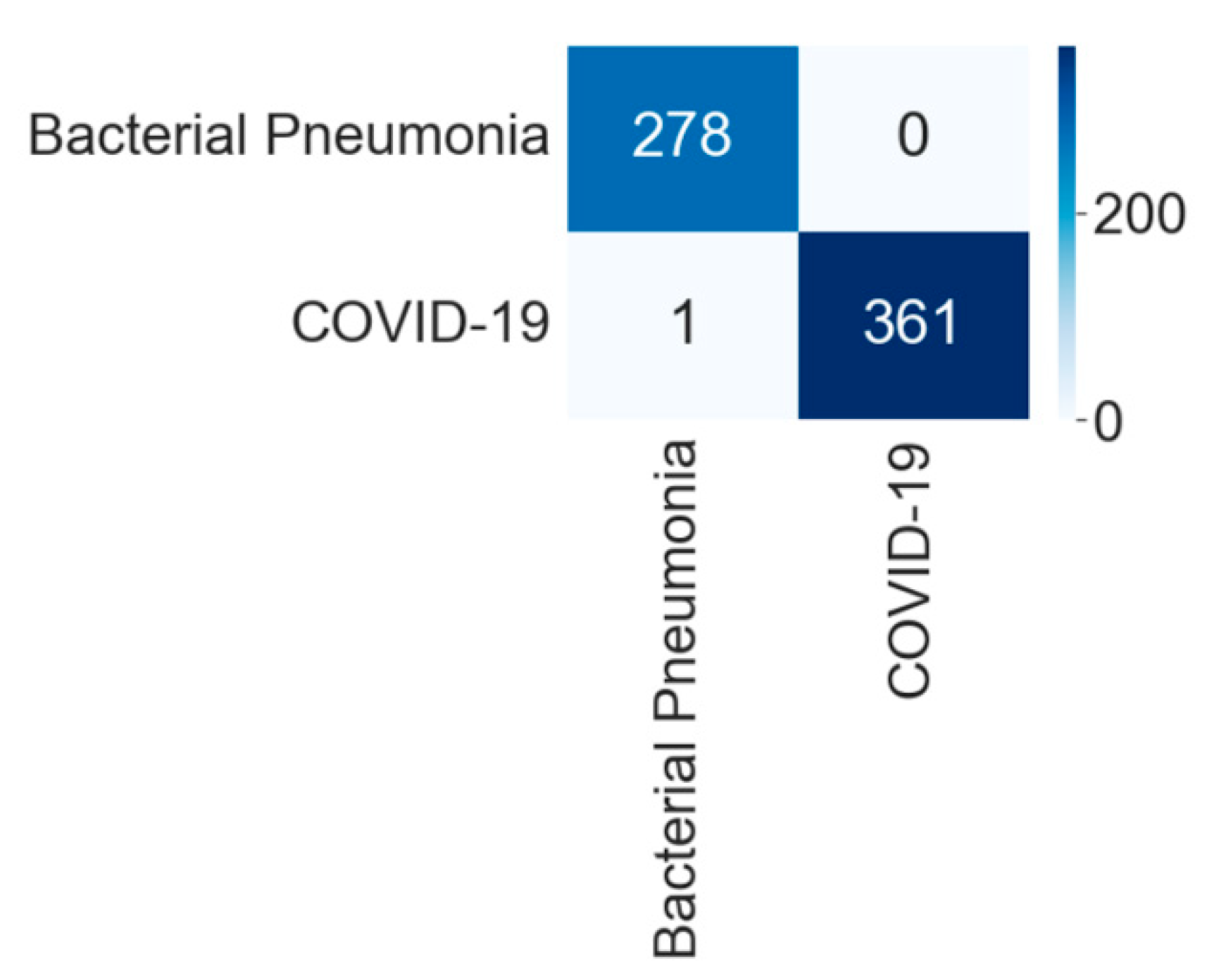
4.1.3. Binary Class Case 3: COVID-19 vs. Bacterial Pneumonia
4.1.4. Binary Class Case 4: COVID-19 and Tuberculosis
4.2. Three-Class Classification into Viral Diseases
4.3. Five-Class Classification into Viral and Bacterial Diseases
5. Performance Evaluation
6. Scientific Validation
7. Discussion
7.1. Principal Findings
7.2. Benchmarking
7.3. A Special Note on Multiclass Frameworks for Pneumonia Classification
7.4. Strengths, Weaknesses, and Extensions
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviation
| AI | Artificial intelligence |
| AUC | Area-under-the-curve |
| BP | Bacterial pneumonia |
| CAD | Computer-aided diagnosis |
| CNN | Convolution neural network |
| COV | Coronavirus |
| CT | Computed tomography |
| CXR | Chest X-ray |
| DL | Deep learning |
| DNN | Deep neural network |
| ESD | Ensemble subspace discriminant |
| FC | Fully connected |
| GPU | Graphics processing unit |
| JPEG | Joint photographic expert group |
| ML | Machine learning |
| Nasnet | Neural search architecture network |
| PNG | Portable network graphics |
| RAM | Random-access memory |
| ReLU | Rectified linear unit |
| ResNet | Residual neural network |
| RNA | Ribonucleic acid |
| ROC | Receiver operating characteristic |
| RT-PCR | Reverse transcriptase polymerase chain reaction |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| TB | Tuberculosis |
| VGG | Visual geometry group |
| VP | Viral pneumonia |
| WHO | World health organization |
| 2-D | Two-dimensional |
References
- Baig, A.M. Neurological manifestations in COVID-19 caused by SARS-CoV-2. CNS Neurosci. Ther. 2020, 26, 499–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Cucinotta, D.; Vanelli, M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020, 91, 157–160. [Google Scholar] [CrossRef] [PubMed]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Pal, M.; Berhanu, G.; Desalegn, C.; Kandi, V. Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Update. Cureus 2020, 12, e7423. [Google Scholar] [CrossRef] [Green Version]
- Saba, L.; Gerosa, C.; Fanni, D.; Marongiu, F.; La Nasa, G.; Caocci, G.; Barcellona, D.; Balestrieri, A.; Coghe, F.; Orru, G.; et al. Molecular pathways triggered by COVID-19 in different organs: ACE2 receptor-expressing cells under attack? A review. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12609–12622. [Google Scholar]
- Cau, R.; Bassareo, P.P.; Mannelli, L.; Suri, J.S.; Saba, L. Imaging in COVID-19-related myocardial injury. Int. J. Cardiovasc. Imaging 2021, 37, 1349–1360. [Google Scholar] [CrossRef]
- Fanni, D.; Cerrone, G.; Saba, L.; Demontis, R.; Congiu, T.; Piras, M.; Gerosa, C.; Suri, J.; Coni, P.; Caddori, A.; et al. Thrombotic sinus-oiditis and local diffuse intrasinusoidal coagulation in the liver of subjects affected by COVID-19: The evidence from histology and scanning electron microscopy. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 5904–5912. [Google Scholar]
- Viswanathan, V.; Puvvula, A.; Jamthikar, A.D.; Saba, L.; Johri, A.M.; Kotsis, V.; Khanna, N.N.; Dhanjil, S.K.; Majhail, M.; Misra, D.P.; et al. Bidirectional link between diabetes mellitus and coronavirus disease 2019 leading to cardiovascular disease: A narrative review. World J. Diabetes 2021, 12, 215–237. [Google Scholar] [CrossRef]
- Cau, R.; Pacielli, A.; Fatemeh, H.; Vaudano, P.; Arru, C.; Crivelli, P.; Stranieri, G.; Suri, J.S.; Mannelli, L.; Conti, M.; et al. Complications in COVID-19 patients: Characteristics of pulmonary embolism. Clin. Imaging 2021, 77, 244–249. [Google Scholar] [CrossRef]
- Gerosa, C.; Faa, G.; Fanni, D.; Manchia, M.; Suri, J.; Ravarino, A.; Barcellona, D.; Pichiri, G.; Coni, P.; Congiu, T.; et al. Fetal pro-gramming of COVID-19: May the barker hypothesis explain the susceptibility of a subset of young adults to develop severe disease. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 5876–5884. [Google Scholar] [PubMed]
- Symptoms of COVID-19. Available online: https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html (accessed on 8 January 2022).
- Koh, H.K.; Geller, A.C.; VanderWeele, T.J. Deaths from COVID-19. JAMA 2021, 325, 1334. [Google Scholar] [CrossRef] [PubMed]
- Woolf, S.H.; Chapman, D.A.; Sabo, R.T.; Weinberger, D.M.; Hill, L. Excess Deaths From COVID-19 and Other Causes, March-April 2020. JAMA 2020, 324, 510–513. [Google Scholar] [CrossRef]
- Faust, J.S.; Del Rio, C. Assessment of Deaths From COVID-19 and From Seasonal Influenza. JAMA Intern. Med. 2020, 180, 1045. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, G. COVID-19: New UK variant may be linked to increased death rate, early data indicate. BMJ 2021, 372, n230. [Google Scholar] [CrossRef] [PubMed]
- Ciminelli, G.; Garcia-Mandicó, S. COVID-19 in Italy: An Analysis of Death Registry Data. J. Public Health 2020, 42, 723–730. [Google Scholar] [CrossRef]
- WHO. Coronavirus (COVID-19) Dashboard. 2022. Available online: https://covid19.who.int/ (accessed on 7 February 2022).
- World Health Organization. Omicron. Available online: https://www.who.int/news/item/28-11-2021-update-on-omicron (accessed on 17 January 2022).
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [Green Version]
- Cau, R.; Falaschi, Z.; Paschè, A.; Danna, P.; Arioli, R.; Arru, C.D.; Zagaria, D.; Tricca, S.; Suri, J.S.; Karla, M.K.; et al. CT findings of COVID-19 pneumonia in ICU-patients. J. Public Health Res. 2021, 10, 2270. [Google Scholar] [CrossRef]
- Wikramaratna, P.S.; Paton, R.S.; Ghafari, M.; Lourenço, J. Estimating the false-negative test probability of SARS-CoV-2 by RT-PCR. Eurosurveillance 2020, 25, 2000568. [Google Scholar] [CrossRef]
- Li, Y.; Yao, L.; Li, J.; Chen, L.; Song, Y.; Cai, Z.; Yang, C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020, 92, 903–908. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Wang, Y.-C.; Shen, C.-F.; Cheng, C.-M. Point-of-Care RNA-Based Diagnostic Device for COVID-19. Diagnostics 2020, 10, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, M.-Y.; Lee, E.Y.; Yang, J.; Yang, F.; Li, X.; Wang, H.; Lui, M.M.-S.; Lo, C.S.-Y.; Leung, B.; Khong, P.-L.; et al. Imaging Profile of the COVID-19 Infection: Radiologic Findings and Literature Review. Radiol. Cardiothorac. Imaging 2020, 2, e200034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Liu, F.; Li, J.; Zhang, T.; Wang, D.; Lan, W. Clinical and CT imaging features of the COVID-19 pneumonia: Focus on pregnant women and children. J. Infect. 2020, 80, e7–e13. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.; Bernheim, A.; Mei, X.; Zhang, N.; Huang, M.; Zeng, X.; Cui, J.; Xu, W.; Yang, Y.; Fayad, Z.A.; et al. CT imaging features of 2019 novel coronavirus (2019–nCoV). Radiology 2020, 295, 202–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroft, L.J.; van der Velden, L.; Girón, I.H.; Roelofs, J.J.; de Roos, A.; Geleijns, J. Added Value of Ultra–low-dose Computed Tomography, Dose Equivalent to Chest X-Ray Radiography, for Diagnosing Chest Pathology. J. Thorac. Imaging 2019, 34, 179–186. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Wang, L.; Deng, X.; Liang, R.; Su, M.; He, C.; Hu, L.; Su, Y.; Ren, J.; Yu, F.; et al. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020, 92, 408–417. [Google Scholar] [CrossRef]
- Hosseiny, M.; Kooraki, S.; Gholamrezanezhad, A.; Reddy, S.; Myers, L. Radiology Perspective of Coronavirus Disease 2019 (COVID-19): Lessons from Severe Acute Respiratory Syndrome and Middle East Respiratory Syndrome. Am. J. Roentgenol. 2020, 214, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Salehi, S.; Abedi, A.; Balakrishnan, S.; Gholamrezanezhad, A. Coronavirus Disease 2019 (COVID-19): A Systematic Review of Imaging Findings in 919 Patients. Am. J. Roentgenol. 2020, 215, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Suri, J.S.; Biswas, M.; Kuppili, V.; Saba, L.; Edla, D.R.; Suri, H.S.; Cuadrado-Godia, E.; Laird, J.R.; Marinhoe, R.T.; Sanches, J.M.; et al. State-of-the-art review on deep learning in medical imaging. Front. Biosci. 2019, 24, 392–426. [Google Scholar] [CrossRef] [PubMed]
- Saba, L.; Biswas, M.; Kuppili, V.; Godia, E.C.; Suri, H.S.; Edla, D.R.; Omerzu, T.; Laird, J.R.; Khanna, N.N.; Mavrogeni, S.; et al. The present and future of deep learning in radiology. Eur. J. Radiol. 2019, 114, 14–24. [Google Scholar] [CrossRef]
- Jena, B.; Saxena, S.; Nayak, G.K.; Saba, L.; Sharma, N.; Suri, J.S. Artificial intelligence-based hybrid deep learning models for image classification: The first narrative review. Comput. Biol. Med. 2021, 137, 104803. [Google Scholar] [CrossRef]
- Singh, M.; Verm, A.; Sharma, N. An Optimized Cascaded Stochastic Resonance for the Enhancement of Brain MRI. IRBM 2018, 39, 334–342. [Google Scholar] [CrossRef]
- Singh, M.; Venkatesh, V.; Verma, A.; Sharma, N. Segmentation of MRI data using multi-objective antlion based improved fuzzy c-means. Biocybern. Biomed. Eng. 2020, 40, 1250–1266. [Google Scholar] [CrossRef]
- Singh, M.; Verma, A.; Sharma, N. Bat optimization based neuron model of stochastic resonance for the enhancement of MR images. Biocybern. Biomed. Eng. 2017, 37, 124–134. [Google Scholar] [CrossRef]
- Singh, M.; Verma, A.; Sharma, N. Optimized Multistable Stochastic Resonance for the Enhancement of Pituitary Microadenoma in MRI. IEEE J. Biomed. Health Inform. 2017, 22, 862–873. [Google Scholar] [CrossRef]
- Hussain, M.; Bird, J.J.; Faria, D.R. A Study on CNN Transfer Learning for Image Classification. In Proceedings of the UK Workshop on Computational Intelligence, Nottingham, UK, 5–7 September 2018; Springer: Cham, Switzerland, 2018; Volume 840, pp. 191–202. [Google Scholar] [CrossRef]
- Lee, H.; Kwon, H. Going Deeper With Contextual CNN for Hyperspectral Image Classification. IEEE Trans. Image Process. 2017, 26, 4843–4855. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Li, W.; Du, Q. Diverse Region-Based CNN for Hyperspectral Image Classification. IEEE Trans. Image Process. 2018, 27, 2623–2634. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xue, B.; Zhang, M.; Yen, G.G.; Lv, J. Automatically Designing CNN Architectures Using the Genetic Algorithm for Image Classification. IEEE Trans. Cybern. 2020, 50, 3840–3854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Nayak, G.K.; Saba, L.; Kalra, M.; Suri, J.S.; Saxena, S. An artificial intelligence framework and its bias for brain tumor segmentation: A narrative review. Comput. Biol. Med. 2022, 105273. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Pan, W.; Xiang, X.; Tan, Y.; Hou, G. A biological image classification method based on improved CNN. Ecol. Inform. 2020, 58, 101093. [Google Scholar] [CrossRef]
- Wei, Y.; Xia, W.; Lin, M.; Huang, J.; Ni, B.; Dong, J.; Zhao, Y.; Yan, S. HCP: A flexible CNN framework for multi-label image classifi-cation. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 38, 1901–1907. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Cai, W.; Wang, X.; Zhou, Y.; Feng, D.D.; Chen, M. Medical image classification with convolutional neural network. In Proceedings of the 2014 13th International Conference on Control Automation Robotics & Vision (ICARCV), Singapore, 10–12 December 2014; pp. 844–848. [Google Scholar]
- Lei, X.; Pan, H.; Huang, X. A Dilated CNN Model for Image Classification. IEEE Access 2019, 7, 124087–124095. [Google Scholar] [CrossRef]
- Ren, X.; Guo, H.; Li, S.; Wang, S.; Li, J. A Novel Image Classification Method with CNN-XGBoost Model. In International Workshop on Digital Watermarking; Springer: Cham, Switzerland, 2017; pp. 378–390. [Google Scholar]
- Sultana, F.; Sufian, A.; Dutta, P. Advancements in Image Classification using Convolutional Neural Network. In Proceedings of the 2018 Fourth International Conference on Research in Computational Intelligence and Communication Networks (ICRCICN), Kolkata, India, 22–23 November 2018; pp. 122–129. [Google Scholar]
- Yu, S.; Jia, S.; Xu, C. Convolutional neural networks for hyperspectral image classification. Neurocomputing 2017, 219, 88–98. [Google Scholar] [CrossRef]
- Zhang, C.; Pan, X.; Li, H.; Gardiner, A.; Sargent, I.; Hare, J.; Atkinson, P.M. A hybrid MLP-CNN classifier for very fine resolution remotely sensed image classification. ISPRS J. Photogramm. Remote Sens. 2018, 140, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Pei, Y.; Huang, Y.; Zou, Q.; Zhang, X.; Wang, S. Effects of image degradation and degradation removal to CNN-based image classification. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 1239–1253. [Google Scholar] [CrossRef]
- Tandel, G.S.; Balestrieri, A.; Jujaray, T.; Khanna, N.N.; Saba, L.; Suri, J.S. Multiclass magnetic resonance imaging brain tumor classi-fication using artificial intelligence paradigm. Comput. Biol. Med. 2020, 122, 103804. [Google Scholar] [CrossRef]
- Tandel, G.S.; Biswas, M.; Kakde, O.G.; Tiwari, A.; Suri, H.S.; Turk, M.; Laird, J.R.; Asare, C.K.; Ankrah, A.A.; Khanna, N.N.; et al. A Review on a Deep Learning Perspective in Brain Cancer Classification. Cancers 2019, 11, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, S.; Verma, A.; Sharma, N. Automatic segmentation of brain tumour in MR images using an enhanced deep learning approach. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2021, 9, 121–130. [Google Scholar] [CrossRef]
- Bhatia, S.; Sinha, Y.; Goel, L. Lung Cancer Detection: A Deep Learning Approach. In Advances in Manufacturing, Production Management and Process Control; Springer: Berlin/Heidelberg, Germany, 2018; pp. 699–705. [Google Scholar]
- Gao, F.; Wu, T.; Li, J.; Zheng, B.; Ruan, L.; Shang, D.; Patel, B. SD-CNN: A shallow-deep CNN for improved breast cancer diagnosis. Comput. Med. Imaging Graph. 2018, 70, 53–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.; Islam, N.; Jan, Z.; Din, I.U.; Rodrigues, J.J.P.C. A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recognit. Lett. 2019, 125, 1–6. [Google Scholar] [CrossRef]
- Feng, C.; ElAzab, A.; Yang, P.; Wang, T.; Zhou, F.; Hu, H.; Xiao, X.; Lei, B. Deep Learning Framework for Alzheimer’s Disease Diagnosis via 3D-CNN and FSBi-LSTM. IEEE Access 2019, 7, 63605–63618. [Google Scholar] [CrossRef]
- Konstantonis, G.; Singh, K.V.; Sfikakis, P.P.; Jamthikar, A.D.; Kitas, G.D.; Gupta, S.K.; Saba, L.; Verrou, K.; Khanna, N.N.; Ruzsa, Z.; et al. Cardiovascular disease detection using machine learning and carotid/femoral arterial imaging frameworks in rheumatoid arthritis patients. Rheumatol. Int. 2022, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Boi, A.; Jamthikar, A.; Saba, L.; Gupta, D.; Sharma, A.; Loi, B.; Laird, J.R.; Khanna, N.N.; Suri, J.S. A Survey on Coronary Atherosclerotic Plaque Tissue Characterization in Intravascular Optical Coherence Tomography. Curr. Atheroscler. Rep. 2018, 20, 33. [Google Scholar] [CrossRef]
- Jain, P.K.; Sharma, N.; Saba, L.; Paraskevas, K.I.; Kalra, M.K.; Johri, A.; Laird, J.R.; Nicolaides, A.N.; Suri, J.S. Unseen Artificial Intelligence—Deep Learning Paradigm for Segmentation of Low Atherosclerotic Plaque in Carotid Ultrasound: A Multicenter Cardiovascular Study. Diagnostics 2021, 11, 2257. [Google Scholar] [CrossRef]
- Jain, P.K.; Sharma, N.; Giannopoulos, A.A.; Saba, L.; Nicolaides, A.; Suri, J.S. Hybrid deep learning segmentation models for atherosclerotic plaque in internal carotid artery B-mode ultrasound. Comput. Biol. Med. 2021, 136, 104721. [Google Scholar] [CrossRef]
- Jain, P.K.; Gupta, S.; Bhavsar, A.; Nigam, A.; Sharma, N. Localization of common carotid artery transverse section in B-mode ultrasound images using faster RCNN: A deep learning approach. Med. Biol. Eng. Comput. 2020, 58, 471–482. [Google Scholar] [CrossRef]
- Saba, L.; Jain, P.K.; Suri, H.S.; Ikeda, N.; Araki, T.; Singh, B.K.; Nicolaides, A.; Shafique, S.; Gupta, A.; Laird, J.R.; et al. Plaque Tissue Morphology-Based Stroke Risk Stratification Using Carotid Ultrasound: A Polling-Based PCA Learning Paradigm. J. Med. Syst. 2017, 41, 31. [Google Scholar] [CrossRef] [PubMed]
- Araki, T.; Jain, P.K.; Suri, H.S.; Londhe, N.D.; Ikeda, N.; El-Baz, A.; Shrivastava, V.K.; Saba, L.; Nicolaides, A.; Shafique, S.; et al. Stroke Risk Stratification and its Validation using Ultrasonic Echolucent Carotid Wall Plaque Morphology: A Machine Learning Paradigm. Comput. Biol. Med. 2017, 80, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Jaiswal, A.K.; Tiwari, P.; Kumar, S.; Gupta, D.; Khanna, A.; Rodrigues, J.J. Identifying pneumonia in chest X-rays: A deep learning approach. Measurement 2019, 145, 511–518. [Google Scholar] [CrossRef]
- Varshni, D.; Thakral, K.; Agarwal, L.; Nijhawan, R.; Mittal, A. Pneumonia detection using CNN based feature extraction. In Proceedings of the IEEE International Conference on Electrical, Computer and Communication Technologies (ICECCT), Coimbatore, India, 20–22 February 2019; pp. 1–7. [Google Scholar]
- GM, H.; Gourisaria, M.K.; Rautaray, S.S.; Pandey, M.A. Pneumonia detection using CNN through chest X-ray. J. Eng. Sci. Technol. 2021, 16, 861–876. [Google Scholar]
- Labhane, G.; Pansare, R.; Maheshwari, S.; Tiwari, R.; Shukla, A. Detection of Pediatric Pneumonia from Chest X-Ray Images using CNN and Transfer Learning. In Proceedings of the 2020 3rd International Conference on Emerging Technologies in Computer Engineering: Machine Learning and Internet of Things (ICETCE), Jaipur, India, 7–8 February 2020; pp. 85–92. [Google Scholar]
- Zhu, Y.; Chen, Y.; Lu, Z.; Pan, S.J.; Xue, G.R.; Yu, Y.; Yang, Q. Heterogeneous transfer learning for image classification. In Proceedings of the Twenty-Fifth AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 7–11 August 2011; Volume 25, pp. 1304–1309. [Google Scholar]
- Saba, L.; Agarwal, M.; Patrick, A.; Puvvula, A.; Gupta, S.K.; Carriero, A.; Laird, J.R.; Kitas, G.D.; Johri, A.M.; Balestrieri, A.; et al. Six artificial intelligence paradigms for tissue characterisation and classification of non-COVID-19 pneumonia against COVID-19 pneumonia in computed tomography lungs. Int. J. Comput. Assist. Radiol. Surgery 2021, 16, 423–434. [Google Scholar] [CrossRef]
- Shaha, M.; Pawar, M. Transfer Learning for Image Classification. In Proceedings of the 2018 Second International Conference on Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 29–31 March 2018; pp. 656–660. [Google Scholar]
- Han, D.; Liu, Q.; Fan, W. A new image classification method using CNN transfer learning and web data augmentation. Expert Syst. Appl. 2018, 95, 43–56. [Google Scholar] [CrossRef]
- Krishna, S.T.; Kalluri, H.K. Deep learning and transfer learning approaches for image classification. Int. J. Recent Technol. Eng. 2019, 7, 427–432. [Google Scholar]
- Kaur, T.; Gandhi, T.K. Deep convolutional neural networks with transfer learning for automated brain image classification. Mach. Vis. Appl. 2020, 31, 20. [Google Scholar] [CrossRef]
- Xiang, Q.; Wang, X.; Li, R.; Zhang, G.; Lai, J.; Hu, Q. Fruit Image Classification Based on MobileNetV2 with Transfer Learning Technique. In Proceedings of the 3rd International Conference on Computer Science and Application Engineering—CSAE, Sanya, China, 22–24 October 2019; Volume 121, pp. 1–7. [Google Scholar] [CrossRef]
- Zhong, C.; Mu, X.; He, X.; Wang, J.; Zhu, M. SAR Target Image Classification Based on Transfer Learning and Model Compression. IEEE Geosci. Remote Sens. Lett. 2018, 16, 412–416. [Google Scholar] [CrossRef]
- Rostami, M.; Kolouri, S.; Eaton, E.; Kim, K. Deep Transfer Learning for Few-Shot SAR Image Classification. Remote Sens. 2019, 11, 1374. [Google Scholar] [CrossRef] [Green Version]
- Akilan, T.; Wu, Q.J.; Yang, Y.; Safaei, A. Fusion of transfer learning features and its application in image classification. In Proceedings of the 2017 IEEE 30th Canadian Conference on Electrical and Computer Engineering (CCECE), Windsor, ON, Canada, 30 April–3 March 2017; pp. 1–5. [Google Scholar]
- Deng, C.; Xue, Y.; Liu, X.; Li, C.; Tao, D. Active Transfer Learning Network: A Unified Deep Joint Spectral–Spatial Feature Learning Model for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2018, 57, 1741–1754. [Google Scholar] [CrossRef] [Green Version]
- Ouhami, M.; Es-Saady, Y.; El Hajji, M.; Hafiane, A.; Canals, R.; El Yassa, M. Deep Transfer Learning Models for Tomato Disease Detection. In Proceedings of the International Conference on Image and Signal Processing, Marrakesh, Morocco, 4–6 June 2020; pp. 65–73. [Google Scholar]
- Chowdhury, M.E.H.; Rahman, T.; Khandakar, A.; Mazhar, R.; Kadir, M.A.; Bin Mahbub, Z.; Islam, K.R.; Khan, M.S.; Iqbal, A.; Al Emadi, N.; et al. Can AI Help in Screening Viral and COVID-19 Pneumonia? IEEE Access 2020, 8, 132665–132676. [Google Scholar] [CrossRef]
- El-Din Hemdan, E.; Shouman, M.A.; Karar, M.E. COVIDX-Net: A Framework of Deep Learning Classifiers to Diagnose COVID-19 in X-Ray Images. arXiv 2020, arXiv:2003.11055. [Google Scholar]
- Hussain, E.; Hasan, M.; Rahman, A.; Lee, I.; Tamanna, T.; Parvez, M.Z. CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images. Chaos Solitons Fractals 2021, 142, 110495. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Gupta, M.; Taneja, S.; Hemanth, D.J. Deep learning based detection and analysis of COVID-19 on chest X-ray images. Appl. Intell. 2021, 51, 1690–1700. [Google Scholar] [CrossRef]
- Mahdy, L.N.; Ezzat, K.A.; Elmousalami, H.H.; Ella, H.A.; Hassanien, A.E. Automatic X-ray COVID-19 Lung Image Classification System based on Multi-Level Thresholding and Support Vector Machine. MedRxiv 2020, 8, 20047787. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Mpesiana, T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef] [Green Version]
- Sethy, P.; Behera, S.K.; Ratha, P.K.; Biswas, P. Detection of Coronavirus Disease (COVID-19) Based on Deep Features and Support Vector Machine. 2020. Available online: www.preprints.org (accessed on 15 January 2022).
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Khan, A.; Shah, J.L.; Bhat, M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef]
- Wang, L.; Wong, A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. arXiv 2020, arXiv:2003.09871. [Google Scholar] [CrossRef] [PubMed]
- Afshar, P.; Heidarian, S.; Naderkhani, F.; Oikonomou, A.; Plataniotis, K.N.; Mohammadi, A. COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recognit. Lett. 2020, 138, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Martinez, C.; Visuña, L.; Khandhar, H.; Bhatt, C.; Carretero, J. Detection and analysis of COVID-19 in medical images using deep learning techniques. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.R.; Nayak, D.R.; Sinha, U.; Arora, V.; Pachori, R.B. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: A comprehensive study. Biomed. Signal Process. Control 2021, 64, 102365. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, A.; Bhaik, D.; Kumar, S.; Thakur, P.; Sharma, R.; Pachori, R.B. A deep learning based approach for automatic detection of COVID-19 cases using chest X-ray images. Biomed. Signal Process. Control 2021, 71, 103182. [Google Scholar] [CrossRef]
- Deb, S.D.; Jha, R.K.; Jha, K.; Tripathi, P.S. A multi model ensemble based deep convolution neural network structure for detection of COVID19. Biomed. Signal Process. Control 2021, 71, 103126. [Google Scholar] [CrossRef]
- Nikolaou, V.; Massaro, S.; Fakhimi, M.; Stergioulas, L.; Garn, W. COVID-19 diagnosis from chest x-rays: Developing a simple, fast, and accurate neural network. Health Inf. Sci. Syst. 2021, 9, 1–11. [Google Scholar] [CrossRef]
- Oh, Y.; Park, S.; Ye, J.C. Deep Learning COVID-19 Features on CXR Using Limited Training Data Sets. IEEE Trans. Med. Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Al-Timemy, A.H.; Khushaba, R.N.; Mosa, Z.M.; Escudero, J. An Efficient Mixture of Deep and Machine Learning Models for COVID-19 and Tuberculosis Detection Using X-ray Images in Resource Limited Settings. Available online: https://www.worldometers.info/coronavirus/ (accessed on 17 January 2022).
- COVID-19 Radiography Database. Available online: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database (accessed on 1 October 2021).
- Tuberculosis (TB) Chest X-ray Database. Available online: https://www.kaggle.com/tawsifurrahman/tuberculosis-tb-chest-xray-dataset (accessed on 1 October 2021).
- Kaggle’s Chest X-ray Images (Pneumonia) Dataset. 2020. Available online: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia (accessed on 1 October 2021).
- Rahman, T.; Khandakar, A.; Qiblawey, Y.; Tahir, A.; Kiranyaz, S.; Kashem, S.B.A.; Islam, M.T.; Al Maadeed, S.; Zughaier, S.M.; Khan, M.S.; et al. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput. Biol. Med. 2021, 132, 104319. [Google Scholar] [CrossRef]
- Labeled Optical Coherence Tomography (OCT) and Chest X-ray Images for Classification. Available online: https://data.mendeley.com/datasets/rscbjbr9sj/2 (accessed on 1 August 2021).
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef]
- Rahman, T.; Khandakar, A.; Kadir, M.A.; Islam, K.R.; Islam, K.F.; Mazhar, R.; Hamid, T.; Islam, M.T.; Kashem, S.; Bin Mahbub, Z.; et al. Reliable Tuberculosis Detection Using Chest X-ray With Deep Learning, Segmentation and Visualization. IEEE Access 2020, 8, 191586–191601. [Google Scholar] [CrossRef]
- Skandha, S.S.; Gupta, S.K.; Saba, L.; Koppula, V.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M.; et al. 3-D optimized classification and characterization artificial intelligence paradigm for cardiovascular/stroke risk stratification using carotid ultrasound-based delineated plaque: Atheromatic™ 2.0. Comput. Biol. Med. 2020, 125, 103958. [Google Scholar] [CrossRef] [PubMed]
- Saba, L.; Sanagala, S.S.; Gupta, S.K.; Koppula, V.K.; Johri, A.M.; Sharma, A.M.; Kolluri, R.; Bhatt, D.L.; Nicolaides, A.; Suri, J.S. Ultra-sound-based internal carotid artery plaque characterization using deep learning paradigm on a supercomputer: A cardiovas-cular disease/stroke risk assessment system. Int. J. Cardiovasc. Imaging 2021, 37, 1511–1528. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Saba, L.; Gupta, S.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M.; Sfikakis, P.P.; et al. Wilson disease tissue classification and characterization using seven artificial intelligence models embedded with 3D optimi-zation paradigm on a weak training brain magnetic resonance imaging datasets: A supercomputer application. Med. Biol. Eng. Comput. 2021, 59, 511–533. [Google Scholar] [CrossRef] [PubMed]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Face95. Libor Spacek’s Facial Images Databases. Available online: https://cmp.felk.cvut.cz/~spacelib/faces/faces95.html (accessed on 8 December 2021).
- Peng, C.; Liu, Y.; Zhang, X.; Kang, Z.; Chen, Y.; Chen, C.; Cheng, Q. Learning discriminative representation for image classification. Knowl.-Based Syst. 2021, 233, 107517. [Google Scholar] [CrossRef]
- Melo-Oliveira, M.E.; Sá-Caputo, D.; Bachur, J.A.; Paineiras-Domingos, L.L.; Sonza, A.; Lacerda, A.C.; Mendonça, V.; Seixas, A.; Taiar, R.; Bernardo-Filho, M. Reported quality of life in countries with cases of COVID-19: A systematic review. Expert Rev. Respir. Med. 2021, 15, 213–220. [Google Scholar] [CrossRef]
- Sapkota, N.; Karwowski, W.; Davahli, M.R.; Al-Juaid, A.; Taiar, R.; Murata, A.; Wrobel, G.; Marek, T. The Chaotic Behavior of the Spread of Infection During the COVID-19 Pandemic in the United States and Globally. IEEE Access 2021, 9, 80692–80702. [Google Scholar] [CrossRef]
- Davahli, M.R.; Karwowski, W.; Fiok, K.; Murata, A.; Sapkota, N.; Farahani, F.V.; Al-Juaid, A.; Marek, T.; Taiar, R. The COVID-19 Infection Diffusion in the US and Japan: A Graph-Theoretical Approach. Biology 2022, 11, 125. [Google Scholar] [CrossRef]
- Zhang, C.; Benz, P.; Argaw, D.M.; Lee, S.; Kim, J.; Rameau, F.; Bazin, J.-C.; Kweon, I.S. ResNet or DenseNet? Introducing Dense Shortcuts to ResNet. In Proceedings of the 2021 IEEE Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–8 January 2021; pp. 3549–3558. [Google Scholar]
- He, G.; Ping, A.; Wang, X.; Zhu, Y. Alzheimer’s disease diagnosis model based on three-dimensional full convolutional DenseNet. In Proceedings of the 2019 10th International Conference on Information Technology in Medicine and Education (ITME), Qingdao, China, 23–25 August 2019; pp. 13–17. [Google Scholar]
- Ruiz, J.; Mahmud, M.; Modasshir; Kaiser, M.S.; Initiative, F. 3D DenseNet Ensemble in 4-Way Classification of Alzheimer’s Disease. In Proceedings of the Brain Informatics: 13th International Conference, BI 2020, Padua, Italy, 19 September 2020; Springer: Cham, Switzerland, 2020; pp. 85–96. [Google Scholar]
- Iandola, F.; Moskewicz, M.; Karayev, S.; Girshick, R.; Darrell, T.; Keutzer, K. Densenet: Implementing efficient convnet descriptor pyramids. arXiv 2014, arXiv:1404.1869. [Google Scholar]
- Sudeep, P.; Palanisamy, P.; Rajan, J.; Baradaran, H.; Saba, L.; Gupta, A.; Suri, J.S. Speckle reduction in medical ultrasound images using an unbiased non-local means method. Biomed. Signal Process. Control 2016, 28, 1–8. [Google Scholar] [CrossRef]
- Sanagala, S.S.; Nicolaides, A.; Gupta, S.K.; Koppula, V.K.; Saba, L.; Agarwal, S.; Johri, A.M.; Kalra, M.S.; Suri, J.S. Ten Fast Transfer Learning Models for Carotid Ultrasound Plaque Tissue Characterization in Augmentation Framework Embedded with Heatmaps for Stroke Risk Stratification. Diagnostics 2021, 11, 2109. [Google Scholar] [CrossRef] [PubMed]
- Kusakunniran, W.; Borwarnginn, P.; Sutassananon, K.; Tongdee, T.; Saiviroonporn, P.; Karnjanapreechakorn, S.; Siriapisith, T. COVID-19 detection and heatmap generation in chest x-ray images. J. Med. Imaging 2021, 8, 014001. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Liu, H.; Gu, Y.; Guo, X.; Li, H.; Li, L.; Wu, Z.; Liu, M.; Tao, L. Fast automated detection of COVID-19 from medical images using convolutional neural networks. Commun. Biol. 2021, 4, 1–13. [Google Scholar] [CrossRef]
- Kuppili, V.; Biswas, M.; Sreekumar, A.; Suri, H.S.; Saba, L.; Edla, D.R.; Marinhoe, R.T.; Sanches, J.M.; Suri, J.S. Extreme learning machine framework for risk stratification of fatty liver disease using ultrasound tissue characterization. J. Med. Syst. 2017, 41, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Rajaraman, S.; Siegelman, J.; Alderson, P.O.; Folio, L.S.; Folio, L.R.; Antani, S.K. Iteratively Pruned Deep Learning Ensembles for COVID-19 Detection in Chest X-Rays. IEEE Access 2020, 8, 115041–115050. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhang, X.; Sun, J. Channel Pruning for Accelerating Very Deep Neural Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Adedigba, A.P.; Adeshina, S.A.; Aina, O.E.; Aibinu, A.M. Optimal hyperparameter selection of deep learning models for COVID-19 chest X-ray classification. Intell. Med. 2021, 5, 100034. [Google Scholar] [CrossRef]
- Zandehshahvar, M.; van Assen, M.; Maleki, H.; Kiarashi, Y.; De Cecco, C.N.; Adibi, A. Toward understanding COVID-19 pneumonia: A deep-learning-based approach for severity analysis and monitoring the disease. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- El-Baz, A.; Gimel’farb, G.; Suri, J.S. Stochastic Modeling for Medical Image Analysis; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]


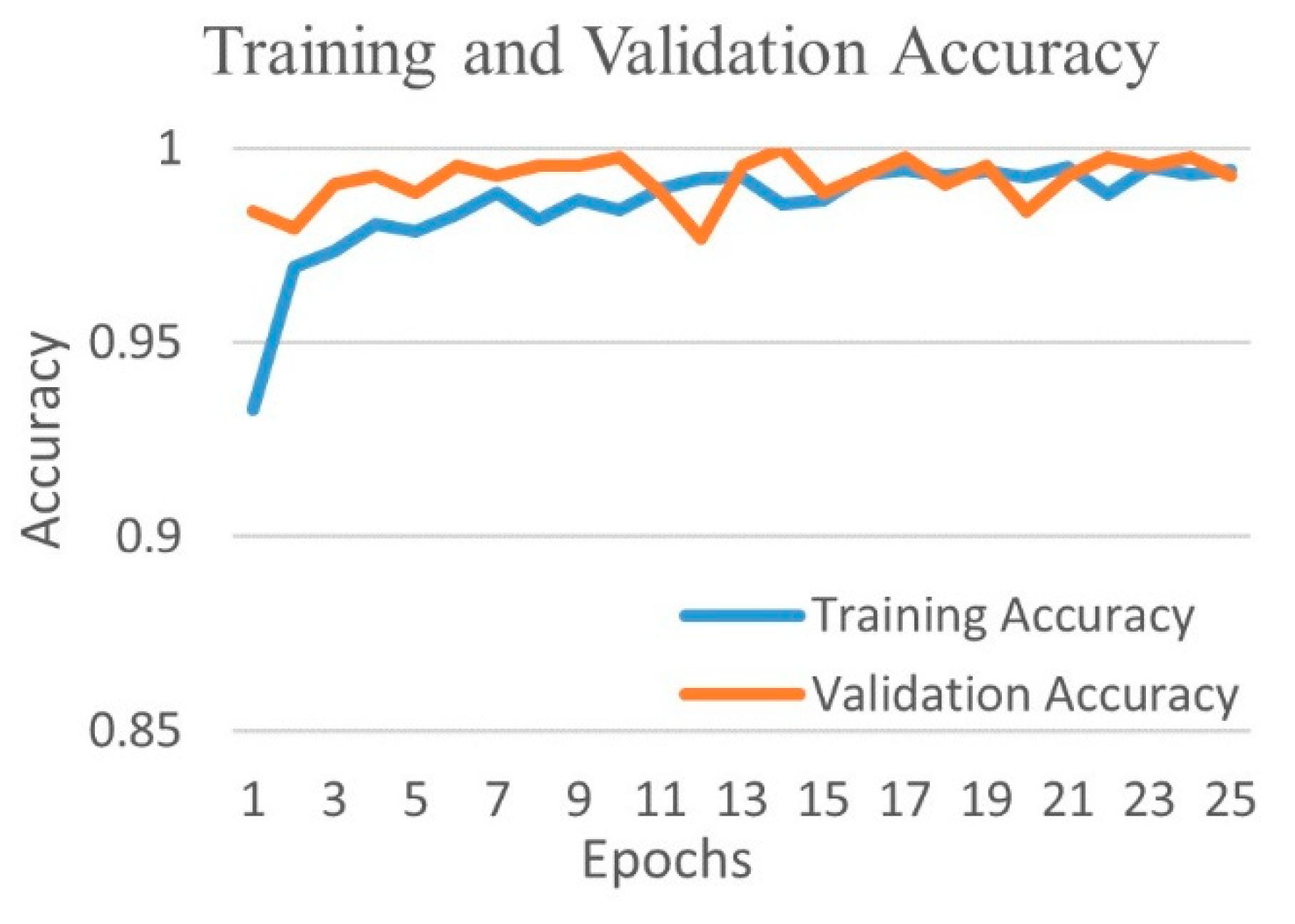
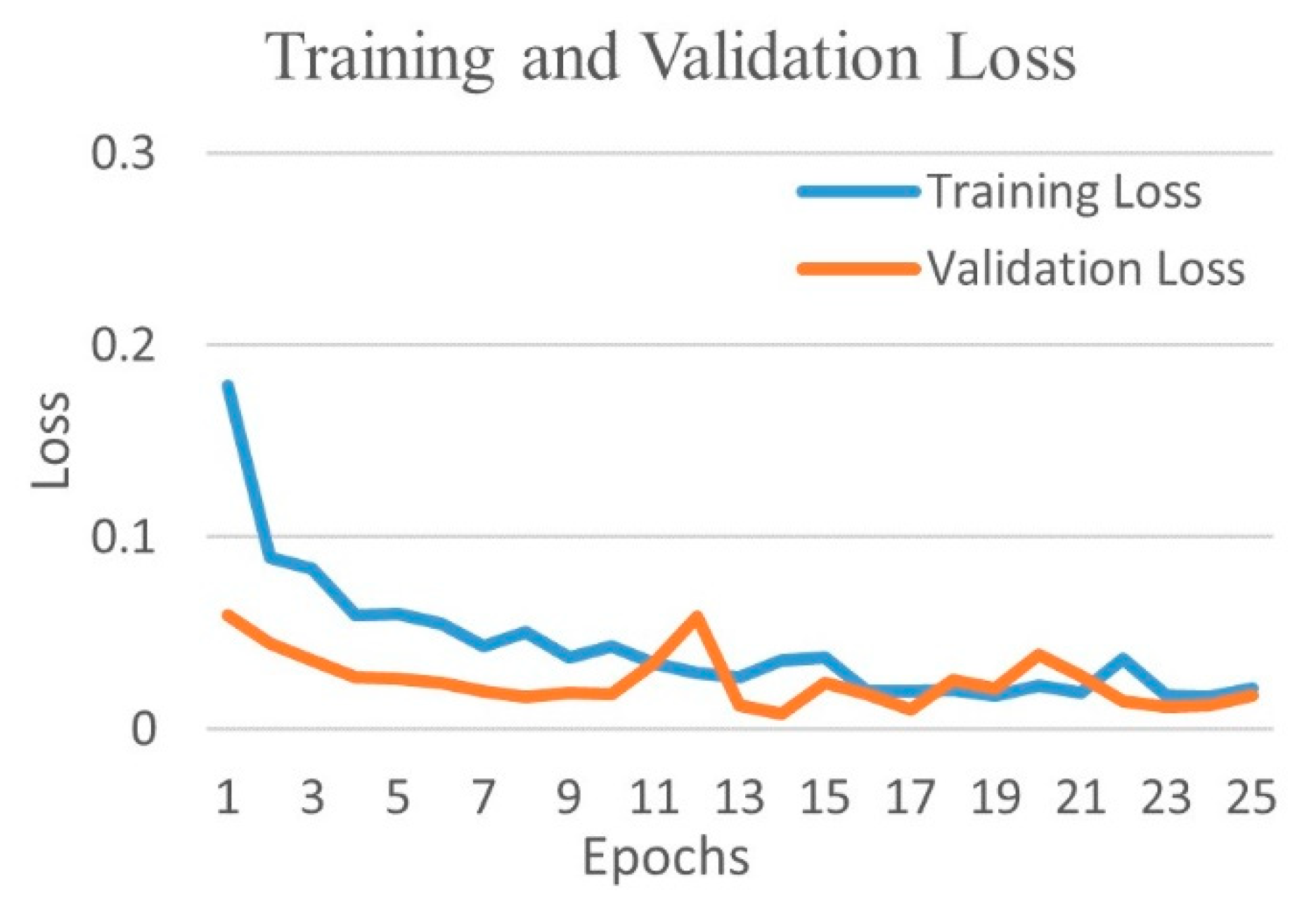


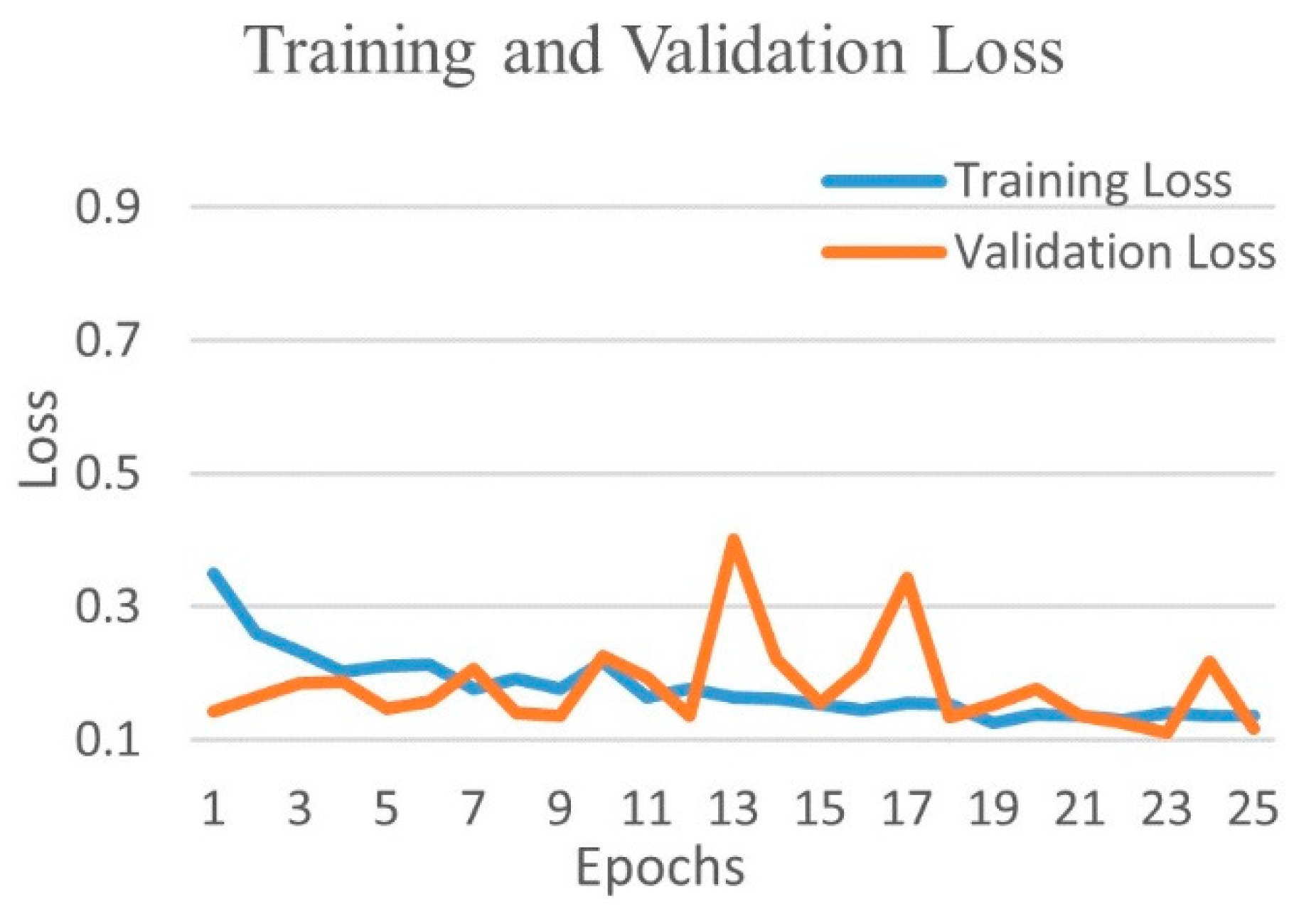
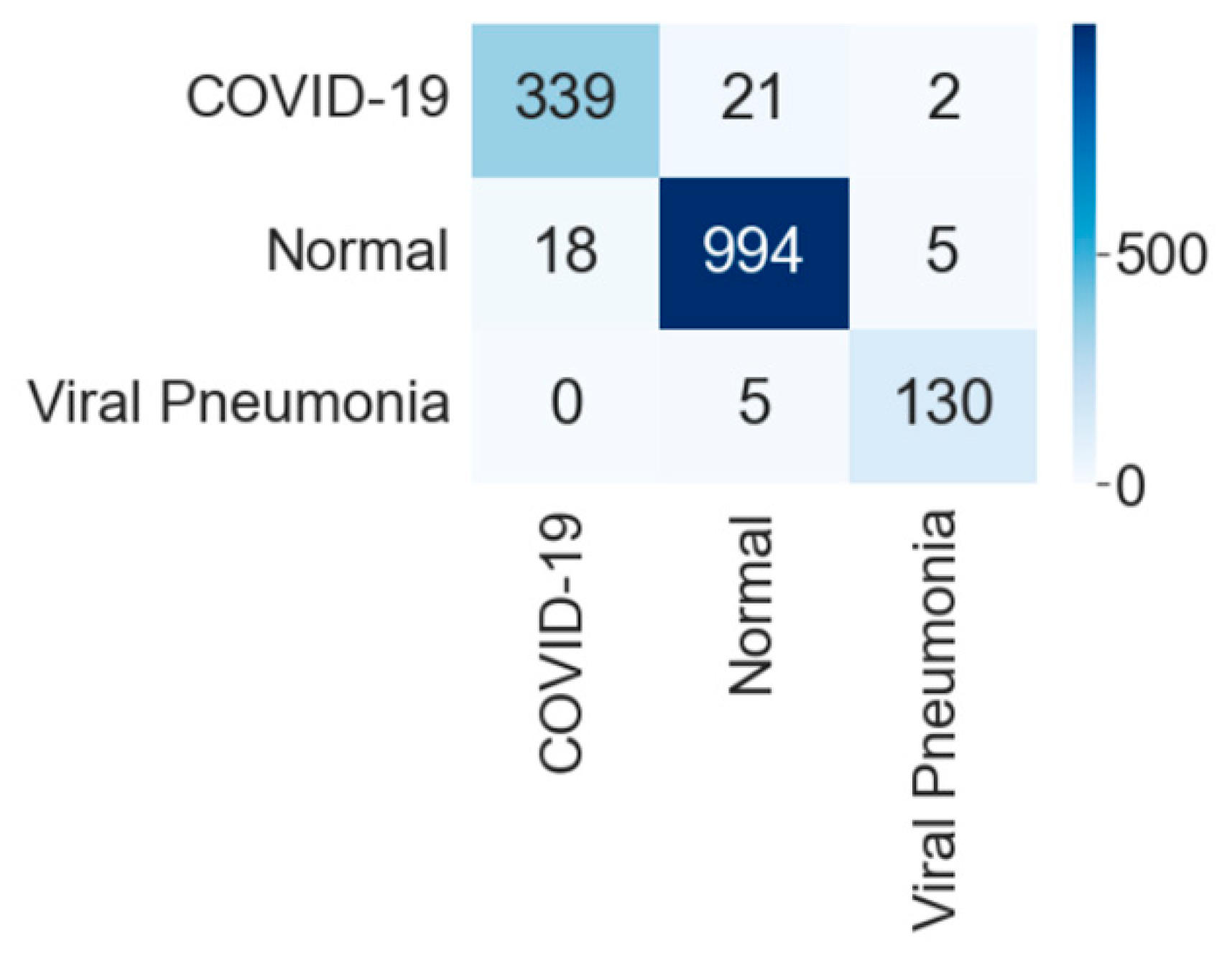
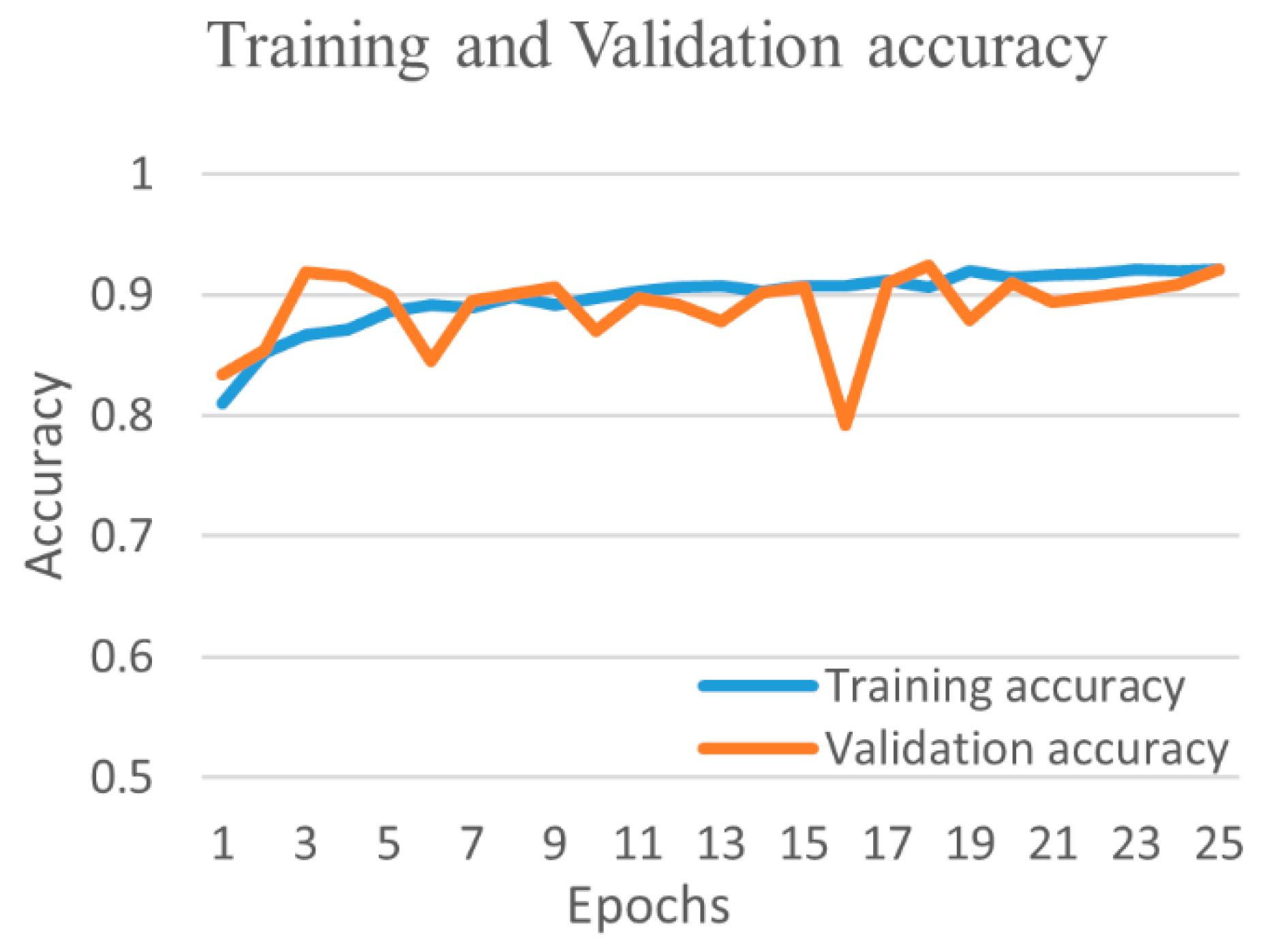

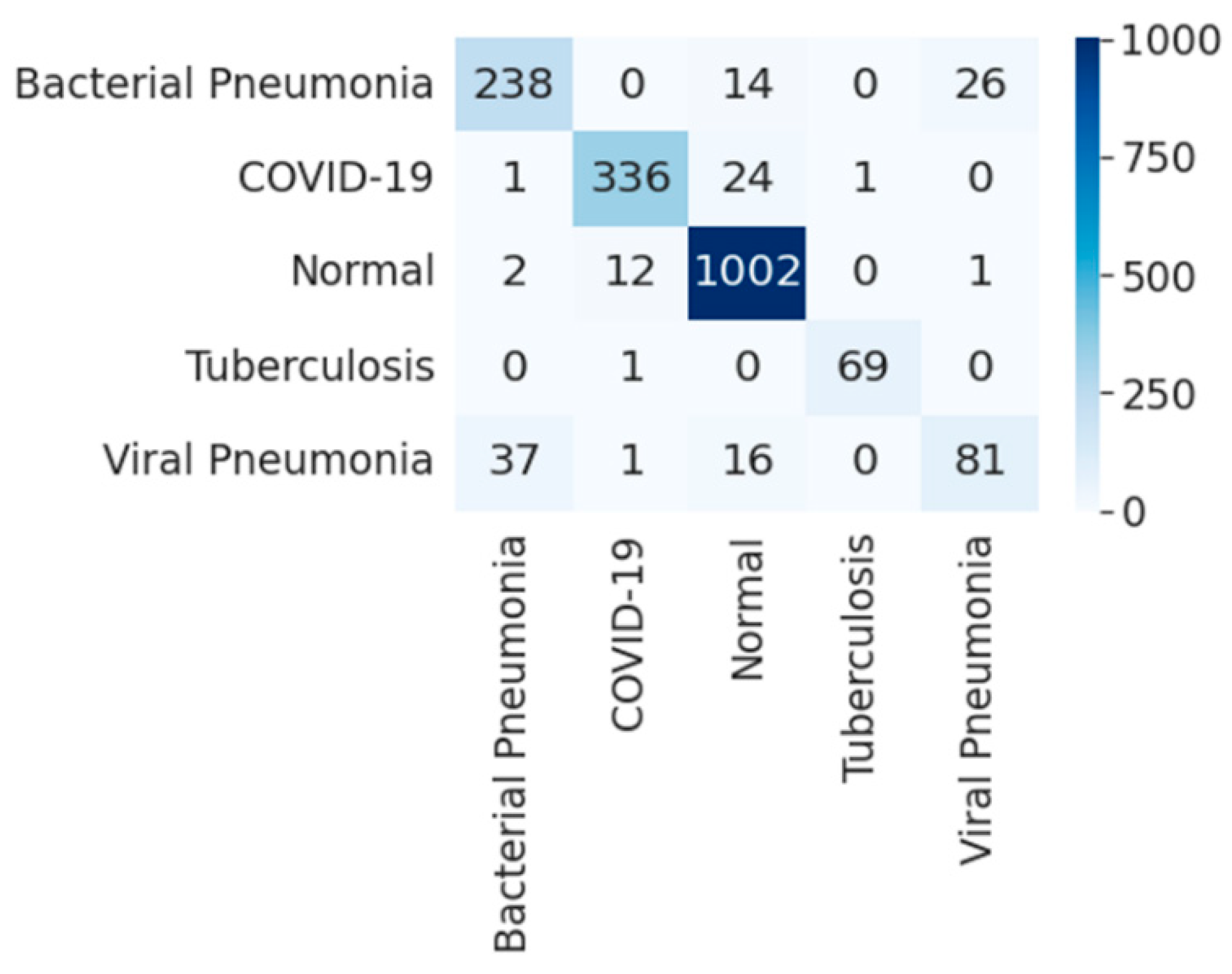
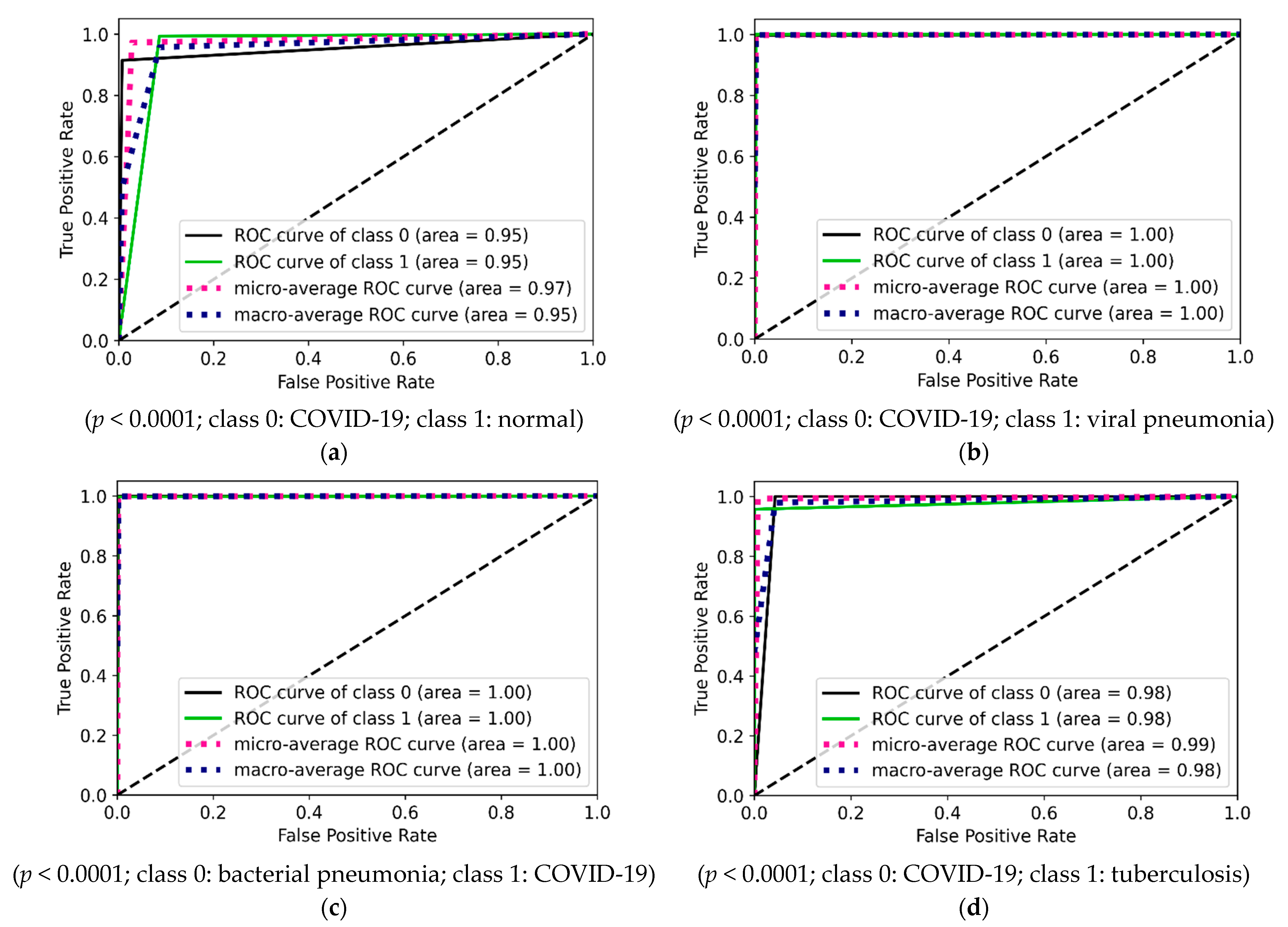
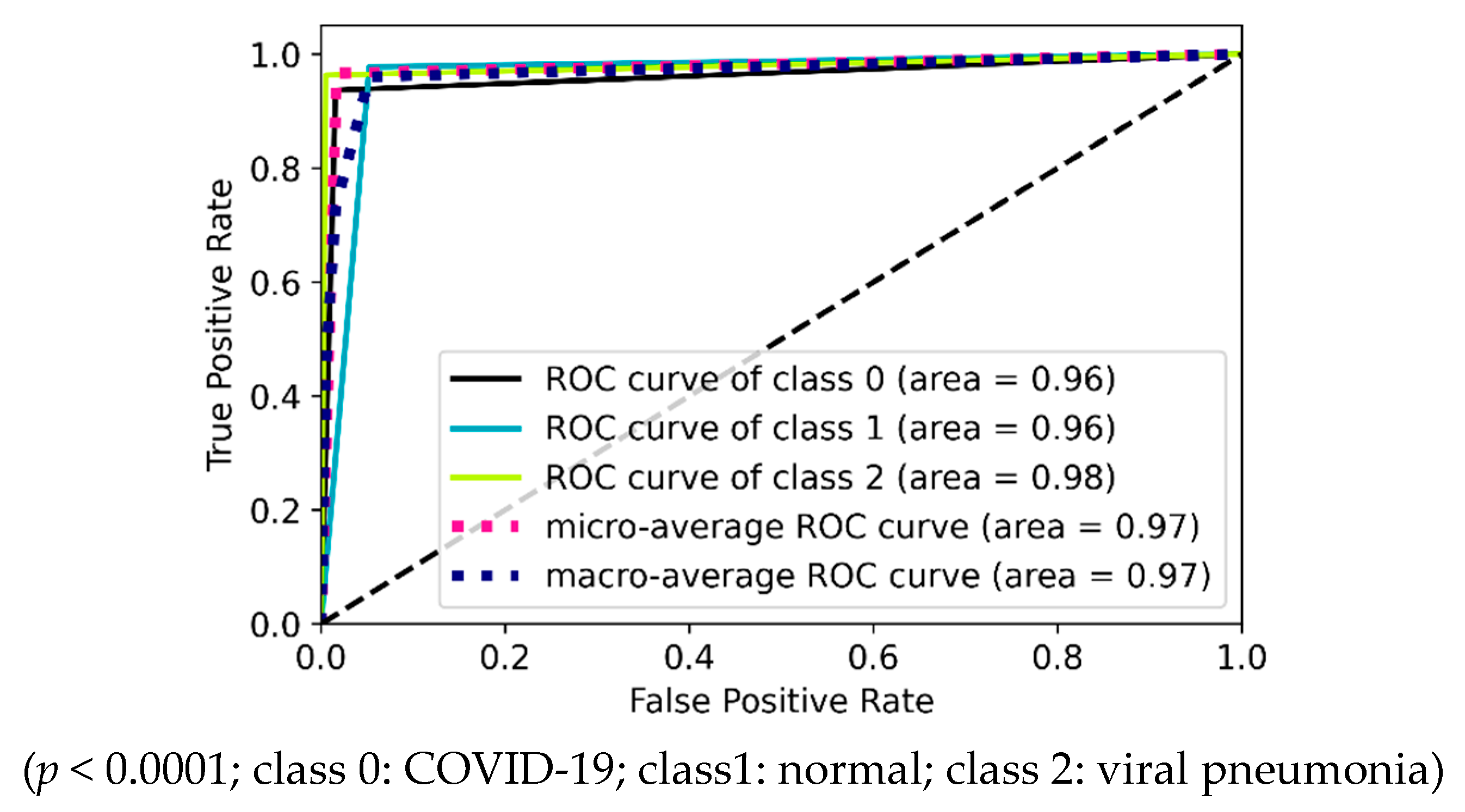


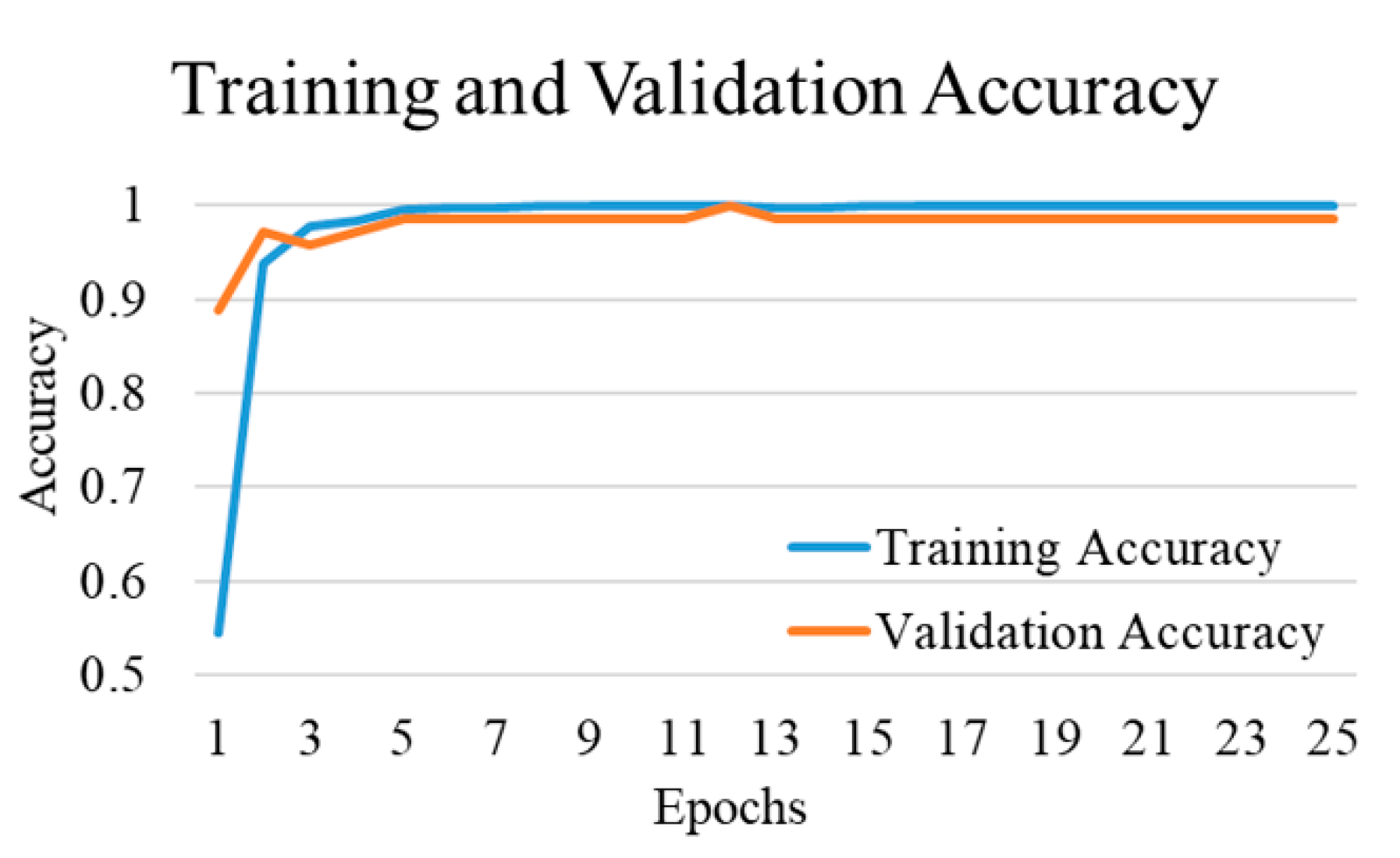
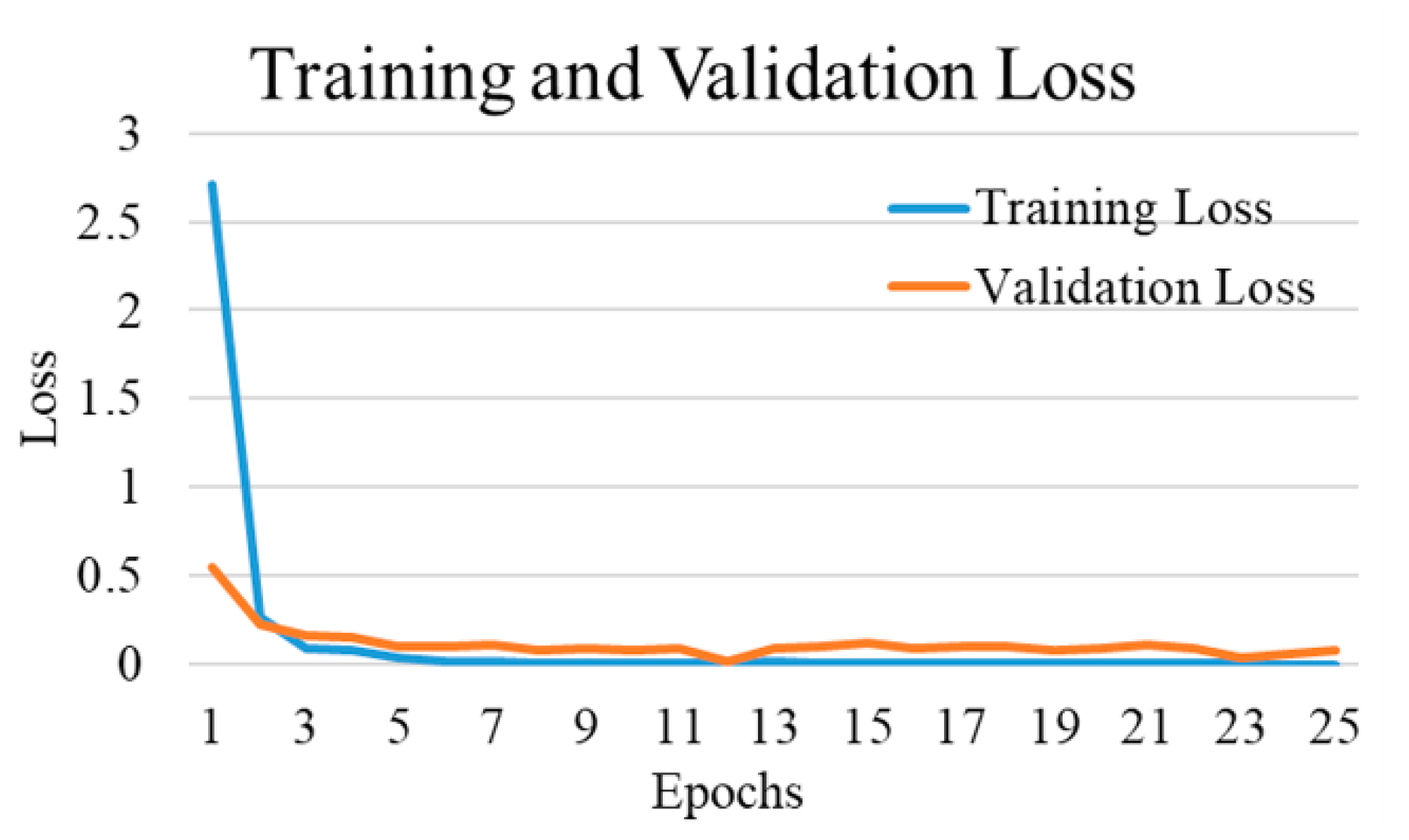
| Experimental Steps | Normal | COVID-19 | Viral Pneumonia | Bacterial Pneumonia | Tuberculosis | Total |
|---|---|---|---|---|---|---|
| Training | 8133 | 2887 | 1075 | 2224 | 560 | 14,879 |
| Validation | 1017 | 362 | 135 | 278 | 70 | 1862 |
| Testing | 1017 | 362 | 135 | 278 | 70 | 1862 |
| CNN Models | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| VGG16 | 97.24 | 97.26 | 97.24 | 97.21 |
| VGG19 | 94.85 | 94.94 | 94.85 | 94.72 |
| Xception | 88.69 | 90.03 | 88.69 | 87.58 |
| InceptionV3 | 93.33 | 93.32 | 93.33 | 93.32 |
| DenseNet201 | 96.01 | 96.00 | 96.01 | 95.96 |
| NasnetMobile | 92.39 | 92.60 | 92.39 | 92.06 |
| ResNet152 | 78.75 | 82.85 | 78.75 | 73.02 |
| CNN Models | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| VGG16 | 99.60 | 99.60 | 99.60 | 99.60 |
| VGG19 | 99.20 | 99.20 | 99.20 | 99.19 |
| Xception | 99.40 | 99.40 | 99.40 | 99.40 |
| InceptionV3 | 98.99 | 99.01 | 98.99 | 99.00 |
| Densenet201 | 99.40 | 99.40 | 99.40 | 99.40 |
| NasnetMobile | 99.80 | 99.80 | 99.80 | 99.80 |
| Resnet152 | 97.79 | 97.80 | 97.79 | 97.77 |
| CNN Models | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| VGG16 | 99.22 | 99.22 | 99.22 | 99.22 |
| VGG19 | 98.75 | 98.76 | 98.75 | 98.75 |
| Xception | 99.06 | 99.08 | 99.06 | 99.06 |
| InceptionV3 | 99.53 | 99.53 | 99.53 | 99.53 |
| Densenet201 | 99.84 | 99.84 | 99.84 | 99.84 |
| NasnetMobile | 99.53 | 99.53 | 99.53 | 99.53 |
| Resnet152 | 98.59 | 98.60 | 98.59 | 98.59 |
| CNN Models | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| VGG16 | 99.31 | 99.31 | 99.31 | 99.30 |
| VGG19 | 99.07 | 99.07 | 99.07 | 99.07 |
| Xception | 99.07 | 99.07 | 99.07 | 99.07 |
| InceptionV3 | 98.38 | 98.47 | 98.38 | 98.40 |
| Densenet201 | 98.84 | 98.88 | 98.84 | 98.85 |
| NasnetMobile | 93.75 | 95.15 | 93.75 | 94.09 |
| Resnet152 | 91.20 | 92.25 | 91.20 | 91.56 |
| CNN Models | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| VGG16 | 96.63 | 96.63 | 96.63 | 96.63 |
| VGG19 | 91.94 | 92.49 | 91.94 | 91.63 |
| Xception | 91.68 | 91.64 | 91.68 | 91.54 |
| InceptionV3 | 92.54 | 92.47 | 92.54 | 92.43 |
| Densenet201 | 95.51 | 95.61 | 95.51 | 95.44 |
| NasnetMobile | 92.93 | 93.32 | 92.93 | 92.96 |
| Resnet152 | 77.21 | 84.70 | 77.21 | 78.57 |
| CNN Models | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| VGG16 | 92.70 | 92.41 | 92.70 | 92.47 |
| VGG19 | 89.04 | 90.37 | 89.04 | 87.00 |
| Xception | 83.35 | 84.83 | 83.35 | 80.61 |
| InceptionV3 | 84.00 | 85.54 | 84.00 | 83.44 |
| Densenet201 | 89.10 | 89.80 | 89.10 | 88.42 |
| NasnetMobile | 87.76 | 88.05 | 87.76 | 86.65 |
| Resnet152 | 74.70 | 76.80 | 74.70 | 71.60 |
| CNN Models | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| VGG16 | 98.61 | 99.07 | 98.61 | 98.52 |
| VGG19 | 96.53 | 97.45 | 96.53 | 96.34 |
| Xception | 93.06 | 93.75 | 93.06 | 92.18 |
| InceptionV3 | 95.83 | 97.22 | 95.83 | 95.56 |
| DenseNet201 | 96.53 | 97.69 | 96.53 | 96.30 |
| NasnetMobile | 93.06 | 95.60 | 93.06 | 92.82 |
| ResNet152 | 75.69 | 76.50 | 75.69 | 80.13 |
| Author and Year | Method and Models | Number of Images Used | Classification Accuracy | AUC 1 | |||
|---|---|---|---|---|---|---|---|
| Two-Class | Three-Class 2 | Four-Class 3 | Five-Class 4 | ||||
| Nayak et al. (2020) [100] | Method: CNN with transfer learning Model: ResNet-34 | C 5: 203 Total: 406 | C 5 & N 6: 98.33% | NA 7 | NA | NA | C & N: 0.98 |
| Choudhury et al. (2020) [88] | Method: CNN with transfer learning Model: CheXNet | C: 423 Total: 3487 | NA | 97.74% | NA | NA | NA |
| Jain et al. (2020) [91] | Method: CNN with transfer learning Model: Xception | C: 490 Total: 6432 | NA | 97.97% | NA | NA | NA |
| Bhattacharyya et al. (2021) [101] | Method: ML 8 + DL 9 DL model: VGG-19 ML model: Random Forest | C: 342 Total: 1029 | NA | 96.6% | NA | NA | NA |
| Nikolaou et al. (2021) [103] | Method: CNN with transfer learning Model: EfficientNetB0 | C: 3616 Total: 15,153 | C & N: 95% | 93% | NA | NA | NA |
| Yang et al. (2021) [99] | Method: CNN with transfer learning Model: VGG16 | C: 3616 Total: 8461 | C & N: 98% C & VP 10: 99% | 97% | NA | NA | NA |
| Khan et al. (2020) [96] | Method: deep learning Model: Coronet (novel CNN) | C: 284 Total: 1251 | NA | 95% | 89.6% | NA | NA |
| Hussain et al. (2020) [90] | Method: deep learning Model: CoroDet (novel CNN) | C: 500 Total: 2100 | C & N: 99.1% | 94.2% | 91.2% | NA | NA |
| Oh et al. (2020) [104] | Method: CNN with transfer learning Model: ResNet-18 | C: 180 Total: 502 | NA | NA | 88.9% | NA | NA |
| Timemy et al. (2021) [105] | Method: ML + DL DL model: ResNet-50 ML model: ESD 11 | C: 435 Total: 2186 | NA | NA | NA | 91.6% | NA |
| Proposed work (Nillmani et al.) | Method: CNN with transfer learning Model: VGG16, NasnetMobile, DenseNet201 | C: 3611 Total: 18,603 | C & N: 97.24% 12 C & VP: 99.80% 13 C & BP 14: 99.84% 15 C & T 16: 99.31% 12 | 96.63% 12 | NA | 92.70% 12 | C & N: 0.95 12 C & VP: 1.0 13 C & BP: 1.0 15 C & T: 0.98 12 Three-class 2: 0.97 12 Five-class 4: 0.92 12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nillmani; Jain, P.K.; Sharma, N.; Kalra, M.K.; Viskovic, K.; Saba, L.; Suri, J.S. Four Types of Multiclass Frameworks for Pneumonia Classification and Its Validation in X-ray Scans Using Seven Types of Deep Learning Artificial Intelligence Models. Diagnostics 2022, 12, 652. https://doi.org/10.3390/diagnostics12030652
Nillmani, Jain PK, Sharma N, Kalra MK, Viskovic K, Saba L, Suri JS. Four Types of Multiclass Frameworks for Pneumonia Classification and Its Validation in X-ray Scans Using Seven Types of Deep Learning Artificial Intelligence Models. Diagnostics. 2022; 12(3):652. https://doi.org/10.3390/diagnostics12030652
Chicago/Turabian StyleNillmani, Pankaj K. Jain, Neeraj Sharma, Mannudeep K. Kalra, Klaudija Viskovic, Luca Saba, and Jasjit S. Suri. 2022. "Four Types of Multiclass Frameworks for Pneumonia Classification and Its Validation in X-ray Scans Using Seven Types of Deep Learning Artificial Intelligence Models" Diagnostics 12, no. 3: 652. https://doi.org/10.3390/diagnostics12030652
APA StyleNillmani, Jain, P. K., Sharma, N., Kalra, M. K., Viskovic, K., Saba, L., & Suri, J. S. (2022). Four Types of Multiclass Frameworks for Pneumonia Classification and Its Validation in X-ray Scans Using Seven Types of Deep Learning Artificial Intelligence Models. Diagnostics, 12(3), 652. https://doi.org/10.3390/diagnostics12030652








