AUE-Net: Automated Generation of Ultrasound Elastography Using Generative Adversarial Network
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genaraitve Adversarial Networks and Pix2pix
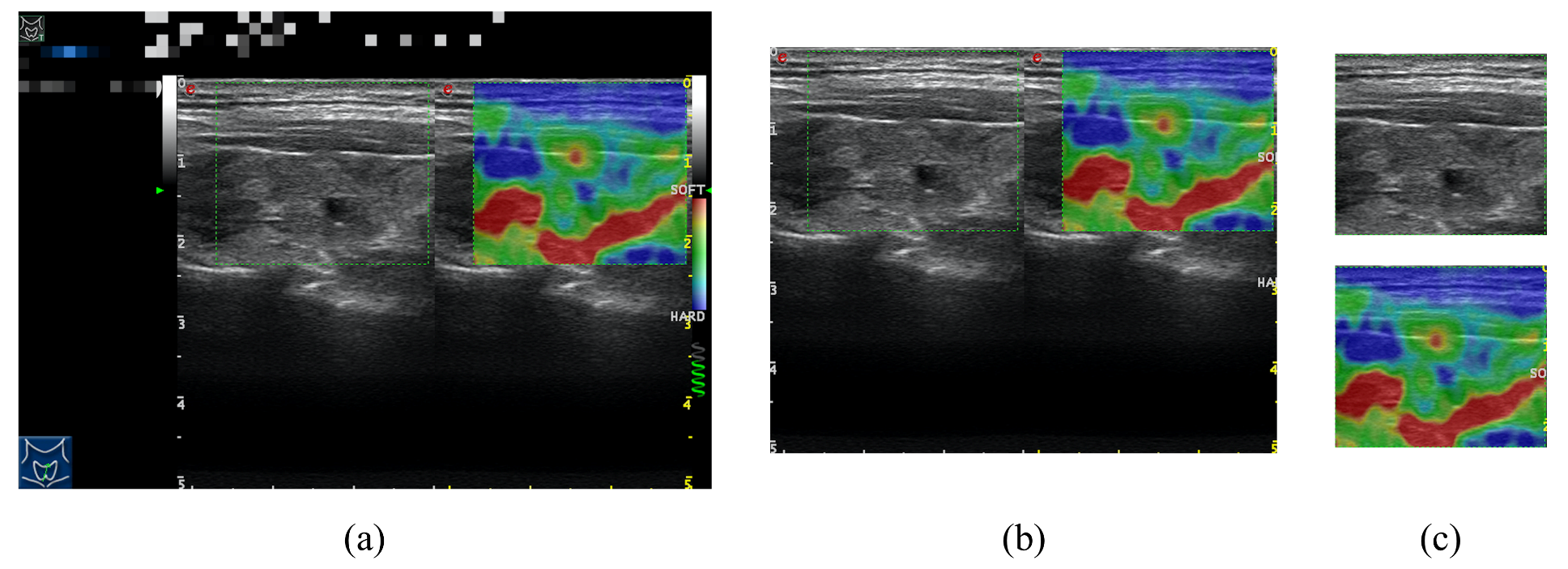
2.2. Datasets
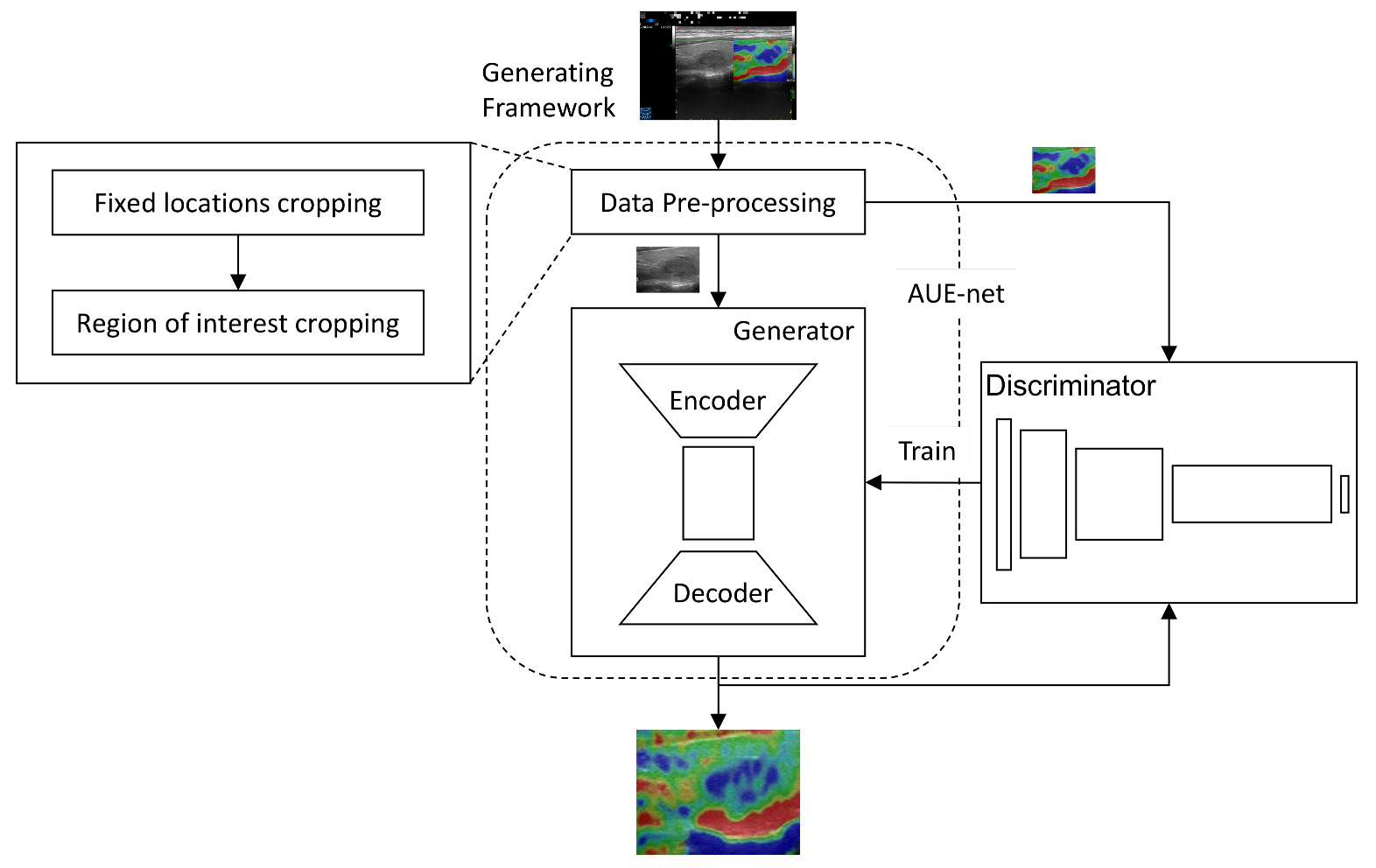
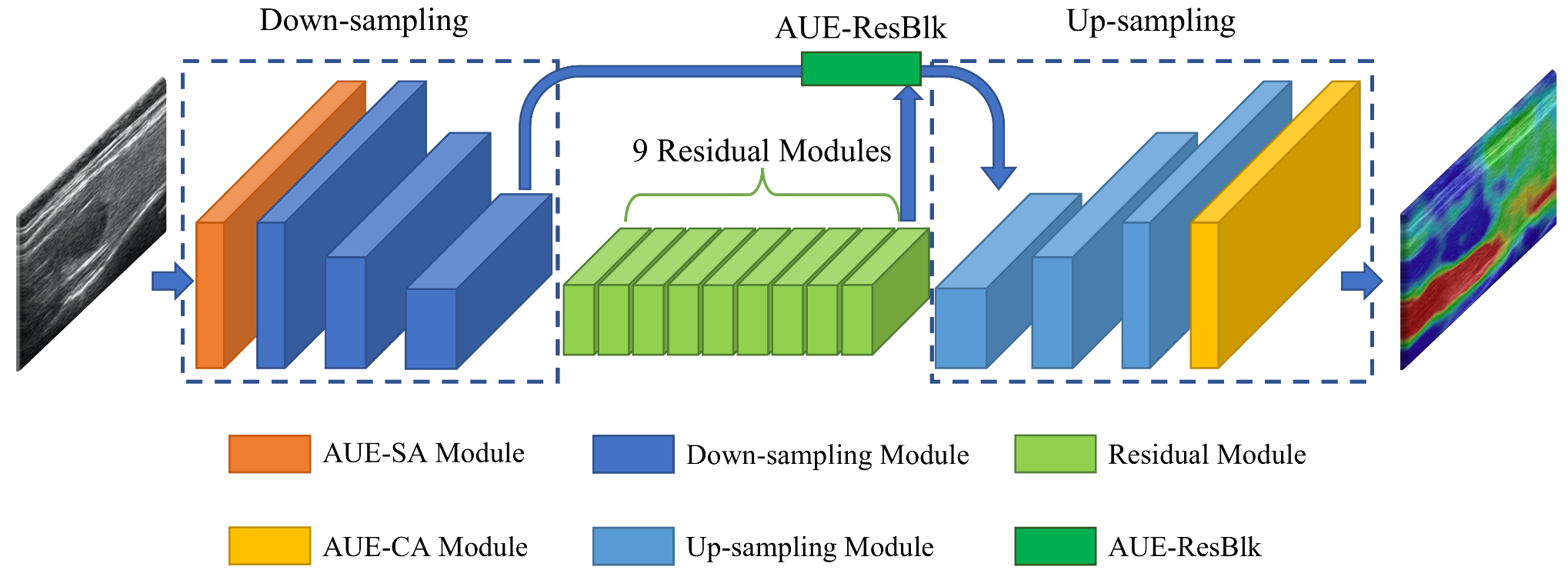
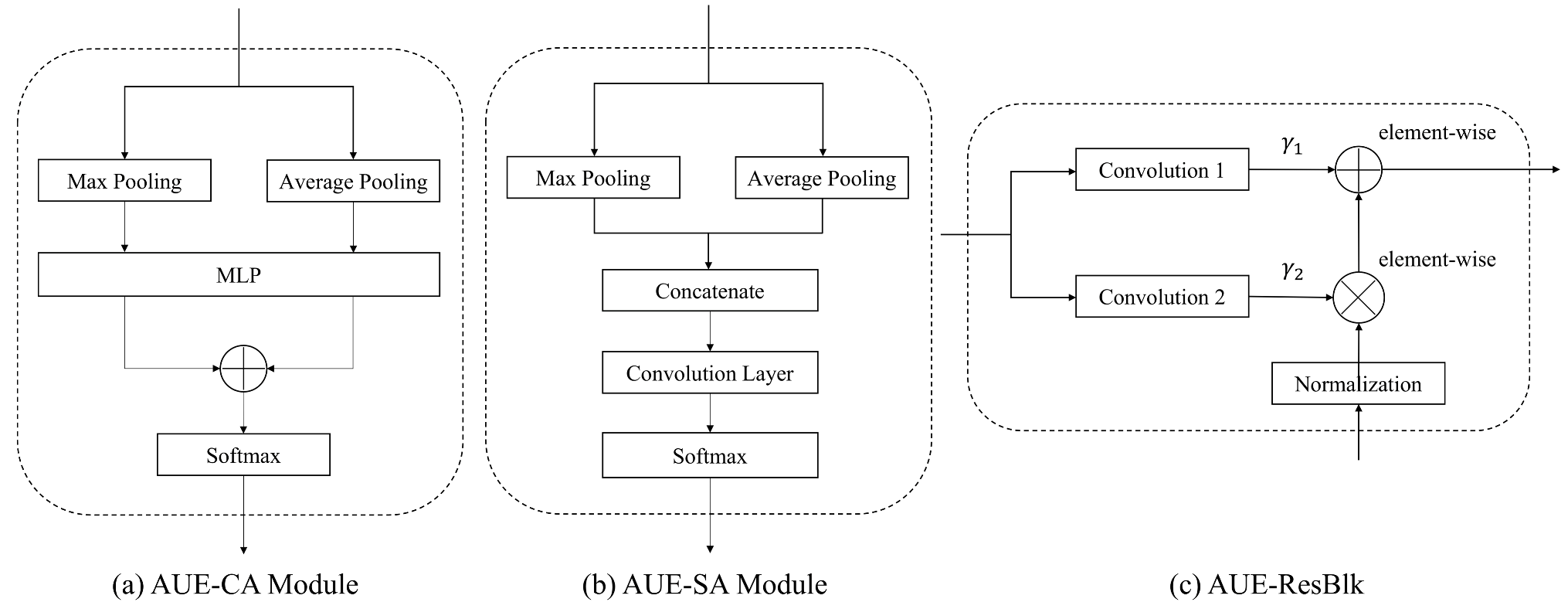
2.3. Network Architecture
2.4. Implementation Details
2.5. Evaluation Metrics
3. Results
3.1. Visual Comparisons
3.2. Quantitative Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| STIR | Short Tau Inversion Recovery |
| GAN | Generative adversarial network |
| ROI | regions of interest |
| PSNR | Peak signal-to-noise ratio |
| FNAC | Fine-needle aspiration cytology |
| SE | Strain elastography |
| I2IT | Image-to-image translation |
| SSIM | Structural similarity index measure |
| MSE | Mean-square error |
| FID | Frechet Inception distance |
References
- Wilhelm, S. Evaluation of thyroid incidentaloma. Surg. Clin. N. Am. 2014, 94, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.F.; Patel, N.J.; Caserta, M.P.; Robbin, M.L. Thyroid ultrasound: Diffuse and nodular disease. Radiol. Clin. 2020, 58, 1041–1057. [Google Scholar] [CrossRef] [PubMed]
- Barr, R.G. Shear wave liver elastography. Abdom. Radiol. 2018, 43, 800–807. [Google Scholar] [CrossRef] [PubMed]
- Barr, R.G.; Ferraioli, G.; Palmeri, M.L.; Goodman, Z.D.; Garcia-Tsao, G.; Rubin, J.; Garra, B.; Myers, R.P.; Wilson, S.R.; Rubens, D. Elastography assessment of liver fibrosis: Society of radiologists in ultrasound consensus conference statement. Radiology 2015, 276, 845–861. [Google Scholar] [CrossRef] [Green Version]
- Correas, J.M.; Tissier, A.M.; Khairoune, A.; Khoury, G.; Eiss, D.; Hélénon, O. Ultrasound elastography of the prostate: State of the art. Diagn. Interv. Imaging 2013, 94, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Onur, M.R.; Poyraz, A.K.; Bozgeyik, Z.; Onur, A.R.; Orhan, I. Utility of semiquantitative strain elastography for differentiation between benign and malignant solid renal masses. J. Ultrasound Med. 2015, 34, 639–647. [Google Scholar] [CrossRef]
- Zhao, C.K.; Xu, H.X. Ultrasound elastography of the thyroid: Principles and current status. Ultrasonography 2019, 38, 106. [Google Scholar] [CrossRef]
- Sigrist, R.M.; Liau, J.; El Kaffas, A.; Chammas, M.C.; Willmann, J.K. Ultrasound elastography: Review of techniques and clinical applications. Theranostics 2017, 7, 1303. [Google Scholar] [CrossRef]
- Soni, M.; Gomathi, S.; Kumar, P.; Churi, P.P.; Mohammed, M.A.; Salman, A.O. Hybridizing Convolutional Neural Network for Classification of Lung Diseases. Int. J. Swarm Intell. Res. (IJSIR) 2022, 13, 1–15. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Al-Khateeb, B.; Rashid, A.N.; Ibrahim, D.A.; Abd Ghani, M.K.; Mostafa, S.A. Neural network and multi-fractal dimension features for breast cancer classification from ultrasound images. Comput. Electr. Eng. 2018, 70, 871–882. [Google Scholar] [CrossRef]
- Hussein, I.J.; Burhanuddin, M.A.; Mohammed, M.A.; Benameur, N.; Maashi, M.S.; Maashi, M.S. Fully-automatic identification of gynaecological abnormality using a new adaptive frequency filter and histogram of oriented gradients (HOG). Expert Syst. 2021, e12789. [Google Scholar] [CrossRef]
- Hussein, I.J.; Burhanuddin, M.; Mohammed, M.A.; Elhoseny, M.; Garcia-Zapirain, B.; Maashi, M.S.; Maashi, M.S. Fully Automatic Segmentation of Gynaecological Abnormality Using a New Viola–Jones Model. Comput. Mater. Contin. 2021, 66, 3161–3182. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014, 27. [Google Scholar] [CrossRef]
- Wang, T.C.; Liu, M.Y.; Zhu, J.Y.; Tao, A.; Kautz, J.; Catanzaro, B. High-resolution image synthesis and semantic manipulation with conditional gans. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 8798–8807. [Google Scholar]
- Karras, T.; Laine, S.; Aila, T. A style-based generator architecture for generative adversarial networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 4401–4410. [Google Scholar]
- Haubold, J.; Demircioglu, A.; Theysohn, J.M.; Wetter, A.; Radbruch, A.; Dörner, N.; Schlosser, T.W.; Deuschl, C.; Li, Y.; Nassenstein, K.; et al. Generating Virtual Short Tau Inversion Recovery (STIR) Images from T1-and T2-Weighted Images Using a Conditional Generative Adversarial Network in Spine Imaging. Diagnostics 2021, 11, 1542. [Google Scholar] [CrossRef]
- Gomi, T.; Sakai, R.; Hara, H.; Watanabe, Y.; Mizukami, S. Usefulness of a Metal Artifact Reduction Algorithm in Digital Tomosynthesis Using a Combination of Hybrid Generative Adversarial Networks. Diagnostics 2021, 11, 1629. [Google Scholar] [CrossRef]
- Bermudez, C.; Plassard, A.J.; Davis, L.T.; Newton, A.T.; Resnick, S.M.; Landman, B.A. Learning implicit brain MRI manifolds with deep learning. In Proceedings of the SPIE—Medical Imaging 2018: Image Processing, Houston, TX, USA, 2 March 2018; Volume 10574, p. 105741L. [Google Scholar]
- Nishiyama, D.; Iwasaki, H.; Taniguchi, T.; Fukui, D.; Yamanaka, M.; Harada, T.; Yamada, H. Deep generative models for automated muscle segmentation in computed tomography scanning. PLoS ONE 2021, 16, e0257371. [Google Scholar] [CrossRef]
- Wu, S.; Gao, Z.; Liu, Z.; Luo, J.; Zhang, H.; Li, S. Direct reconstruction of ultrasound elastography using an end-to-end deep neural network. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 374–382. [Google Scholar]
- Kibria, M.G.; Rivaz, H. Gluenet: Ultrasound elastography using convolutional neural network. In Simulation, Image Processing, and Ultrasound Systems for Assisted Diagnosis and Navigation; Springer: Berlin/Heidelberg, Germany, 2018; pp. 21–28. [Google Scholar]
- Wildeboer, R.R.; van Sloun, R.J.; Mannaerts, C.K.; Moraes, P.; Salomon, G.; Chammas, M.; Wijkstra, H.; Mischi, M. Synthetic Elastography Using B-Mode Ultrasound through a Deep Fully Convolutional Neural Network. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2020, 67, 2640–2648. [Google Scholar] [CrossRef]
- Shin, H.C.; Tenenholtz, N.A.; Rogers, J.K.; Schwarz, C.G.; Senjem, M.L.; Gunter, J.L.; Andriole, K.P.; Michalski, M. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In International Workshop on Simulation and Synthesis in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–11. [Google Scholar]
- Johnson, J.; Alahi, A.; Fei-Fei, L. Perceptual losses for real-time style transfer and super-resolution. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 8–16 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 694–711. [Google Scholar]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Park, T.; Liu, M.Y.; Wang, T.C.; Zhu, J.Y. Semantic image synthesis with spatially-adaptive normalization. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 2337–2346. [Google Scholar]
- Liang, J.; Zeng, H.; Zhang, L. High-Resolution Photorealistic Image Translation in Real-Time: A Laplacian Pyramid Translation Network. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 19–25 June 2021; pp. 9392–9400. [Google Scholar]
- Ulyanov, D.; Vedaldi, A.; Lempitsky, V. Instance normalization: The missing ingredient for fast stylization. arXiv 2016, arXiv:1607.08022. [Google Scholar]
- Wang, S.Y.; Bau, D.; Zhu, J.Y. Sketch Your Own GAN. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Nashville, TN, USA, 19–25 June 2021; pp. 14050–14060. [Google Scholar]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein generative adversarial networks. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 214–223. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Regmi, K.; Borji, A. Cross-view image synthesis using conditional gans. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 3501–3510. [Google Scholar]
- Odena, A.; Olah, C.; Shlens, J. Conditional image synthesis with auxiliary classifier gans. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 2642–2651. [Google Scholar]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. Gans trained by a two time-scale update rule converge to a local nash equilibrium. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar] [CrossRef]
- Zhang, R.; Isola, P.; Efros, A.A.; Shechtman, E.; Wang, O. The unreasonable effectiveness of deep features as a perceptual metric. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 586–595. [Google Scholar]
- Rago, T.; Vitti, P. Potential value of elastosonography in the diagnosis of malignancy in thyroid nodules. Q. J. Nucl. Med. Mol. Imaging 2009, 53, 455. [Google Scholar]
- Ueno, E.; Ito, A. Diagnosis of breast cancer by elasticity imaging. Eizo Joho Med. 2004, 36, 2–6. [Google Scholar]
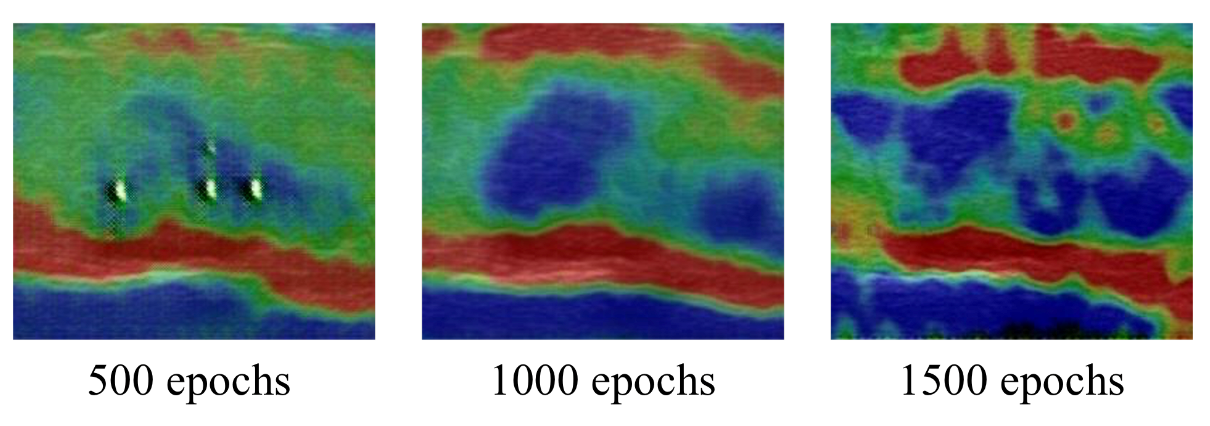
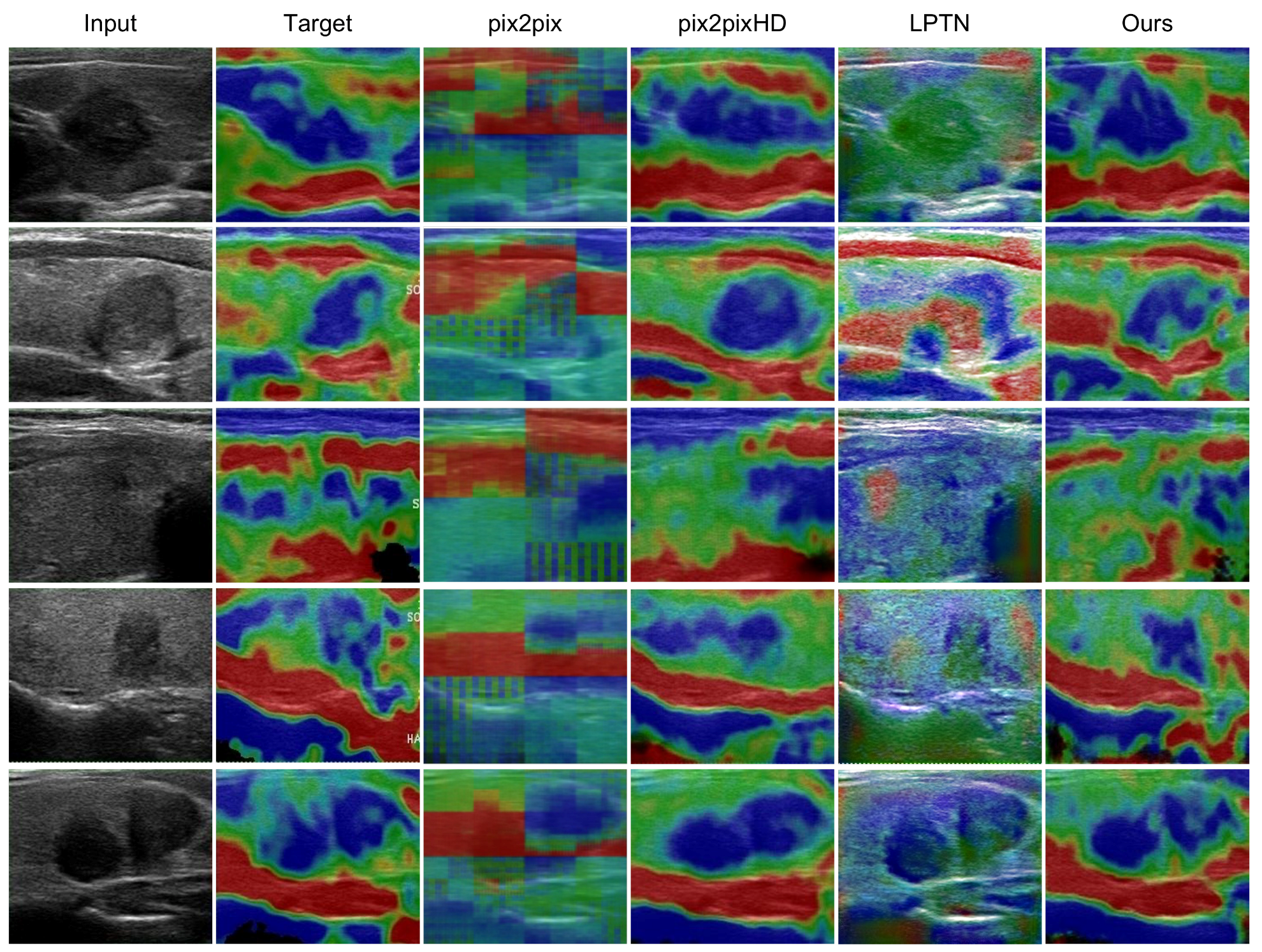
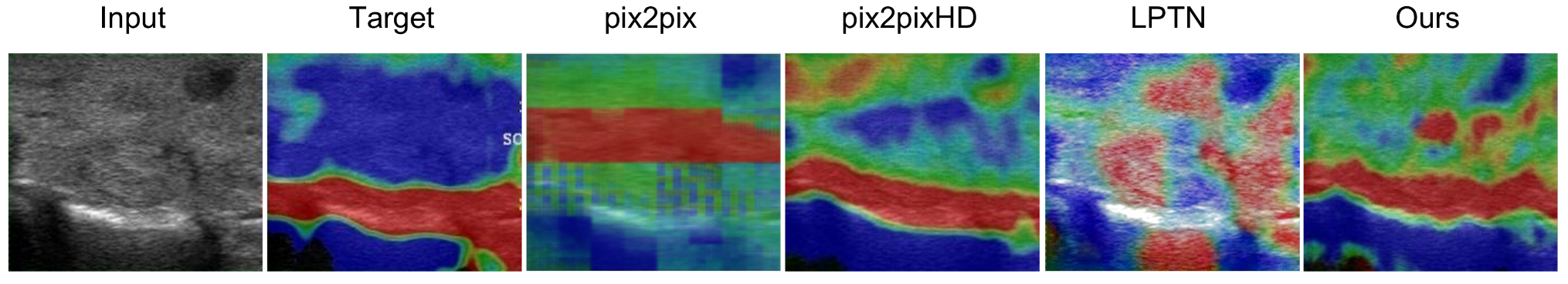
| Items | Information |
|---|---|
| Operating System | Linux Red Hat 4.8.5-36 |
| CPU | IBM(R) POWER9 (3.8 GHz) |
| GPU | NVIDIA Tesla V100 32 G |
| Methods | Year | Highlights |
|---|---|---|
| pix2pix [25] | 2017 | General-purpose solution to image-to-image translation based on cGAN |
| pix2pixHD [14] | 2018 | Synthesizing high-resolution photo-realistic images A novel adversarial loss, new multiscale generator, and discriminator architectures |
| LPTN [27] | 2021 | Laplacian pyramid decomposition and reconstruction Speeding-up the high-resolution photo-realistic I2IT tasks |
| AUE-net | 2021 | AUE attention modules, AUE residual blocks and color loss Generating strain elastography images from conventional ultrasound images |
| Methods | PSNR ↑ | SSIM ↑ | FID ↓ | Score Acc ↑ |
|---|---|---|---|---|
| pix2pix [25] | 12.688 | 0.281 | 205.38 | 30.8% |
| pix2pixHD [14] | 28.651 | 0.456 | 56.71 | 75.4% |
| LPTN [27] | 10.879 | 0.364 | 155.24 | 32.3% |
| Ours | 28.736 | 0.499 | 51.09 | 84.4% |
| Correct Amount of 80 ↑ | Score Accuracy ↑ | |
|---|---|---|
| Specialist 1 | 67 | 83.75% |
| Specialist 2 | 68 | 85.00% |
| Mean | 67.5 | 84.38% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Zhao, J.; Long, X.; Luo, Q.; Wang, R.; Ding, X.; Shen, C. AUE-Net: Automated Generation of Ultrasound Elastography Using Generative Adversarial Network. Diagnostics 2022, 12, 253. https://doi.org/10.3390/diagnostics12020253
Zhang Q, Zhao J, Long X, Luo Q, Wang R, Ding X, Shen C. AUE-Net: Automated Generation of Ultrasound Elastography Using Generative Adversarial Network. Diagnostics. 2022; 12(2):253. https://doi.org/10.3390/diagnostics12020253
Chicago/Turabian StyleZhang, Qingjie, Junjuan Zhao, Xiangmeng Long, Quanyong Luo, Ren Wang, Xuehai Ding, and Chentian Shen. 2022. "AUE-Net: Automated Generation of Ultrasound Elastography Using Generative Adversarial Network" Diagnostics 12, no. 2: 253. https://doi.org/10.3390/diagnostics12020253
APA StyleZhang, Q., Zhao, J., Long, X., Luo, Q., Wang, R., Ding, X., & Shen, C. (2022). AUE-Net: Automated Generation of Ultrasound Elastography Using Generative Adversarial Network. Diagnostics, 12(2), 253. https://doi.org/10.3390/diagnostics12020253






