3D Convolutional Neural Network-Based Denoising of Low-Count Whole-Body 18F-Fluorodeoxyglucose and 89Zr-Rituximab PET Scans
Abstract
:1. Introduction
2. Materials and Methods
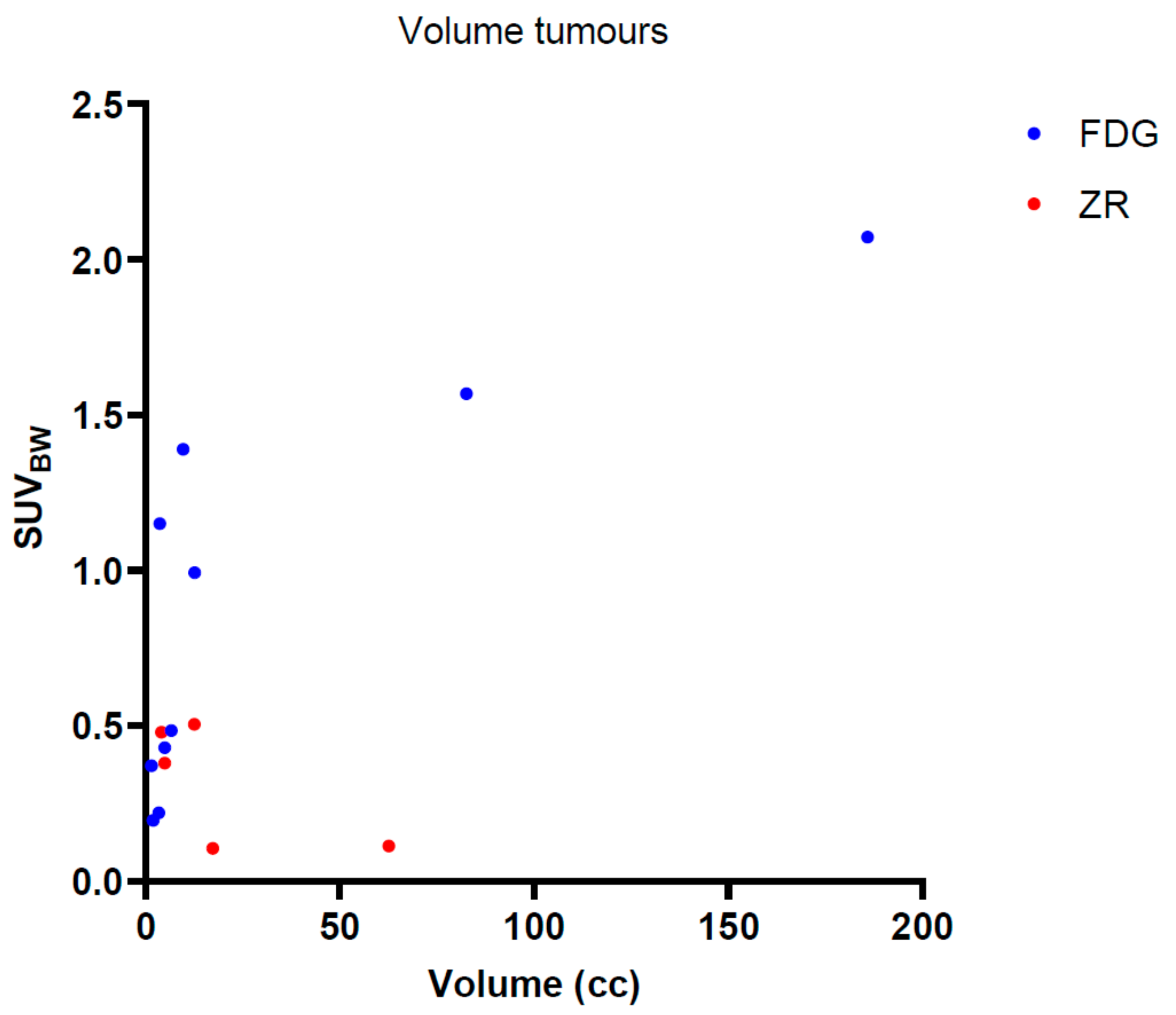
2.1. Participants
2.2. Data Acquisition
2.3. Image Processing
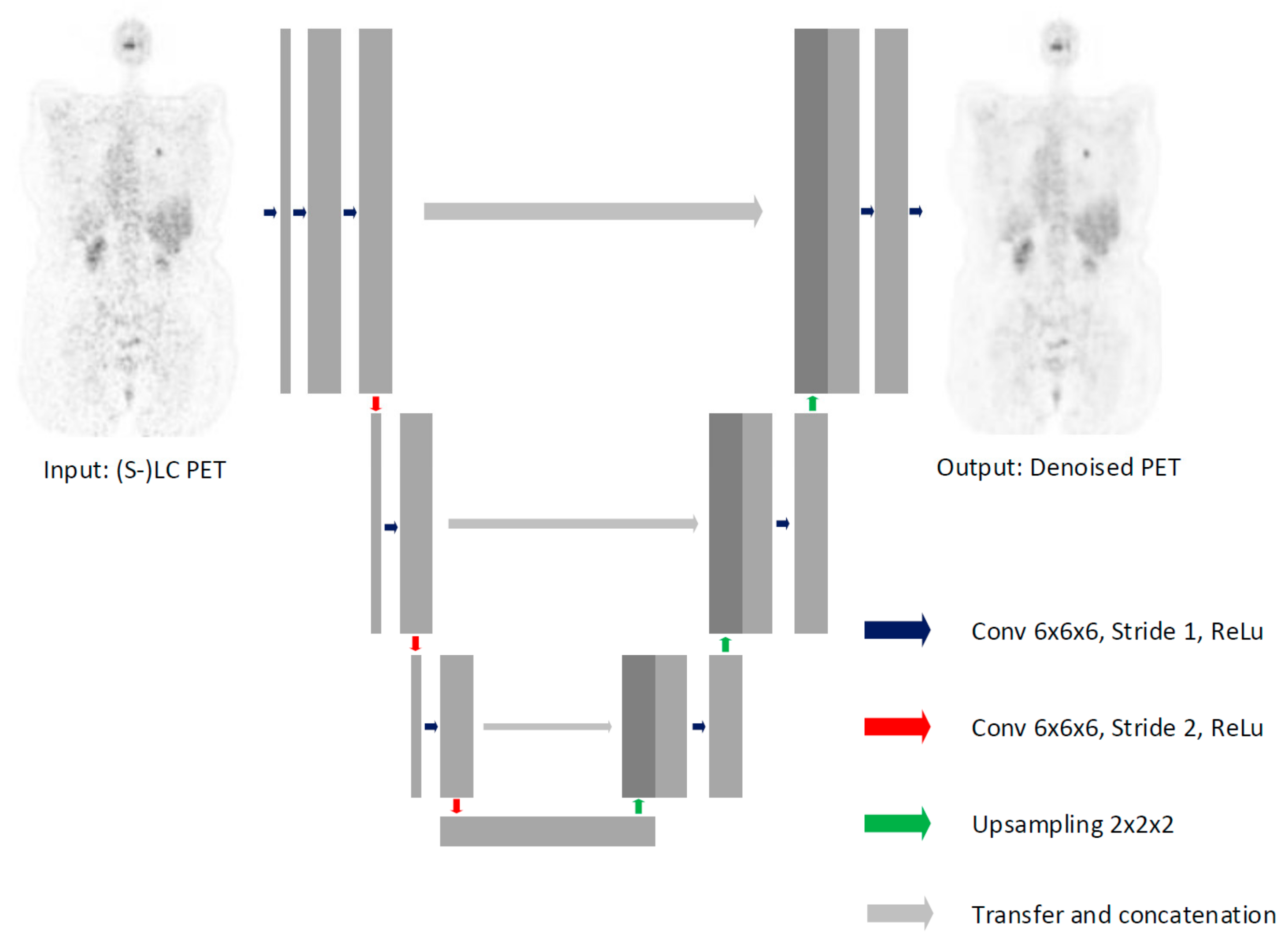
2.4. Model Architecture
2.5. Model Performance
2.5.1. Quantitative Performance
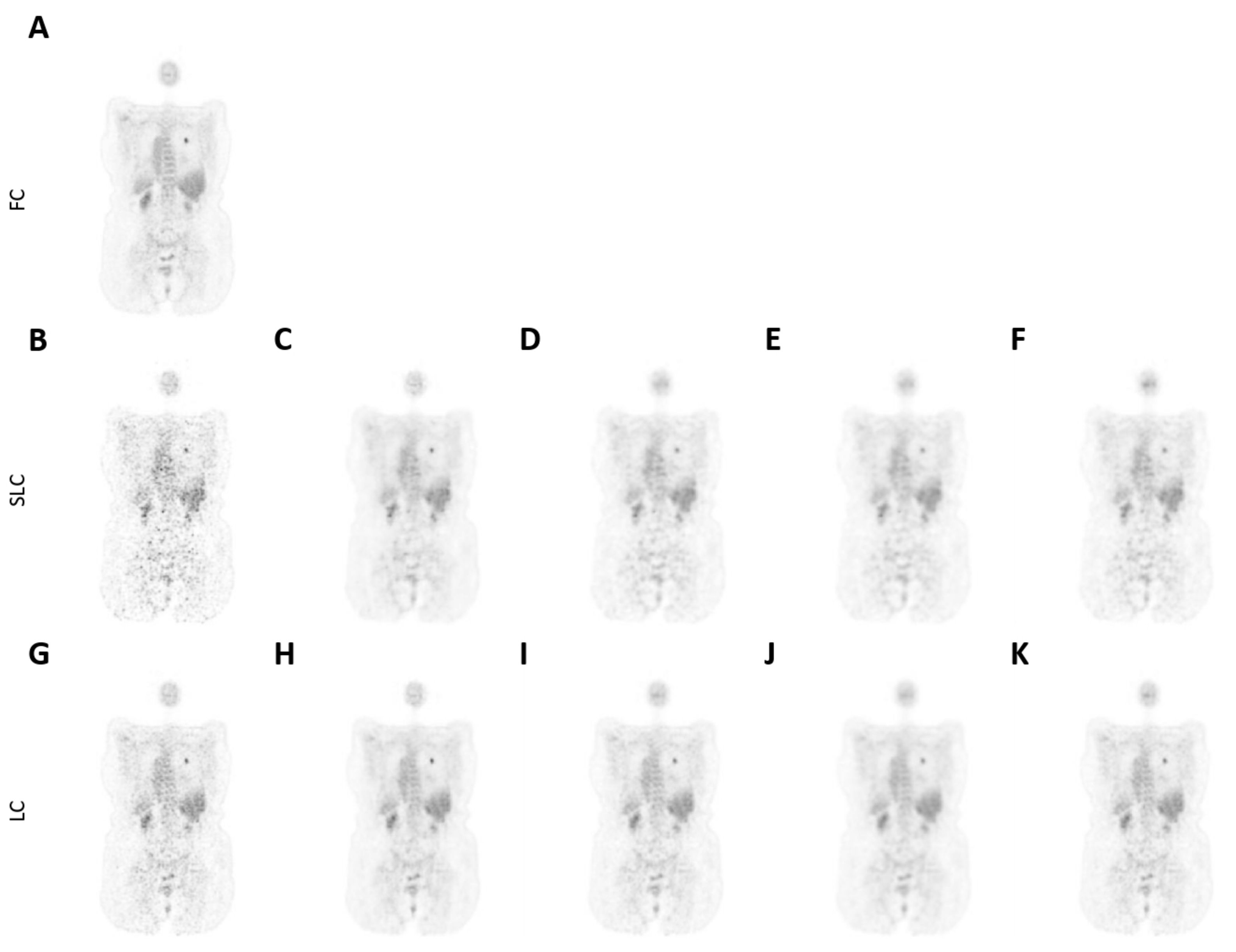
2.5.2. Qualitative Performance
3. Results
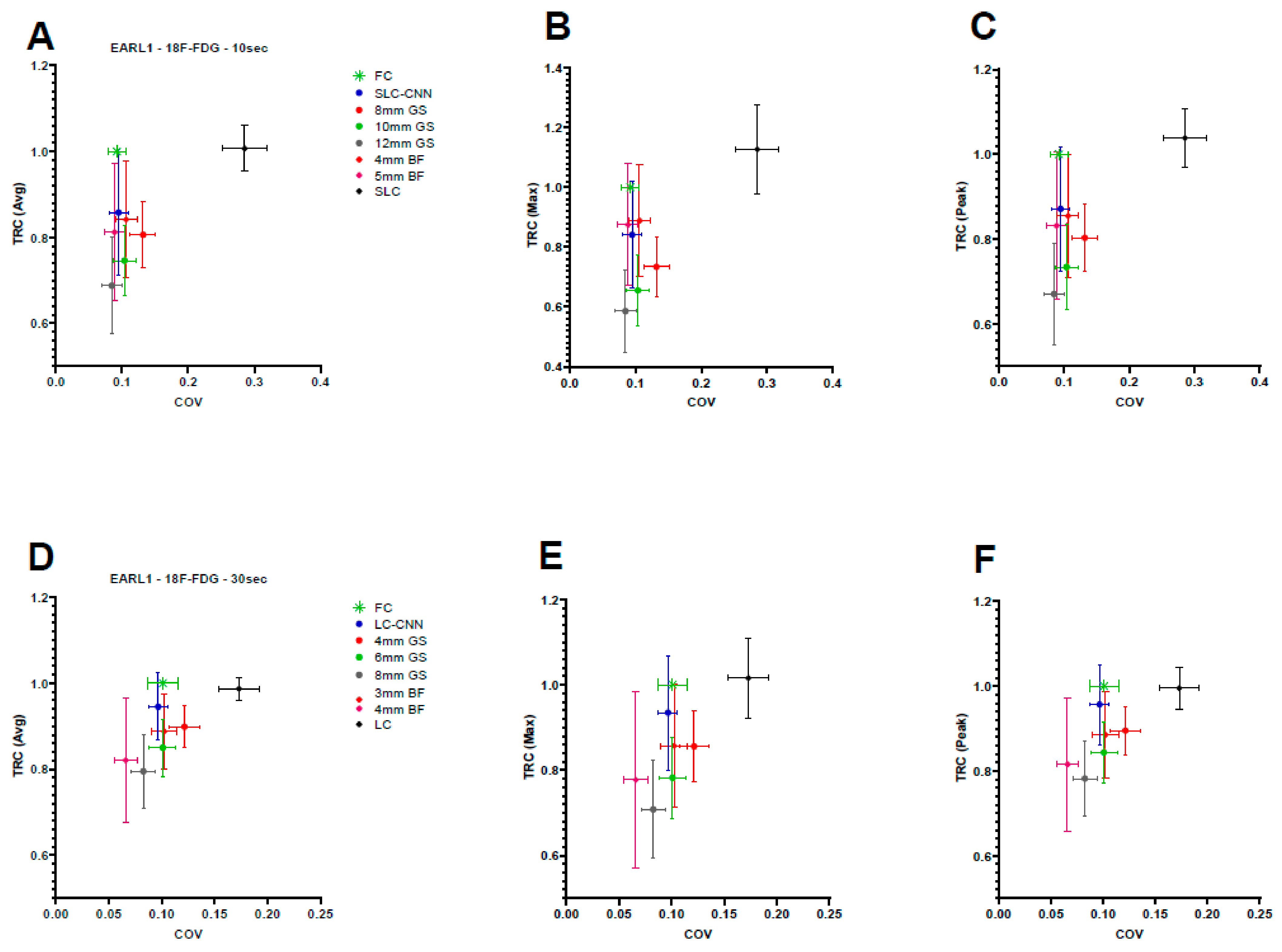
3.1. Quantitative Assessment
3.1.1. 18F-FDG
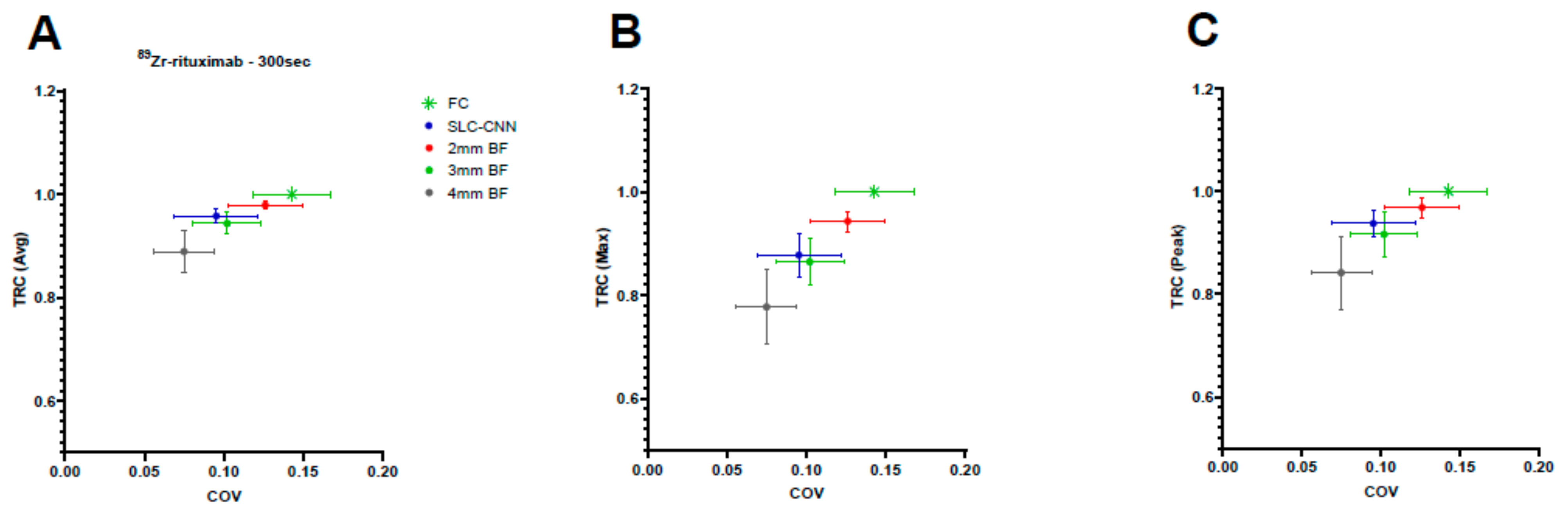
3.1.2. 89Zr-Rituximab
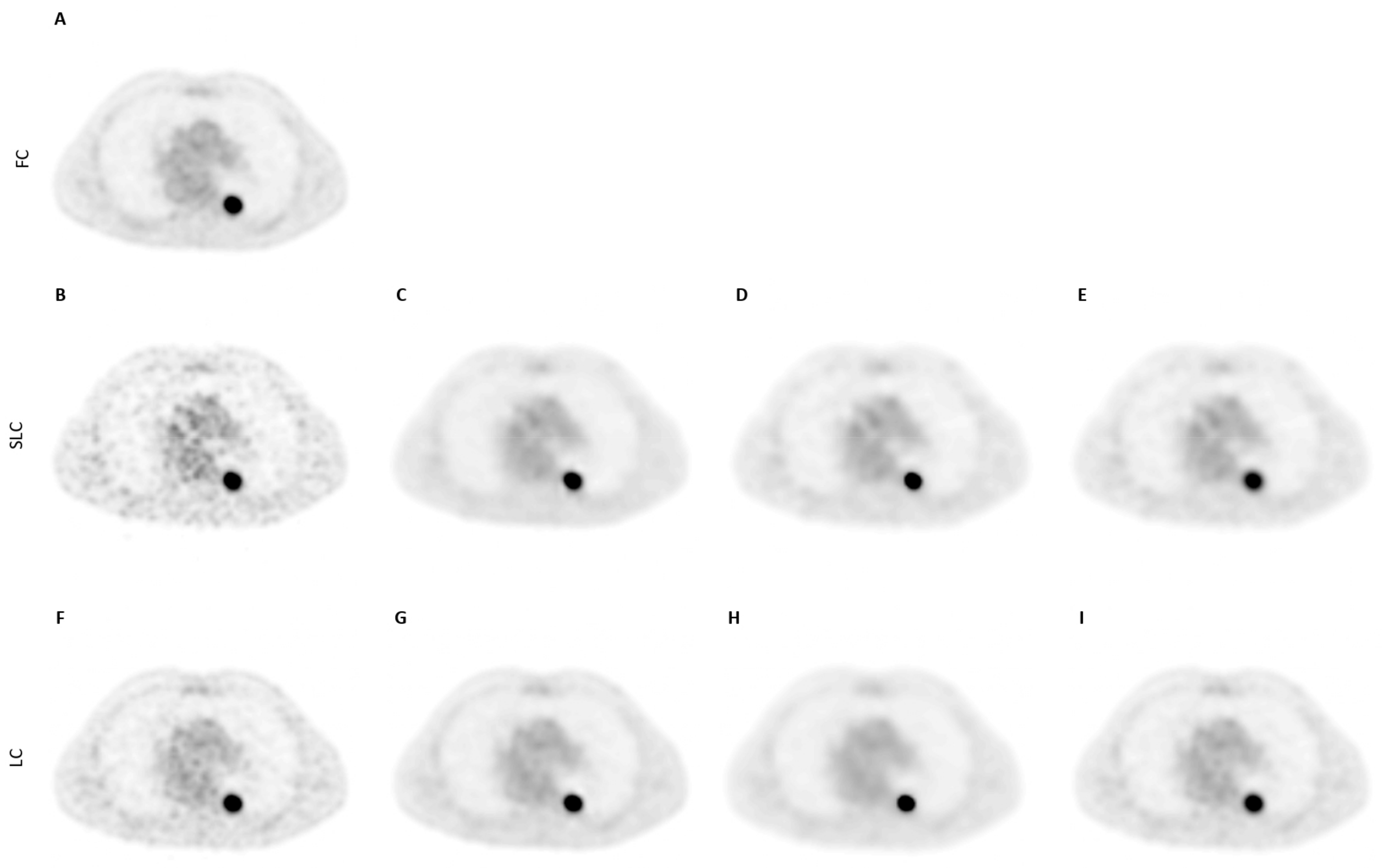
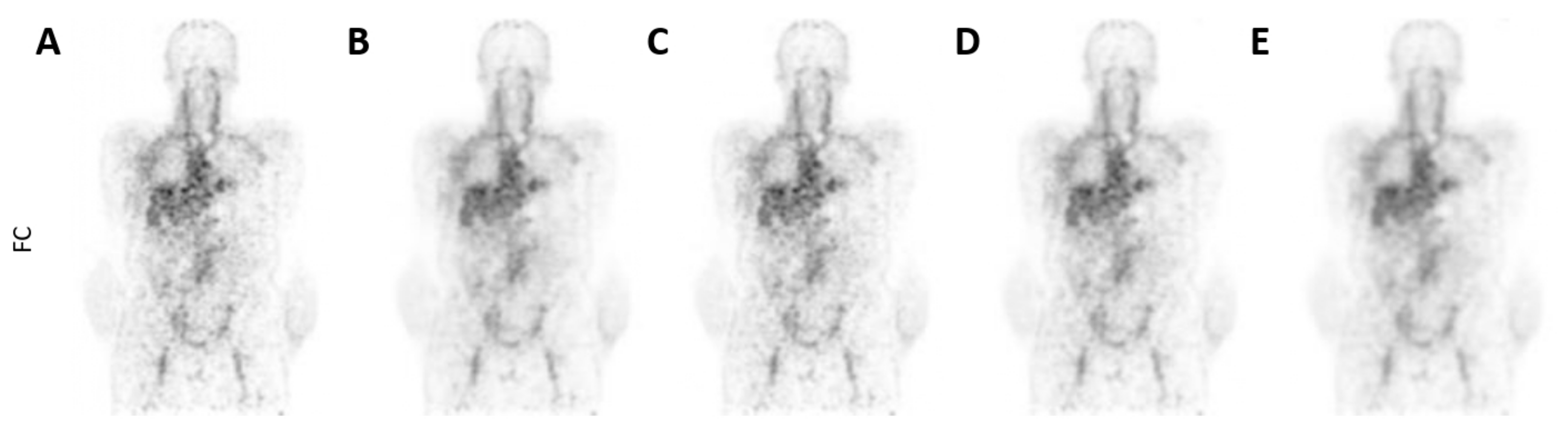
3.2. Qualitative Assessment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| TRC–Average | TRC–Maximum | TRC–3DPeak | COV | |
|---|---|---|---|---|
| FC | 0.09 | |||
| SLC | 0.99 | 1.11 | 1.03 | 0.29 |
| GS (12; 10; 8 mm) | [0.69 *; 0.74 *; 0.81 **] | [0.59 *; 0.66 *; 0.74] | [0.68 *; 0.74 *; 0.80] | [0.09; 0.11; 0.14] |
| BF (5; 4 mm) | [0.79; 0.82] | [0.85; 0.87] | [0.82; 0.84] | [0.01; 0.11] |
| SLC-CNN | 0.86 | 0.85 | 0.87 | 0.10 |
| LC | 0.98 | 1.02 | 0.99 | 0.17 |
| GS (8; 6; 4 mm) | [0.80 *; 0.85 **; 0.90] | [0.71 *; 0.79 *; 0.86 *] | [0.79 *; 0.84 *; 0.90 *] | [0.09; 0.11; 0.13] |
| BF (4; 3 mm) | [0.81 **; 0.88] | [0.78; 0.87] | [0.82 **; 0.89] | [0.07; 0.11] |
| LC-CNN | 0.95 | 0.94 | 0.96 | 0.10 |
| TRC–Average | TRC–Maximum | TRC–3Dpeak | COV | |
|---|---|---|---|---|
| FC | 0.14 | |||
| BF (4; 3; 2 mm) | [0.88 *; 0.94 *; 0.98] | [0.78 **; 0.86; 0.94 *] | [0.84 **; 0.92 **; 0.97 *] | [0.07; 0.10; 0.13] |
| SLC-CNN | 0.96 | 0.88 | 0.94 | 0.10 |
Appendix B
Appendix B.1. Image Processing
Appendix B.2. CNN Architecture
Appendix B.3. Model Performance
References
- Almuhaideb, A.; Papathanasiou, N.; Bomanji, J. 18F-FDG PET/CT imaging in oncology. Ann. Saudi Med. 2011, 31, 3–13. (In English) [Google Scholar] [CrossRef] [Green Version]
- Budak, E.; Çok, G.; Akgün, A. The Contribution of Fluorine (18)F-FDG PET/CT to Lung Cancer Diagnosis, Staging and Treatment Planning. Mol. Imaging Radionucl. Ther. 2018, 27, 73–80. (In English) [Google Scholar] [CrossRef]
- Verhagen, A.; Bootsma, G.; Tjan-Heijnen, V.; Van Der Wilt, G.; Cox, A.; Brouwer, M.; Corstens, F.; Oyen, W. FDG-PET in staging lung cancer: How does it change the algorithm? Lung Cancer 2004, 44, 175–181. (In English) [Google Scholar] [CrossRef] [PubMed]
- Jauw, Y.W.; O’Donoghue, J.A.; Zijlstra, J.M.; Hoekstra, O.S.; Menke-Van Der Houven, C.W.; Morschhauser, F.; Carrasquillo, J.A.; Zweegman, S.; Pandit-Taskar, N.; Lammertsma, A.A.; et al. (89)Zr-Immuno-PET: Toward a Noninvasive Clinical Tool to Measure Target Engagement of Therapeutic Antibodies In Vivo. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2019, 60, 1825–1832. (In English) [Google Scholar] [CrossRef]
- Tsutsui, Y.; Awamoto, S.; Himuro, K.; Umezu, Y.; Baba, S.; Sasaki, M. Characteristics of Smoothing Filters to Achieve the Guideline Recommended Positron Emission Tomography Image without Harmonization. Asia Ocean J. Nucl. Med. Biol. 2018, 6, 15–23. (In English) [Google Scholar] [CrossRef] [PubMed]
- Soret, M.; Bacharach, S.L.; Buvat, I. Partial-Volume Effect in PET Tumor Imaging. J. Nucl. Med. 2007, 48, 932–945. [Google Scholar] [CrossRef] [Green Version]
- Hofheinz, F.; Langner, J.; Beuthien-Baumann, B.; Oehme, L.; Steinbach, J.; Kotzerke, J.; Van den Hoff, J. Suitability of bilateral filtering for edge-preserving noise reduction in PET. EJNMMI Res. 2011, 1, 23. (In English) [Google Scholar] [CrossRef] [Green Version]
- de Vries, B.M.; Golla, S.S.V.; Ebenau, J.; Verfaillie, S.C.J.; Timmers, T.; Heeman, F.; Cysouw, M.C.F.; van Berckel, B.N.M.; van der Flier, W.M.; Yaqub, M.; et al. Classification of negative and positive 18F-florbetapir brain PET studies in subjective cognitive decline patients using a convolutional neural network. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 721–728. [Google Scholar] [CrossRef]
- Liu, M.; Cheng, D.; Yan, W. Classification of Alzheimer’s Disease by Combination of Convolutional and Recurrent Neural Networks Using FDG-PET Images. Front. Neuroinformatics 2018, 12, 35. (In English) [Google Scholar] [CrossRef] [Green Version]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Gong, K.; Guan, J.; Liu, C.-C.; Qi, J. PET Image Denoising Using a Deep Neural Network Through Fine Tuning. IEEE Trans. Radiat. Plasma Med. Sci. 2018, 3, 153–161. [Google Scholar] [CrossRef]
- Hashimoto, F.; Ohba, H.; Ote, K.; Teramoto, A.; Tsukada, H. Dynamic PET Image Denoising Using Deep Convolutional Neural Networks Without Prior Training Datasets. IEEE Access 2019, 7, 96594–96603. [Google Scholar] [CrossRef]
- Cui, J.; Gong, K.; Guo, N.; Wu, C.; Meng, X.; Kim, K.; Zheng, K.; Wu, Z.; Fu, L.; Xu, B.; et al. PET image denoising using unsupervised deep learning. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2780–2789. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Ibaraki, M.; Nemoto, M.; Watabe, H.; Kimura, Y. A review on AI in PET imaging. Ann. Nucl. Med. 2022, 36, 133–143. [Google Scholar] [CrossRef]
- Schaefferkoetter, J.; Yan, J.; Ortega, C.; Sertic, A.; Lechtman, E.; Eshet, Y.; Metser, U.; Veit-Haibach, P. Convolutional neural networks for improving image quality with noisy PET data. EJNMMI Res. 2020, 10, 105. [Google Scholar] [CrossRef] [PubMed]
- Spuhler, K.; Serrano-Sosa, M.; Cattell, R.; DeLorenzo, C.; Huang, C. Full-count PET recovery from low-count image using a dilated convolutional neural network. Med. Phys. 2020, 47, 4928–4938. [Google Scholar] [CrossRef]
- Chen, K.T.; Gong, E.; Macruz, F.B.D.C.; Xu, J.; Boumis, A.; Khalighi, M.; Poston, K.L.; Sha, S.J.; Greicius, M.D.; Mormino, E.; et al. Ultra–Low-Dose 18F-Florbetaben Amyloid PET Imaging Using Deep Learning with Multi-Contrast MRI Inputs. Radiology 2019, 290, 649–656. [Google Scholar] [CrossRef]
- Lu, W.; A Onofrey, J.; Lu, Y.; Shi, L.; Ma, T.; Liu, Y.; Liu, C. An investigation of quantitative accuracy for deep learning based denoising in oncological PET. Phys. Med. Biol. 2019, 64, 165019. [Google Scholar] [CrossRef]
- Pfaehler, E.; Mesotten, L.; Zhovannik, I.; Pieplenbosch, S.; Thomeer, M.; Vanhove, K.; Adriaensens, P.; Boellaard, R. Plausibility and redundancy analysis to select FDG-PET textural features in non-small cell lung cancer. Med. Phys. 2021, 48, 1226–1238. (In English) [Google Scholar] [CrossRef]
- Jauw, Y.W.S.; Zijlstra, J.M.; De Jong, D.; Vugts, D.J.; Zweegman, S.; Hoekstra, O.S.; Van Dongen, G.A.M.S.; Huisman, M.C. Performance of 89Zr-Labeled-Rituximab-PET as an Imaging Biomarker to Assess CD20 Targeting: A Pilot Study in Patients with Relapsed/Refractory Diffuse Large B Cell Lymphoma. PLoS ONE 2017, 12, e0169828. (In English) [Google Scholar] [CrossRef]
- Makris, N.E.; Boellaard, R.; Visser, E.P.; de Jong, J.R.; Vanderlinden, B.; Wierts, R.; van der Veen, B.J.; Greuter, H.J.; Vugts, D.J.; van Dongen, G.A.; et al. Multicenter harmonization of 89Zr PET/CT performance. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2014, 55, 264–267. [Google Scholar] [CrossRef] [Green Version]
- Pfaehler, E.; Jong, J.; Dierckx, R.; van Velden, F.; Boellaard, R. SMART (SiMulAtion and ReconsTruction) PET: An efficient PET simulation-reconstruction tool. EJNMMI Phys. 2018, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, J. A Gentle Introduction to Pooling Layers for Convolutional Neural Networks. Deep Learning for Computer Vision. Available online: https://machinelearningmastery.com/pooling-layers-for-convolutional-neural-networks/ (accessed on 18 December 2021).
- Chen, J.; Shen, Y. The effect of kernel size of CNNs for lung nodule classification. In Proceedings of the 2017 9th International Conference on Advanced Infocomm Technology (ICAIT), Chengdu, China, 22–24 November 2017; pp. 340–344. [Google Scholar] [CrossRef]
- Boellaard, R. Quantitative oncology molecular analysis suite: ACCURATE. J. Nucl. Med. 2018, 59, 1753. [Google Scholar]
- Boellaard, R. Accurate: An Oncology PET/CT Quantitative Analysis Tool. Available online: https://doi.org/10.5281/zenodo.3908203 (accessed on 19 January 2022).
- Hou, W.; Wang, W.; Liu, R.; Lu, T. Cropout: A General Mechanism for Reducing Overfitting on Convolutional Neural Networks. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–8. [Google Scholar] [CrossRef]
- Johnson, J.M.; Khoshgoftaar, T.M. Survey on deep learning with class imbalance. J. Big Data 2019, 6, 27. [Google Scholar] [CrossRef]
- Horé, A.; Ziou, D. Image Quality Metrics: PSNR vs. SSIM. In Proceedings of the 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 2366–2369. [Google Scholar] [CrossRef]
| 18F-FDG (n = 20) NSCLC–Amsterdam UMC | 18F-FDG (n = 40) NSCLC–LPC | 89Zr-Rituximab (n = 5)Non-Hodgkin Lymphoma | |
|---|---|---|---|
| Male/female (n) | 11/9 | 25/15 | 3/2 |
| Injected dose (MBq) | 259.4 ± 43.8 | 298.2 ± 49.4 | 73.7 ± 0.3 |
| Tumour volume (cubic centimeter (cc))–Test data | 29.2 ± 54.3 | - | 25.6 ± 51.3 |
| GS (FWHM) | BF (FWHM; SUV) | |
|---|---|---|
| 18F-FDG PET | ||
| SLC | 8 mm, 10 mm and 12 mm | 4 mm and 5 mm; SUV2.5 |
| LC | 4 mm, 6 mm and 8 mm | 3 mm and 4 mm; SUV2.5 |
| 89Zr-rituximab PET | 6 mm, 8 mm and 10 mm | 2 mm, 3 mm and 4 mm; SUV2.5 |
| SLC | LC | FC | |||||
|---|---|---|---|---|---|---|---|
| Metrics [1–5: Low–High] | SLC-CNN | BF-4 mm | BF-5 mm | LC-CNN | BF-3 mm | BF-4 mm | |
| Level of noise | 3.0–3.2 | 4.0–4.0 * | 3.6–4.0 * | 1.8–2.0 | 2.6–2.0 ** | 2.2–3.8 * | 1–1 * |
| Tumour detectability | 2.0–3.0 | 2.2–3.0 | 2.0–3.0 | 4.0–4.0 | 3.6–4.0 | 2.8–3.0 * | 5–5 * |
| Overall scan quality | 2.4–2.6 | 1.6–2.0 * | 1.8–2.0 * | 4.4–4.0 | 3.8–4.0 | 2.8–2.2 * | 5–5 * |
| Clinically acceptable? [%] | 0–80 | 0–0 * | 0–0 * | 100–100 | 80–100 | 20–0 * | 100–100 |
| Best scan (1/2/3/4) | 2–2 | 3–3 | 4–4 | 2–3 | 3–2 | 4–4 | 1–1 |
| Metrics [1–5: Low–High] | SLC-CNN | BF-2 mm | BF-3 mm | BF-4 mm | FC |
|---|---|---|---|---|---|
| Level of noise | 2.4–2.6 | 3.8–4.6 * | 2.8–2.6 | 1.4–1.2 * | 4.6–4.8 * |
| Tumour detectability | 3.4–3.8 | 4.4–4.0 * | 2.4–2.4 * | 1.4–1.4 * | 4.6–4.2 * |
| Overall scan quality | 3.8–3.8 | 3.6–3.8 | 3.4–3.0 * | 2.0–1.4 * | 3.4–3.0 * |
| Clinical acceptable? [%] | 100–100 | 100–100 | 80–80 | 0–0 * | 80–80 |
| Best scan (1/2/3/4/5) | 1–1 | 3–2 | 2–4 | 4–5 | 3–3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Vries, B.M.; Golla, S.S.V.; Zwezerijnen, G.J.C.; Hoekstra, O.S.; Jauw, Y.W.S.; Huisman, M.C.; van Dongen, G.A.M.S.; Menke-van der Houven van Oordt, W.C.; Zijlstra-Baalbergen, J.J.M.; Mesotten, L.; et al. 3D Convolutional Neural Network-Based Denoising of Low-Count Whole-Body 18F-Fluorodeoxyglucose and 89Zr-Rituximab PET Scans. Diagnostics 2022, 12, 596. https://doi.org/10.3390/diagnostics12030596
de Vries BM, Golla SSV, Zwezerijnen GJC, Hoekstra OS, Jauw YWS, Huisman MC, van Dongen GAMS, Menke-van der Houven van Oordt WC, Zijlstra-Baalbergen JJM, Mesotten L, et al. 3D Convolutional Neural Network-Based Denoising of Low-Count Whole-Body 18F-Fluorodeoxyglucose and 89Zr-Rituximab PET Scans. Diagnostics. 2022; 12(3):596. https://doi.org/10.3390/diagnostics12030596
Chicago/Turabian Stylede Vries, Bart M., Sandeep S. V. Golla, Gerben J. C. Zwezerijnen, Otto S. Hoekstra, Yvonne W. S. Jauw, Marc C. Huisman, Guus A. M. S. van Dongen, Willemien C. Menke-van der Houven van Oordt, Josée J. M. Zijlstra-Baalbergen, Liesbet Mesotten, and et al. 2022. "3D Convolutional Neural Network-Based Denoising of Low-Count Whole-Body 18F-Fluorodeoxyglucose and 89Zr-Rituximab PET Scans" Diagnostics 12, no. 3: 596. https://doi.org/10.3390/diagnostics12030596
APA Stylede Vries, B. M., Golla, S. S. V., Zwezerijnen, G. J. C., Hoekstra, O. S., Jauw, Y. W. S., Huisman, M. C., van Dongen, G. A. M. S., Menke-van der Houven van Oordt, W. C., Zijlstra-Baalbergen, J. J. M., Mesotten, L., Boellaard, R., & Yaqub, M. (2022). 3D Convolutional Neural Network-Based Denoising of Low-Count Whole-Body 18F-Fluorodeoxyglucose and 89Zr-Rituximab PET Scans. Diagnostics, 12(3), 596. https://doi.org/10.3390/diagnostics12030596






