Role of MBL2 Polymorphisms in Sepsis and Survival: A Pilot Study and In Silico Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. In Silico Analysis
2.2.1. General Information
2.2.2. Analyzing the Effect of Variants on Protein Function
2.2.3. Identifying Variants’ Locations on MBL Protein Domains
2.2.4. Analyzing Variants Impact on Protein Stability
2.2.5. Analysis of Evolutionary Conservation of MBL Protein Sequences
2.2.6. Analyzing Structural Impacts of Variants
2.3. Study Design
2.4. Genotyping
2.5. Statistical Analysis
3. Results
3.1. In Silico Analysis
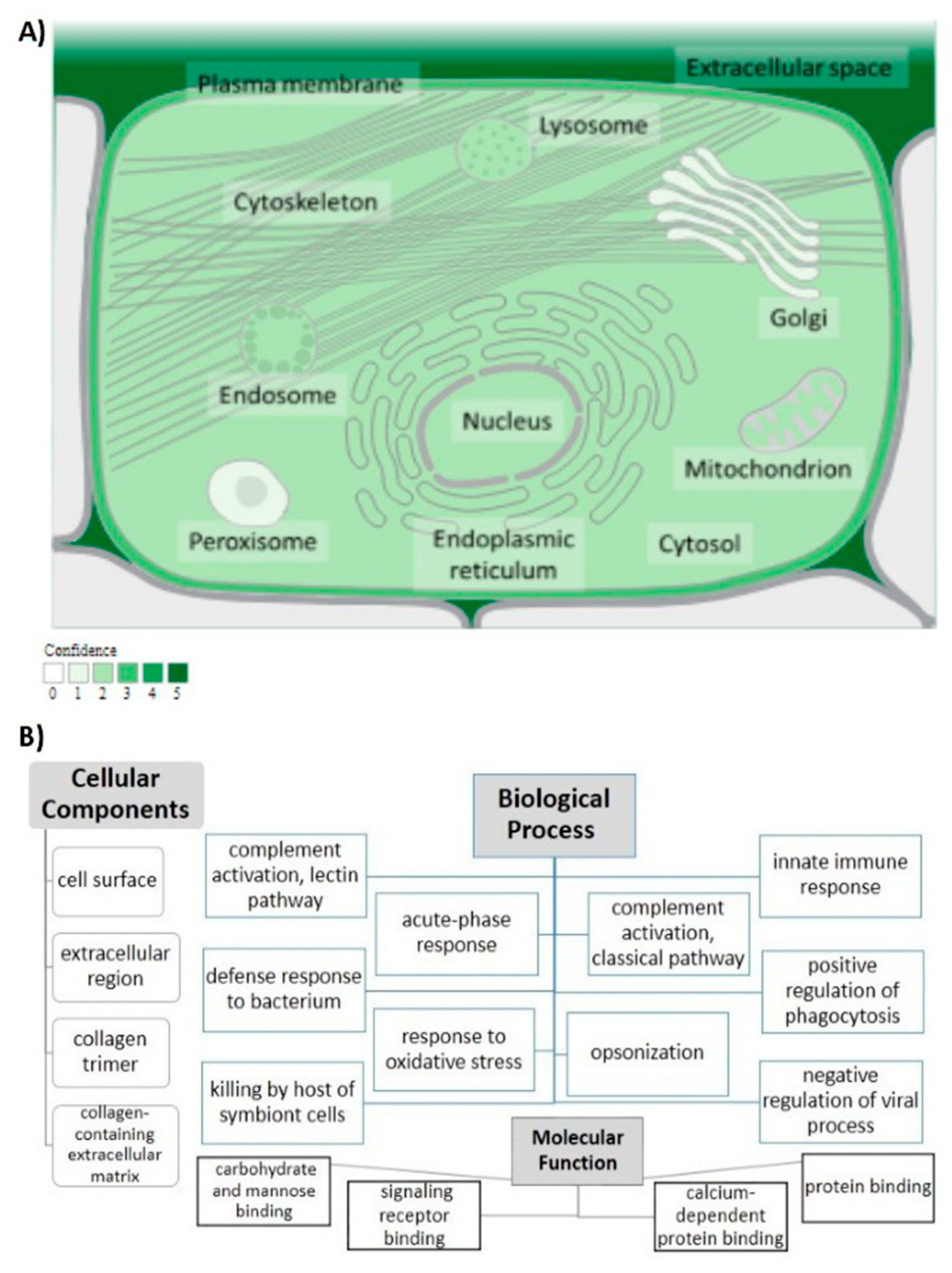
3.1.1. General Information
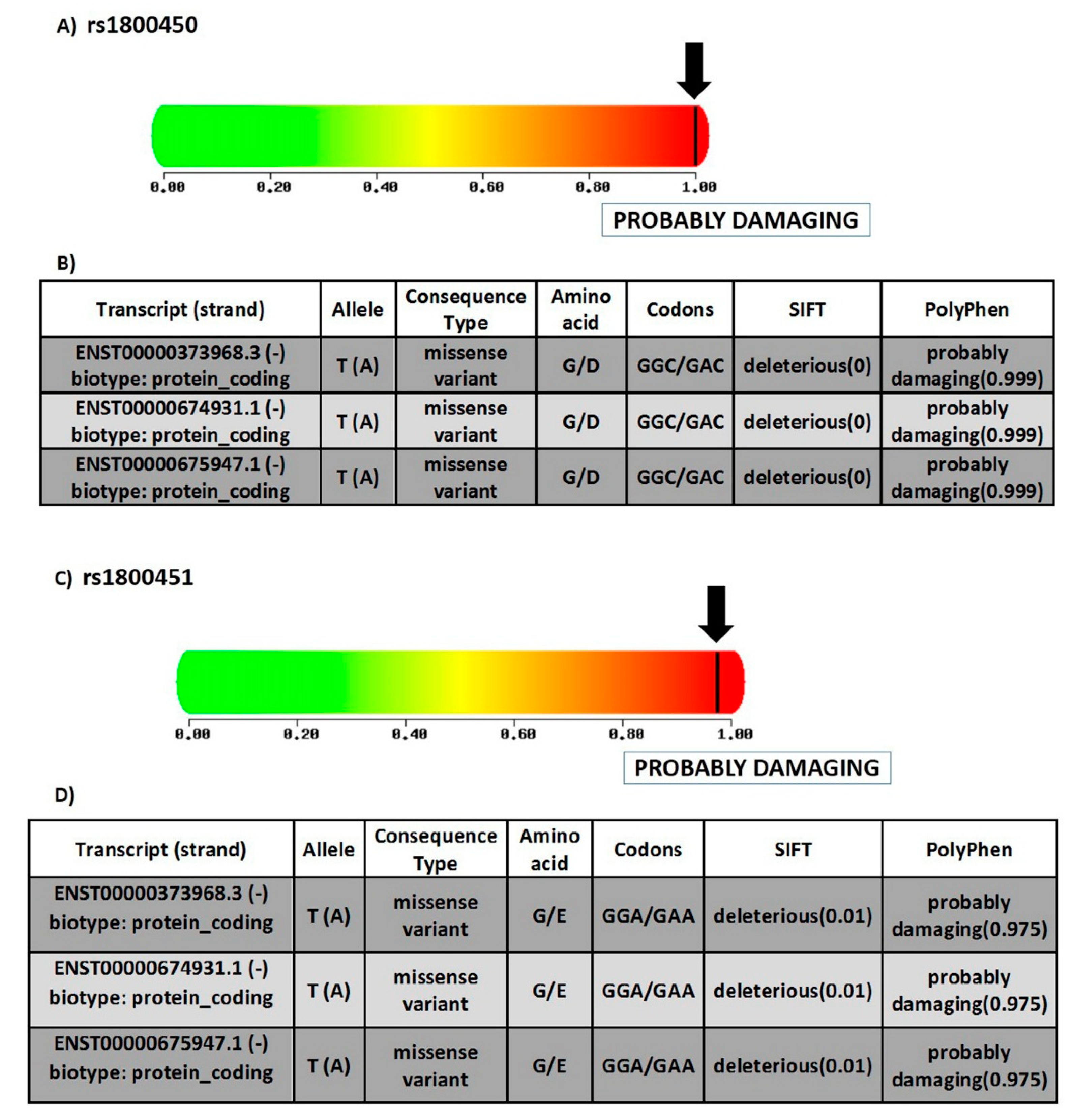
3.1.2. Prediction of SNPs Impact on MBL Protein Function
3.1.3. Determining Variants’ Locations on Protein Domains
3.1.4. Predicting MBL Protein Stability with rs1800450 and rs1800451 SNPs
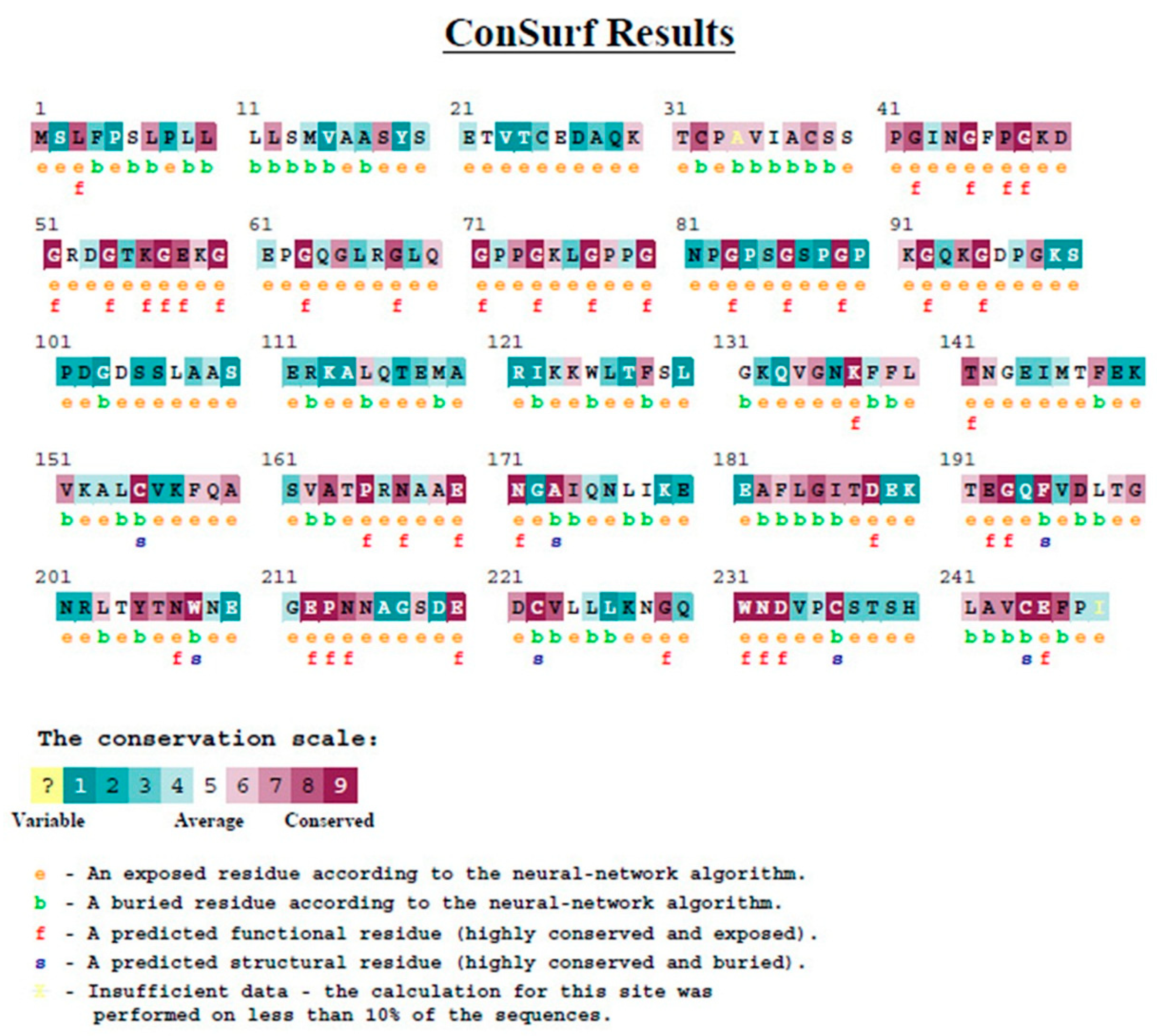
3.1.5. Evolutionary Conservation Analysis
3.1.6. Analyzing Structural Effects of MBL Variants
3.2. Study Population
3.3. Genotype Analysis
3.4. Polymorphisms and Clinical Characteristics
3.5. Survival Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Nachtigall, I.; Tamarkin, A.; Tafelski, S.; Weimann, A.; Rothbart, A.; Heim, S.; Wernecke, K.D.; Spies, C. Polymorphisms of the toll-like receptor 2 and 4 genes are associated with faster progression and a more severe course of sepsis in critically ill patients. J. Int. Med. Res. 2013, 42, 93–110. [Google Scholar] [CrossRef] [PubMed]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef]
- Fleischmann-Struzek, C.; Mellhammar, L.; Rose, N.; Cassini, A.; Rudd, K.E.; Schlattmann, P.; Allegranzi, B.; Reinhart, K. Incdence and mortality of hospital- and ICU-treated sepsis: Results from an updated and expanded systematic review and meta-analysis. Intensiv. Care Med. 2020, 46, 1552–1562. [Google Scholar] [CrossRef]
- World Health Organization. Improving the Prevention, Diagnosis and Clinical Management of Sepsis. Available online: https://apps.who.int/iris/bitstream/handle/10665/275646/A70_R7-en.pdf?sequence=1&isAlowed=y (accessed on 2 December 2021).
- David, V.L.; Ercisli, M.F.; Rogobete, A.F.; Boia, E.S.; Horhat, R.; Nitu, R.; Diaconu, M.M.; Pirtea, L.; Ciuca, I.; Horhat, D.I.; et al. Early Prediction of Sepsis Incidence in Critically Ill Patients Using Specific Genetic Polymorphisms. Biochem. Genet. 2016, 55, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Mao, Z.-R.; Pang, K.; Zhang, S.-L.; Li, L. Association of tumor necrosis factor α −308G/A and interleukin-6 −174G/C gene polymorphism with pneumonia-induced sepsis. J. Crit. Care 2015, 30, 920–923. [Google Scholar] [CrossRef]
- Liu, L.; Ning, B. The role of MBL2 gene polymorphism in sepsis incidence. Int. J. Clin. Exp. Pathol. 2015, 8, 15123–15127. [Google Scholar]
- Eddie Ip, W.K.; Takahashi, K.; Ezekowitz, R.A.; Stuart, L.M. Mannose-binding lectin and innate immunity. Immunol. Rev. 2009, 230, 9–21. [Google Scholar] [CrossRef]
- Ramos-Casals, M.; Zeron, P.B.; Soria, N.; Nardi, N.; Vargas, A.; Munoz, S.; Bove, A.; Suarez, B.; Lozano, F. Mannose-binding lectin-low genotypes are associated with milder systemic and immunological disease expression in primary Sjogren’s sydrome. Rheumatology 2008, 48, 65–69. [Google Scholar] [CrossRef]
- Turner, M. The role of mannose-binding lectin in health and disease. Mol. Immunol. 2003, 40, 423–429. [Google Scholar] [CrossRef]
- Worthley, D.L.; Bardy, P.G.; Gordon, D.-L.; Mullighan, C.-G. Mannose-binding lectin and maladies of the bowel and liver. World J. Gastroenterol. 2006, 12, 6420–6428. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, M.H.; Ali, S.S.; Fattouh, A.; Hamza, H.S.; Badr, M.M. MBL2 gene polymorphism rs1800450 and rheumatic fever with and without rheumatic heart disease: An Egyptian pilot study. Pediatr. Rheumatol. 2018, 16, 24. [Google Scholar] [CrossRef] [PubMed]
- Tiyo, B.T.; Vendramini, E.C.L.; Souza, V.; Colli, C.M.; Alves, H.V.; Sell, A.M.; Zucoloto, S.B.P.; Visentainer, J.E.L. Association of MBL2 Exon 1 Polymorphisms With Multibacillary Leprosy. Front. Immunol. 2020, 11, 927. [Google Scholar] [CrossRef] [PubMed]
- Auriti, C.; Prencipe, G.; Moriondo, M.; Bersani, I.; Bertaina, C.; Mondì, V.; Inglese, R. Mannose-Binding Lectin: Biologic Charcteristics and Role in the Susceptibility to Infections and Ischemia-Reperfusion Related Injury in Critically Ill Neonates. J. Immunol. Res. 2017, 2017, 7045630. [Google Scholar] [CrossRef]
- Lipscombe, R.J.; Sumiya, M.; Summerfield, J.A.; Turner, M.W. Distinct physicochemical characteristics of human mannose binding protein expressed by individuals of differing genotype. Immunology 1995, 85, 660–667. [Google Scholar]
- Jacobson, S.; Larsson, P.; Åberg, A.-M.; Johansson, G.; Winsö, O.; Söderberg, S. Levels of mannose-binding lectin (MBL) associates with sepsis-related in-hospital mortality in women. J. Inflamm. 2020, 17, 1–11. [Google Scholar] [CrossRef]
- Turner, M.W. Mannose-Binding Lectin (MBL) in Health and Disease. Immunobiology 1998, 199, 327–339. [Google Scholar] [CrossRef]
- Heitzeneder, S.; Seidel, M.G.; Förster-Waldl, E.; Heitger, A. Mannan-binding lectin deficiency—Good news, bad news, doesn’t matter? Clin. Immunol. 2012, 143, 22–38. [Google Scholar] [CrossRef]
- Zhang, A.-Q.; Yue, C.-L.; Pan, W.; Gao, J.-W.; Zeng, L.; Gu, W.; Jiang, J.-X. Mannose-binding lectin polymorphisms and the risk of sepsis: Evidence from a meta-analysis. Epidemiol. Infect. 2014, 142, 2195–2206. [Google Scholar] [CrossRef]
- Mills, T.C.; Chapman, S.; Hutton, P.; Gordon, A.C.; Bion, J.; Chiche, J.-D.; Holloway, P.A.H.; Stüber, F.; Garrard, C.S.; Hinds, C.J.; et al. Variants in the Mannose-binding Lectin GeneMBL2do not Associate With Sepsis Susceptibility or Survival in a Large European Cohort. Clin. Infect. Dis. 2015, 61, 695–703. [Google Scholar] [CrossRef]
- Hammad, N.M.; El Badawy, N.E.; Nasr, A.M.; Ghramh, H.A.; Al Kady, L.M. Mannose-Binding Lectin Gene Polymorphism and Its Association with Susceptibility to Recurrent Vulvovaginal Candidiasis. BioMed Res. Int. 2018, 2018, 7648152. [Google Scholar] [CrossRef]
- Badawy, M.; Saber, D.; Madani, H.; Mosallam, D.S. Use of Mannose-Binding Lectin Gene Polymorphisms and the Serum MBL Level for the Early Detection of Neonatal Sepsis. J. Pediatr. Genet. 2018, 7, 150–157. [Google Scholar] [CrossRef]
- Sim, N.-L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Tang, H.; Thomas, P. PANTHER-PSEP: Predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics 2016, 32, 2230–2232. [Google Scholar] [CrossRef]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef]
- Capriotti, E.; Calabrese, R.; Fariselli, P.; Martelli, P.L.; Altman, R.B.; Casadio, R. WS-SNPs&GO: A web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genom. 2013, 14, S6. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005, 33, W306–W310. [Google Scholar] [CrossRef] [PubMed]
- Bava, K.A. ProTherm, version 4.0: Thermodynamic database for proteins and mutants. Nucleic Acids Res. 2004, 32, 120D–121D. [Google Scholar] [CrossRef] [PubMed]
- Berezin, C.; Glaser, F.; Rosenberg, J.; Paz, I.; Pupko, T.; Fariselli, P.; Casadio, R.; Ben-Tal, N. ConSeq: The identification of functionally and structurally important residues in protein sequences. Bioinformatics 2004, 20, 1322–1324. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [PubMed]
- Venselaar, H.; Beek, T.A.H.T.; Kuipers, R.K.P.; Hekkelman, M.L.; Vriend, G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinform. 2010, 11, 548. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Tumangger, H.; Jamil, K.F. Contribution of genes polymorphism to susceptibility and outcome of sepsis. Egypt. J. Med Hum. Genet. 2010, 11, 97–103. [Google Scholar] [CrossRef][Green Version]
- Hartz, A.; Pagel, J.; Humberg, A.; Preuss, M.; Schreiter, L.; Rupp, J.; Figge, J.; Karsten, C.M.; Nürnberg, P.; Herting, E.; et al. The association of mannose-binding lectin 2 polymorphisms with outcome in very low birth weight infants. PLoS ONE 2017, 12, e0178032. [Google Scholar] [CrossRef]
- Nasr, M.; Marie, A.; Boghdadi, G.; Elsaid, R.; Salah, E. Role of mannose binding lectin in response to candida antigen immunotherapy of warts. J. Dermatol. Treat. 2021, 32, 376–380. [Google Scholar] [CrossRef]
- Eisen, D.P.; Osthoff, M. If there is an evolutionary selection pressure for the high frequency of MBL2 polymorphisms, what is it? Clin. Exp. Immunol. 2014, 176, 165–171. [Google Scholar] [CrossRef]
- Tereshchenko, S.Y.; Smolnikova, M.V.; Freidin, M.B. Mannose-binding lectin gene polymorphisms in the East Siberia and Russian Arctic populations. Immunogenetics 2020, 72, 347–354. [Google Scholar] [CrossRef]
- Troelsen, L.N.; Garred, P.; Jacobsen, S. Mortality and Predictors of Mortality in Rheumatoid Arthritis—A Role for Mannose-binding Lectin? J. Rheumatol. 2010, 37, 536–543. [Google Scholar] [CrossRef][Green Version]
- Fumagalli, S.; Perego, C.; Zangari, R.; De Blasio, D.; Oggioni, M.; De Nigris, F.; Snider, F.; Garred, P.; Ferrante, A.M.R.; De Simoni, M.-G. Lectin Pathway of Complement Activation Is Associated with Vulnerability of Atherosclerotic Plaques. Front. Immunol. 2017, 8, 288. [Google Scholar] [CrossRef] [PubMed]
- Alonso, D.; Ferreira, A.F.B.; Ribolla, P.; Santos, I.K.F.D.M.; Cruz, M.D.S.; De Carvalho, F.A.; Abatepaulo, A.R.R.; Costa, D.L.; Werneck, G.L.; Farias, T.J.C.; et al. Genotypes of the Mannan-Binding Lectin Gene and Susceptibility to Visceral Leishmaniasis and Clinical Complications. J. Infect. Dis. 2007, 195, 1212–1217. [Google Scholar] [CrossRef] [PubMed]
- Verdu, P.; Barreiro, L.B.; Patin, E.; Gessain, A.; Cassar, O.; Kidd, J.R.; Kidd, K.K.; Behar, D.M.; Froment, A.; Heyer, E.; et al. Evolutionary insights into the high worldwide prevalence of MBL2 deficiency alleles. Hum. Mol. Genet. 2006, 15, 2650–2658. [Google Scholar] [CrossRef] [PubMed]
- Boldt, A.B.; Messias-Reason, I.J.; Meyer, D.; Schrago, C.G.; Lang, F.; Lell, B.; Dietz, K.; Kremsner, P.G.; Petzl-Erler, M.L.; Kun, J.F. Phylogenetic nomenclature and evolution of mannose-binding lectin (MBL2) haplotypes. BMC Genet. 2010, 11, 38. [Google Scholar] [CrossRef]
- Gordon, A.; Waheed, U.; Hansen, T.K.; Hitman, G.A.; Garrard, C.S.; Turner, M.W.; Klein, N.J.; Brett, S.J.; Hinds, C.J. Mannose-binding lectin polymorphisms in severe sepsis: Relationship to levels, incidence, and outcome. Shock 2006, 25, 88–93. [Google Scholar] [CrossRef]
- Huh, J.W.; Song, K.; Yum, J.-S.; Hong, S.-B.; Lim, C.-M.; Koh, Y. Association of mannose-binding lectin-2 genotype and serum levels with prognosis of sepsis. Crit. Care 2009, 13, R176–R179. [Google Scholar] [CrossRef]
- Dahl, M.; Tybjaerg-Hansen, A.; Schnohr, P.; Nordestgaard, B.G. A Population-based Study of Morbidity and Mortality in Mannose-binding Lectin Deficiency. J. Exp. Med. 2004, 199, 1391–1399. [Google Scholar] [CrossRef]
- Van Kempen, G.; Meijvis, S.; Endeman, H.; Vlaminckx, B.; Meek, B.; De Jong, B.; Rijkers, G.; Bos, W.J.W. Mannose-binding lectin andl-ficolin polymorphisms in patients with community-acquired pneumonia caused by intracellular pathogens. Immunology 2016, 151, 81–88. [Google Scholar] [CrossRef]
- El-Behedy, E.M.; Akeel, N.; El Maghraby, H.; Shawky, A. Serum Level and Genetic Polymorphism of Mannose-Binding Lectin in Infants with Neonatal Sepsis at Zagazig University Hospitals. Egypt J. Immunol. 2019, 26, 91–99. [Google Scholar]
- Garnacho-Montero, J.; García-Cabrera, E.; Jiménez-Álvarez, R.; Díaz-Martín, A.; Revuelto-Rey, J.; Aznar-Martín, J.; Garnacho-Montero, C. Genetic variants of the MBL2 gene are associated with mortality in pneumococcal sepsis. Diagn. Microbiol. Infect. Dis. 2012, 73, 39–44. [Google Scholar] [CrossRef]
- De Pascale, G.; Cutuli, S.L.; Pennisi, M.A.; Antonelli, M. The Role of Mannose-Binding Lectin in Severe Sepsis and Septic Shock. Mediat. Inflamm. 2013, 2013, 625803. [Google Scholar] [CrossRef] [PubMed]
- Casanova, J.-L.; Abel, L. Human genetics of infectious diseases: Unique insights into immunological redundancy. Semin. Immunol. 2018, 36, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Roos, A.; Garred, P.; Wildenberg, M.E.; Lynch, N.; Munoz, J.R.; Zuiverloon, T.C.; Bouwman, L.H.; Schlagwein, N.; Houten, F.C.F.V.D.; Faber-Krol, M.C.; et al. Antibody-mediated activation of the classical pathway of complement may compensate for mannose-binding lectin deficiency. Eur. J. Immunol. 2004, 34, 2589–2598. [Google Scholar] [CrossRef] [PubMed]
- Frodsham, A.J. Genetics of infectious diseases. Hum. Mol. Genet. 2004, 13, R187–R194. [Google Scholar] [CrossRef] [PubMed]
- Mayr, F.B.; Yende, S.; Angus, D.C. Epidemiology of severe sepsis. Virulence 2014, 5, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, J.D.; McHugh, B.; Doherty, C.; Smith, M.P.; Govan, J.R.; Kilpatrick, D.C.; Hill, A.T. Mannose-binding lectin deficiency and disease severity in non-cystic fibrosis bronchiectasis: A prospective study. Lancet Respir. Med. 2013, 1, 224–232. [Google Scholar] [CrossRef]
- Mandal, J.; Malla, B.; Steffensen, R.; Costa, L.; Egli, A.; Trendelenburg, M.; Blasi, F.; Kostikas, K.; Welte, T.; Torres, A.; et al. Mannose-binding lectin protein and its association to clinical outcomes in COPD: A longitudinal study. Respir. Res. 2015, 16, 150. [Google Scholar] [CrossRef]
- Vincent, J.-L.; Rello, J.; Marshall, J.K.; Silva, E.; Anzueto, A.; Martin, C.D.; Moreno, R.; Lipman, J.; Gomersall, C.; Sakr, Y.; et al. International Study of the Prevalence and Outcomes of Infection in Intensive Care Units. JAMA J. Am. Med Assoc. 2009, 302, 2323–2329. [Google Scholar] [CrossRef]
- Vincent, J.-L.; Sakr, Y.; Singer, M.; Martin-Loeches, I.; Machado, F.R.; Marshall, J.C.; Finfer, S.; Pelosi, P.; Brazzi, L.; Aditianingsih, D.; et al. Prevalence and Outcomes of Infection Among Patients in Intensive Care Units in 2017. JAMA J. Am. Med Assoc. 2020, 323, 1478–1487. [Google Scholar] [CrossRef]
- Wong, D.T.; Gomez, M.; McGuire, G.P.; Kavanagh, B. Utilization of intensive care unit days in a Canadian medical-surgical intensive care unit. Crit. Care Med. 1999, 27, 1319–1324. [Google Scholar] [CrossRef]
- Abelha, F.J.; Castro, M.A.; Landeiro, N.M.; Neves, A.M.; Santos, C.C. Mortalidade e o tempo de internação em uma unidade de terapia intensiva cirúrgica. Braz. J. Anesthesiol. 2006, 56, 34–45. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vincent, J.-L.; Jones, G.; David, S.; Olariu, E.; Cadwell, K.K. Frequency and mortality of septic shock in Europe and North America: A systematic review and meta-analysis. Crit. Care 2019, 23, 1–11. [Google Scholar] [CrossRef] [PubMed]




| Variables | All | Control | Infection without Sepsis | Sepsis | p-Value | |
|---|---|---|---|---|---|---|
| Demographic Characteristics | ||||||
| Number | 130 | 39 | 53 | 38 | ||
| Age, years | median (IQR) | 60.0 (22.3) | 55.0 (29.0) | 59.0 (33.5) | 65.0 (17.3) | 0.005 |
| ≤40 years | 28 (21.5%) | 12 (30.8%) | 14 (26.4%) | 2 (5.3%) | ||
| ≤60 years | 40 (30.8%) | 14 (35.9%) | 15 (28.3%) | 11 (28.9%) | 0.020 | |
| >60 years | 62 (47.7%) | 13 (33.3%) | 24 (45.3%) | 25 (65.8%) | ||
| Sex | Male | 76 (58.5%) | 24 (61.5%) | 33 (62.3%) | 19 (50.0%) | 0.45 |
| Female | 54 (41.5%) | 15 (38.5%) | 20 (37.7%) | 19 (50.0%) | ||
| Vital signs | HR | 90.0 (17.0) | 90.0 (00.0) | 100.0 (24.0) | 90.0 (20.8) | 0.008 |
| MAP | 83.0 (14.0) | 83.0 (00.0) | 83.0 (24.5) | 75.0 (20.0) | 0.047 | |
| Concomitant diseases | ||||||
| Diabetes | positive | 45 (34.6%) | 10 (25.6%) | 17 (32.1%) | 18 (47.4%) | 0.12 |
| Hypertension | positive | 65 (50.0%) | 16 (41.0%) | 28 (52.8%) | 21 (55.3%) | 0.40 |
| Vascular disease | positive | 34 (26.2%) | 4 (10.3%) | 17 (32.1%) | 13 (34.2%) | 0.025 |
| Chronic lung disease | positive | 8 (6.2%) | 2 (5.1%) | 5 (9.4%) | 1 (2.6%) | 0.54 |
| Chronic liver disease | positive | 10 (7.7%) | 2 (5.1%) | 3 (5.7%) | 5 (13.2%) | 0.42 |
| Chronic renal disease | positive | 25 (19.2%) | 5 (12.8%) | 11 (20.8%) | 9 (23.7%) | 0.45 |
| ICU assessment | ||||||
| APACHE score | median (IQR) | 15.0 (7.3) | 12.0 (7.0) | 16.0 (7.0) | 16.5 (6.8) | 0.001 |
| Glasgow scale | median (IQR) | 11.5 (8.0) | 14.0 (9.0) | 9.0 (7.5) | 14.0 (8.0) | 0.057 |
| Length of stay, days | median (IQR) | 13.0 (19.3) | 10.0 (16.0) | 15.0 (18.5) | 13.5 (25.8) | 0.386 |
| Consequence | discharge | 54 (41.5%) | 19 (48.7%) | 21 (39.6%) | 14 (36.8%) | |
| transferred | 6 (4.6%) | 1 (2.6%) | 3 (5.7%) | 2 (5.3%) | 0.82 | |
| death | 70 (53.9%) | 19 (48.7%) | 29 (54.7%) | 22 (57.9%) | ||
| OS, days | median (IQR) | 13.0 (20.5) | 11.0 (15.0) | 15.0 (20.0) | 16.5 (28.3) | 0.424 |
| Admission category | ||||||
| Renal | positive | 4 (3.1%) | 1 (2.6%) | 1 (1.9%) | 2 (5.3%) | 0.69 |
| Cardiovascular | positive | 5 (3.8%) | 2 (5.1%) | 2 (3.8%) | 1 (2.6%) | 1.00 |
| Infection | positive | 24 (18.5%) | 0 (0.0%) | 8 (15.1%) | 16 (42.1%) | 0.000 |
| Neurology | positive | 36 (27.7%) | 13 (33.3%) | 19 (35.8%) | 4 (10.5%) | 0.019 |
| Post-surgical | positive | 19 (14.6%) | 4 (10.3%) | 6 (11.3%) | 9 (23.7%) | 0.17 |
| Respiratory | positive | 14 (10.8%) | 2 (5.1%) | 11 (20.8%) | 1 (2.6%) | 0.013 |
| Trauma | positive | 10 (7.7%) | 7 (17.9%) | 3 (5.7%) | 0 (0.0%) | 0.009 |
| Other causes | positive | 9 (6.9%) | 4 (10.3%) | 2 (3.8%) | 3 (7.9%) | 0.52 |
| Gastrointestinal | positive | 9 (6.9%) | 6 (15.4%) | 1 (1.9%) | 2 (5.3%) | 0.042 |
| Causative Organism | All | Infection without Sepsis | Sepsis | p-Value | Odds Ratio (95% CI) |
|---|---|---|---|---|---|
| Enterobacter spp. | 10 (11.0%) | 4 (7.5%) | 6 (15.8%) | 0.31 | 2.30 (0.60–8.78) |
| Acinetobacter spp. | 11 (12.1%) | 7 (13.2%) | 4 (10.5%) | 0.76 | 0.77 (0.21–2.85) |
| Candida spp. | 3 (3.3%) | 1 (1.9%) | 2 (5.3%) | 0.57 | 2.89 (0.25–33.07) |
| Escherichia coli | 16 (17.6%) | 11 (20.8%) | 5 (13.2%) | 0.35 | 0.58 (0.18–1.83) |
| Gram negative bacilli | 9 (9.9%) | 5 (9.4%) | 4 (10.5%) | 1.00 | 1.13 (0.28–4.52) |
| Klebsiella spp. | 21 (23.1%) | 13 (24.5%) | 8 (21.1%) | 0.70 | 0.82 (0.30–2.23) |
| Pseudomonas spp. | 13 (14.3%) | 6 (11.3%) | 7 (18.4%) | 0.34 | 1.77 (0.54–5.76) |
| Staph spp. | 18 (19.8%) | 12 (22.6%) | 6 (15.8%) | 0.42 | 0.64 (0.22–1.89) |
| Streptococcus spp. | 4 (4.4%) | 3 (5.7%) | 1 (2.6%) | 0.64 | 0.45 (0.05–4.51) |
| Aeromonas spp. | 1 (1.1%) | 1 (1.9%) | 0 (0.0%) | 1.00 | 0.45 (0.02 to 11.46) |
| Proteus spp. | 2 (2.2%) | 1 (1.9%) | 1 (2.6%) | 1.00 | 1.41 (0.09–23.20) |
| Citrobacter spp. | 1 (1.1%) | 1 (1.9%) | 0 (0.0%) | 1.00 | 0.45 (0.02 to 11.46) |
| Serratia spp. | 1 (1.1%) | 1 (1.9%) | 0 (0.0%) | 1.00 | 0.45 (0.02 to 11.46) |
| All | Control | Infection without Sepsis | Sepsis | p-Value | OR (95% CI) | |||
|---|---|---|---|---|---|---|---|---|
| Sepsis Group against Control Group | Sepsis Group against Infection Group | Infection Group against Control Group | ||||||
| Genotype Frequencies | ||||||||
| Rs1800451 | ||||||||
| A/A | 109 (83.8%) | 32 (82.1%) | 45 (84.9%) | 32 (84.2%) | 0.91 | Reference | ||
| A/C | 19 (14.6%) | 7 (17.9%) | 7 (13.2%) | 5 (13.2%) | 0.71 (0.21 to 2.49) | 1.00 (0.29 to 3.45) | 0.71 (0.23 to 2.23) | |
| C/C | 2 (1.6%) | 0 (0.0%) | 1 (1.9%) | 1 (2.6%) | 3.00 (0.12 to 76.40) | 1.41 (0.08 to 23.33) | 2.14 (0.09 to 54.29) | |
| P HWE | 0.90 | 1.00 | 1.00 | 1.00 | ||||
| Rs1800450 | ||||||||
| A/A | 40 (30.8%) | 11 (28.2%) | 15 (28.3%) | 14 (36.8%) | 0.63 | Reference | ||
| A/B | 90 (69.2%) | 28 (71.8%) | 38 (71.7%) | 24 (63.2%) | 0.68 (0.28 to 1.65) | 0.67 (0.26 to 1.76) | 1.00 (0.40 to 2.49) | |
| B/B | 0 | 0 | 0 | 0 | 0.79 (0.01 to 43.12) | 1.07 (0.02 to 57.49) | 0.74 (0.01 to 40.25) | |
| P HWE | 0.000 | 0.012 | 0.002 | 0.039 | ||||
| Allele frequencies | ||||||||
| Rs1800451 | ||||||||
| A | 237 (91.15%) | 71 (91.0%) | 97 (91.5%) | 69 (90.8%) | 0.98 | Reference | ||
| C | 23 (8.85%) | 7 (9.0%) | 9 (8.5%) | 7 (9.2%) | 1.03 (0.34 to 3.09) | 1.09 (0.39 to 3.08) | 0.94 (0.33 to 2.64) | |
| Rs1800450 | ||||||||
| A | 170 (65.4%) | 50 (64.1%) | 68 (64.2%) | 52 (68.4%) | 0.80 | Reference | ||
| B | 90 (34.6%) | 28 (35.9%) | 38 (35.8%) | 24 (31.6%) | 0.82 (0.42 to 1.61) | 0.83 (0.44 to 1.54) | 1.00 (0.54 to 1.84) | |
| Carriage rate | ||||||||
| Rs1800451 | ||||||||
| A | 128 (98.5%) | 39 (100%) | 52 (98.1%) | 37 (97.4%) | 0.07 | Reference | ||
| C | 21 (16.2%) | 7 (17.9%) | 8 (15.1%) | 6 (15.8%) | 0.90 (0.28 to 2.94) | 1.05 (0.34 to 3.29) | 0.86 (0.29 to 2.56) | |
| Rs1800450 | ||||||||
| A | 130 (100%) | 39 (100%) | 53 (100%) | 38 (100%) | 0.17 | Reference | ||
| B | 90 (69.2%) | 28 (71.8%) | 38 (71.7%) | 24 (63.2) | 0.88 (0.43 to 1.78) | 0.88 (0.46 to 1.70) | 1.00 (0.53 to 1.89) | |
| Genotype | Control | Infection without Sepsis | Sepsis | p-Value | OR (95% CI) | |||
|---|---|---|---|---|---|---|---|---|
| Sepsis Group against Control Group | Sepsis Group against Infection Group | Infection Group against Control Group | ||||||
| rs1800451 | ||||||||
| Codominant | A/A | 32 (82.1%) | 45 (84.9%) | 32 (84.2%) | 0.91 | Reference | ||
| A/C | 7 (17.9%) | 7 (13.2%) | 5 (13.2%) | 0.71 (0.21 to 2.49) | 1.00 (0.29 to 3.45) | 0.71 (0.23 to 2.23) | ||
| C/C | 0 (0.0%) | 1 (1.9%) | 1 (2.6%) | 3.00 (0.12 to 76.40) | 1.41 (0.08 to 23.33) | 2.14 (0.09 to 54.29) | ||
| Dominant | A/A | 32 (82.1%) | 45 (84.9%) | 32 (84.2%) | 0.93 | Reference | ||
| A/C-C/C | 7 (17.9%) | 8 (15.1%) | 6 (15.8%) | 0.86 (0.26 to 2.83) | 1.05 (0.33 to 3.34) | 0.81 (0.27 to 2.47) | ||
| Recessive | A/C-A/A | 39 (100%) | 52 (98.1%) | 37 (97.4%) | 0.75 | Reference | ||
| C/C | 0 (0.0%) | 1 (1.9%) | 1 (2.6%) | 3.16 (0.12 to 80.02) | 1.41 (0.09 to 23.20) | 2.26 (0.09 to 56.90) | ||
| Over-dominant | A/A-C/C | 32 (82.1%) | 46 (86.8%) | 33 (86.8%) | 0.78 | Reference | ||
| A/C | 7 (17.9%) | 7 (13.2%) | 5 (13.2%) | 0.69 (0.20 to 2.41) | 1.00 (0.29 to 3.41) | 0.70 (0.22 to 2.18) | ||
| rs1800450 | ||||||||
| Codominant | A/A | 11 (28.2%) | 15 (28.3%) | 14 (36.8%) | 0.63 | Reference | ||
| A/B | 28 (71.8%) | 38 (71.7%) | 24 (63.2%) | 0.68 (0.28 to 1.65) | 0.67 (0.26 to 1.76) | 1.00 (0.40 to 2.49) | ||
| B/B | 0 | 0 | 0 | 0.79 (0.01 to 43.12) | 1.07 (0.02 to 57.49) | 0.74 (0.01 to 40.25) | ||
| Dominant | A/A | 11 (28.2%) | 15 (28.3%) | 14 (36.8%) | 0.63 | Reference | ||
| A/B-B/B | 28 (0.0%) | 38 (0.0%) | 24 (0.0%) | 0.67 (0.26 to 1.76) | 0.68 (0.28 to 1.65) | 1.00 (0.40 to 2.49) | ||
| Recessive | A/A-A/B | 39 (100%) | 53 (100%) | 38 (100%) | 1.00 | Reference | ||
| B/B | 0 | 0 | 0 | 1.03 (0.02 to 53.02) | 1.39 (0.03 to 71.58) | 0.74 (0.01 to 38.02) | ||
| Over-dominant | A/A-B/B | 11 (28.2%) | 15 (28.3%) | 14 (36.8%) | 0.63 | Reference | ||
| A/B | 28 (0.0%) | 38 (0.0%) | 24 (0.0%) | 0.67 (0.26 to 1.76) | 0.68 (0.28 to 1.65) | 1.00 (0.40 to 2.49) | ||
| Variables | Codon 54 (rs1800450) | Codon 57 (rs1800451) | |
| p-Value | p-Value | ||
| Demographic | Age, years | 0.81 | 0.61 |
| Sex | 0.88 | 0.90 | |
| Vital signs | HR, beats/min | 0.36 | 0.47 |
| MAP, mm Hg | 0.81 | 0.45 | |
| SBP, mm Hg | 0.42 | 0.49 | |
| DBP, mm Hg | 0.71 | 0.39 | |
| Concomitant diseases | Diabetes | 0.65 | 0.44 |
| Hypertension | 0.70 | 1.00 | |
| Vascular disease | 0.27 | 0.44 | |
| Chronic lung disease | 0.25 | 1.00 | |
| Chronic liver disease | 1.00 | 1.00 | |
| Chronic renal disease | 0.53 | 1.00 | |
| ICU assessment | APACHE score | 0.75 | 0.80 |
| Glasgow scale | 0.10 | 0.50 | |
| Length of stay | 0.22 | 0.24 | |
| Sepsis | 0.63 | 0.91 | |
| Septic shock | 0.78 | 0.76 | |
| Death | 0.84 | 0.17 | |
| Overall survival | 0.97 | 0.40 | |
| Admission category (cause of admission) | Renal | 0.59 | 1.00 |
| Cardiovascular | 1.00 | 0.59 | |
| Infection | 0.24 | 0.84 | |
| Neurology | 0.19 | 0.69 | |
| Post-surgical | 0.54 | 0.80 | |
| Respiratory | 0.011 | 0.38 | |
| Trauma | 0.72 | 1.00 | |
| Other causes | 1.00 | 0.11 | |
| Gastrointestinal | 1.00 | 0.67 | |
| Laboratory results | WBC, ×103 cells/μL | 0.46 | 0.58 |
| HB, g% | 0.35 | 0.83 | |
| Creatinine, mg/dL | 0.55 | 0.74 | |
| Variables | Codon 54 (rs1800450) | Codon 57 (rs1800451) | |
| p-Value | p-Value | ||
| Causative organism | Enterobacter spp. | 0.50 | 1.00 |
| Acinetobacter spp. | 0.74 | 1.00 | |
| Candida spp. | 1.00 | 0.41 | |
| E. coli | 1.00 | 0.43 | |
| Gram-negative bacilli | 1.00 | 1.00 | |
| Klebsiella spp. | 0.81 | 0.17 | |
| Pseudomonas spp. | 1.00 | 0.75 | |
| Staph spp. | 0.42 | 0.30 | |
| Streptococcus spp. | 0.59 | 0.07 | |
| Aeromonas spp. | 1.00 | 1.00 | |
| Proteus spp. | 0.52 | 1.00 | |
| Citrobacter spp. | 1.00 | 1.00 | |
| Serratia spp. | 1.00 | 1.00 | |
| Type of culture | Blood | 0.40 | 0.18 |
| Sputum | 0.70 | 0.29 | |
| Urine | 0.54 | 1.00 | |
| Pus | 0.19 | 1.00 | |
| CSF | 0.31 | 1.00 | |
| No. of infections | 0.48 | 0.31 | |
| Variables | Overall Comparisons | |||
|---|---|---|---|---|
| Log Rank | Breslow | Tarone–Ware | ||
| Demographic data | Age | 0.154 | 0.247 | 0.180 |
| Sex | 0.701 | 0.542 | 0.582 | |
| Concomitant disease | Diabetes | 0.820 | 0.401 | 0.543 |
| Hypertension | 0.536 | 0.377 | 0.418 | |
| Vascular disease | 0.141 | 0.361 | 0.271 | |
| Chronic liver disease | 0.891 | 0.979 | 0.891 | |
| Chronic lung disease | 0.026 | 0.064 | 0.044 | |
| Chronic renal disease | 0.124 | 0.126 | 0.122 | |
| ICU assessment | APACHE score | 0.308 | 0.261 | 0.256 |
| Glasgow scale | 0.115 | 0.228 | 0.175 | |
| Length of stay | 0.000 | 0.000 | 0.000 | |
| Sepsis | 0.807 | 0.797 | 0.793 | |
| Septic shock | 0.090 | 0.020 | 0.038 | |
| Admission category | Renal | 0.571 | 0.396 | 0.442 |
| Cardiovascular | 0.954 | 0.687 | 0.766 | |
| Infection | 0.018 | 0.011 | 0.014 | |
| Neurology | 0.030 | 0.004 | 0.007 | |
| Post-surgical | 0.273 | 0.279 | 0.264 | |
| Respiratory | 0.454 | 0.865 | 0.672 | |
| Trauma | 0.111 | 0.200 | 0.155 | |
| Other causes | 0.308 | 0.357 | 0.314 | |
| Gastrointestinal | 0.078 | 0.240 | 0.128 | |
| Laboratory results | WBC, x103 cells/μl | 0.165 | 0.062 | 0.080 |
| HB, g% | 0.066 | 0.218 | 0.140 | |
| Creatinine, mg/dL | 0.064 | 0.144 | 0.117 | |
| No. of infections | 0.149 | 0.050 | 0.048 | |
| Molecular analysis | RS1800450 | 0.336 | 0.728 | 0.548 |
| RS1800451 | 0.116 | 0.093 | 0.102 | |
| Variables | HR | 95% CI | p-Value | |
|---|---|---|---|---|
| Demographic data | Age | 1.018 | 1.004–1.034 | 0.015 |
| Sex | 1.262 | 0.753–2.117 | 0.377 | |
| ICU assessment | APACHE core | 1.003 | 0.970–1.036 | 0.866 |
| Glasgow scale | 1.056 | 0.988–1.129 | 0.107 | |
| Septic shock | 2.882 | 1.130–7.347 | 0.027 | |
| Sepsis (sepsis–no sepsis) | 0.455 | 0.191–1.084 | 0.075 | |
| No. of infections | 0.738 | 0.541–1.006 | 0.055 | |
| Molecular analysis | RS1800451 (AA, AC + CC) | 1.599 | 0.742–3.444 | 0.231 |
| RS1800450 (AA, AB + BB) | 1.108 | 0.632–1.940 | 0.720 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Behairy, M.Y.; Abdelrahman, A.A.; Abdallah, H.Y.; Ibrahim, E.E.-D.A.; Hashem, H.R.; Sayed, A.A.; Azab, M.M. Role of MBL2 Polymorphisms in Sepsis and Survival: A Pilot Study and In Silico Analysis. Diagnostics 2022, 12, 460. https://doi.org/10.3390/diagnostics12020460
Behairy MY, Abdelrahman AA, Abdallah HY, Ibrahim EE-DA, Hashem HR, Sayed AA, Azab MM. Role of MBL2 Polymorphisms in Sepsis and Survival: A Pilot Study and In Silico Analysis. Diagnostics. 2022; 12(2):460. https://doi.org/10.3390/diagnostics12020460
Chicago/Turabian StyleBehairy, Mohammed Y., Ali A. Abdelrahman, Hoda Y. Abdallah, Emad El-Deen A. Ibrahim, Hany R. Hashem, Anwar A. Sayed, and Marwa M. Azab. 2022. "Role of MBL2 Polymorphisms in Sepsis and Survival: A Pilot Study and In Silico Analysis" Diagnostics 12, no. 2: 460. https://doi.org/10.3390/diagnostics12020460
APA StyleBehairy, M. Y., Abdelrahman, A. A., Abdallah, H. Y., Ibrahim, E. E.-D. A., Hashem, H. R., Sayed, A. A., & Azab, M. M. (2022). Role of MBL2 Polymorphisms in Sepsis and Survival: A Pilot Study and In Silico Analysis. Diagnostics, 12(2), 460. https://doi.org/10.3390/diagnostics12020460






