Ensemble Model for Diagnostic Classification of Alzheimer’s Disease Based on Brain Anatomical Magnetic Resonance Imaging
Abstract
:1. Introduction
- This study evaluated the performance of various machine learning techniques, i.e., 11 models which lack in the state-of-the-art literature.
- To enhance the diagnostic performance of AD, this article presents an ensemble technique formed by combining SVM kernel master demonstration, along with extreme gradient boosting transitioning sequential model to concur the hidden difficult extractions in the dataset. The proposed architecture (XGB + DT + SVM) uses a Decision Tree to reduce invariability in model demonstration and performance. Overall, the proposed model not only decreases inheriting variance but enhances tough sample prediction using boosting.
- This article performed a comparative analysis of algorithms before and after hyper parameter tuning accomplished through Grid search and the outcomes are validated using k-fold cross-validation.
- To ensure the generalization of the considered models, this article evaluated the efficiency of the same using statistical test, i.e., Friedman’s rank test and t-test.
- Various performance metrics such as receiver operating characteristics (ROC) curve, sensitivity, specificity and log loss are used to compare the proposed model with other ML models.
- This article also performed a comparison of the proposed model with existing state-of-the-art studies to prove its efficiency in detecting AD.
- The article also focuses on the future scope that should be addressed for reliable AD diagnosis.
2. Related Work
3. Materials and Methods
3.1. Framework
3.2. Dataset Description
3.2.1. Pre-Processing Steps
- (a)
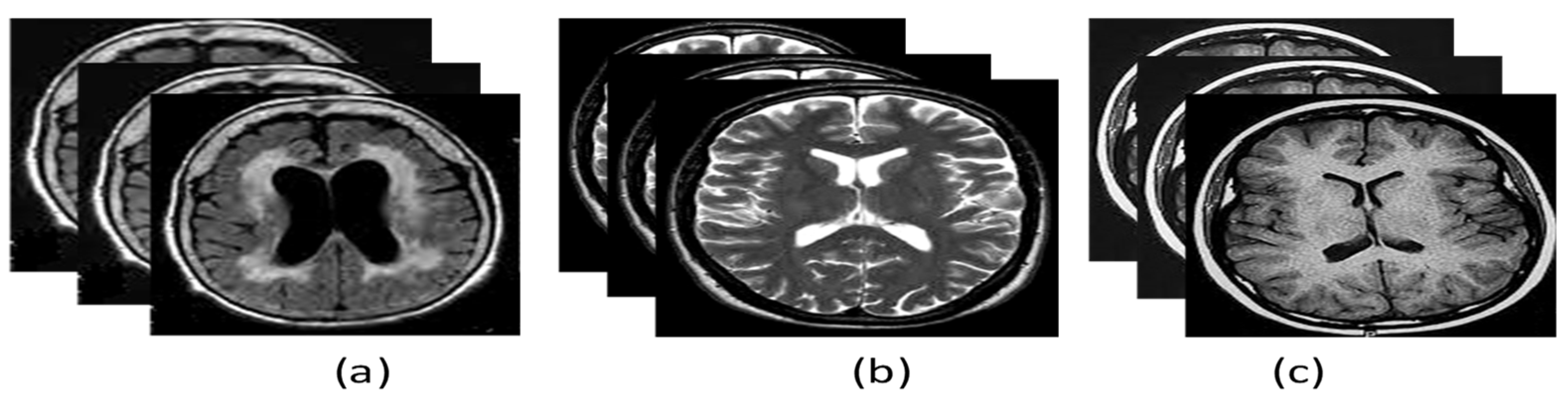
- Data filtering is performed based on the acquisition plane and ADNI phase. We selected the axial acquisition plan of the MR (T1 and T2 weighted) images and ADNI phase 3 for three classes: AD, CN, and MCI as shown in Figure 3.
- (b)
- The images are reshaped to 512 × 512 to provide the best quality pixel.
- (c)
- The labels about the classes of the images are extracted and put into the vector storage. The classes undergo label encoding, i.e., 0 for AD, 1 for MCI and 2 for CN.
- (d)
- The loaded images are standardized and normalized using a standard scaler and min-max scaler as the normalization functions.
- (e)
- Once the preprocessing of the data is completed, the data are split into training and testing to perform validation on unseen data.
- Data: Brain axial T1 and T2 weighted MRI data taken from the ADNI database is labelled into three classes such that AD CN, and MCI along with the preprocessing and data loading pipelines.
- Data training: using VGG-16 [43] as a feature extractor primarily using the illustrations provided by the Imagenet on the ADNI dataset. After Feature Extraction using VGG-16, the extracted blob is then processed and sent to conventional machine learning models and proposed ensemble architecture.
- Parameter optimization: post-training hyperparameter tuning is achieved using GridSearch.
- Data Evaluation: after rigorous training, the models are tested and compared for overfitting. If overfitting is found, then retraining is done with reduced hyper-planes and data sampling. Else, a comparative analysis of the result is evaluated.
3.2.2. Feature Extraction
3.3. Classification Models
3.3.1. Naive Bayes (NB)
3.3.2. Decision Tree (DT)
3.3.3. Random Forest (RF)
3.3.4. K-Nearest Neighbor (K-NN)
3.3.5. Support Vector Machine (RBF Kernel)
3.3.6. Support Vector Machine (Polynomial Kernel)
3.3.7. Support Vector Machine (Sigmoid Kernel)
3.3.8. Gradient Boost (GB)
3.3.9. Extreme Gradient Boosting (XGB)
3.3.10. Multi-Layer Perceptron Neural Network (MLP-NN)
3.4. Proposed Ensemble Learning Approach: XGB + DT + SVM
3.5. Grid Search-Based Optimization on ML Methods and Ensemble Technique
4. Experimental Analysis
4.1. Implementation Setup
4.2. Performance Evaluation Metrics
- (a)
- Accuracy: Classification models can be evaluated based on their accuracy. The fraction of correct predictions made by the model divided by all predictions is referred to as its accuracy as shown in Equation (16).
- (b)
- Sensitivity: The value is represented numerically by dividing the number of correct positive predictions by the total number of positive values, as given in Equation (17).
- (c)
- Specificity: The value is mathematically represented by dividing the number of successful negative predictions by the total number of negative values. Sensitivity and specificity are statistical indicators of test result performance. The specificity is computed using the formula given in Equation (18).
- (d)
- Logarithmic loss (log loss): Using a log loss parameter, classification models that yield a probability value between zero and one may be evaluated for their efficiency. Increasing the logarithmic loss value is associated with increasing the anticipated probability value. The log loss action considers the confidence of the prediction in determining how incorrect classifications can be penalized [59]. The mathematical model used for computing logarithmic loss is shown in Equation (19).
5. Results
5.1. Parameter Optimization Results
5.2. Classification Results
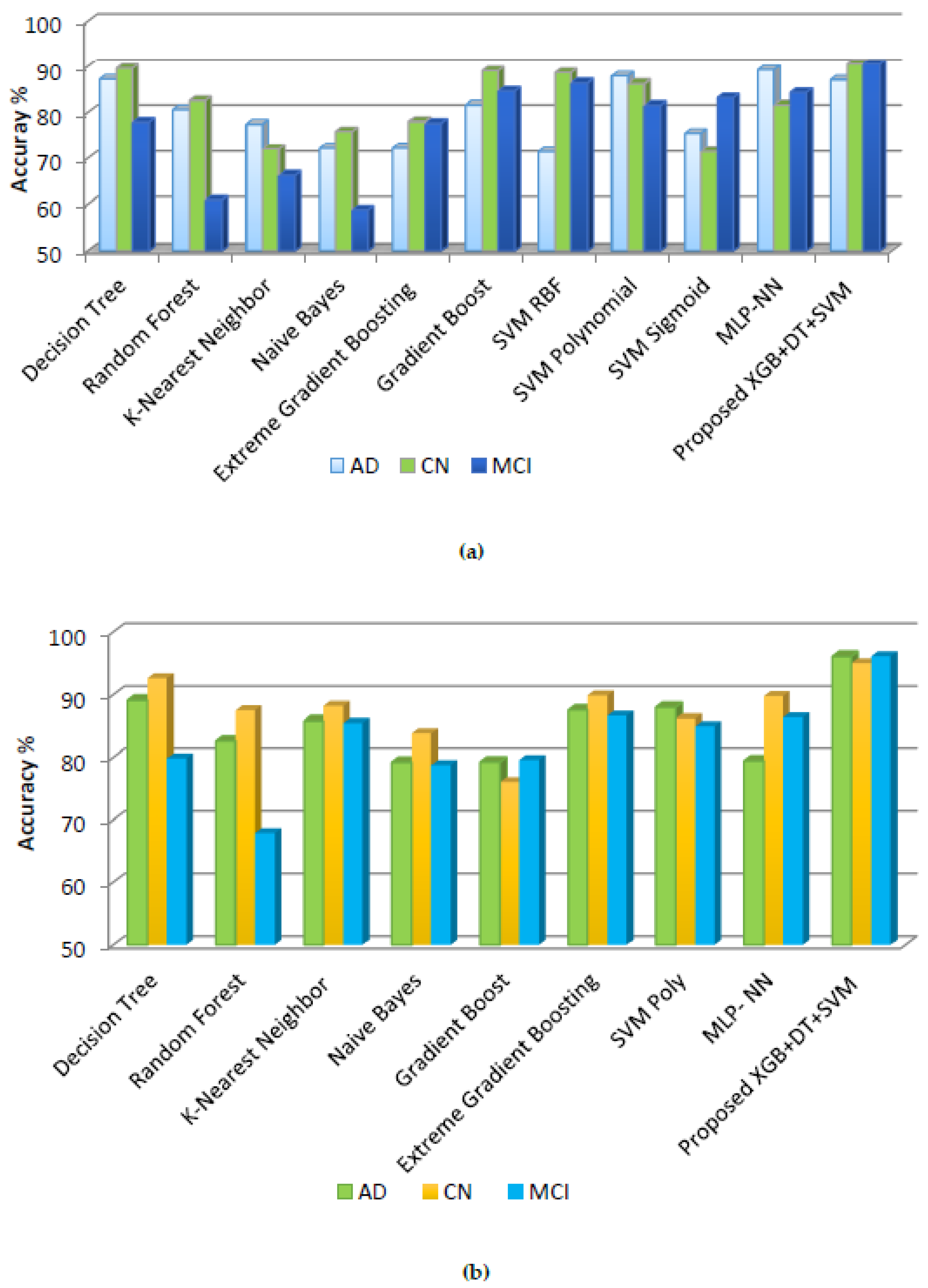
5.2.1. Before Hyperparameter Tuning
5.2.2. After Hyperparameter Tuning
5.3. Statistical Evaluation of Outcomes
5.3.1. Ten-Fold Cross-Validation
5.3.2. Friedman’s Rank Test
5.3.3. T-Test
5.4. ROC Demonstration of Models Classification Abilities
5.5. Comparison with Existing Studies
6. Conclusions
7. Future Scope
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Labels | Acronym | Labels | Acronym |
|---|---|---|---|
| AF | Activation functions | 8L-CNN | 8-layer convolution neural network |
| AIBL | Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing | MIRIAD | Minimal Interval Resonance Imaging in Alzheimer’s Disease dataset |
| LDMIL | landmark-based deep multi-instance learning | VBM | Voxel-based morphometry |
| CLM | Conventional landmark-based morphometry | MMSR | Multi-modal sparse representation |
| SPA | Sparse patch-based approach | DL Algo | Deep learning algorithm |
| FS | Fisher score | RVB | Regional Voxel-based method |
| SUVR | Standardized uptake value ratio | GGML | Graph-guided multitask-task learning |
| N-CK | N-number (combination of multi-kernels) | E-MK | Ensemble multi-kernel |
| RFE | Recursive feature elimination | OASIS | Open Access Series of Imaging Studies |
| RFES | Random forest-based ensemble strategy | GIM | Gini importance measure |
| lMRI | longitudinal-MRI | RFLD | Regression forest-based landmark detection model |
| AAL | Automated Anatomical Labeling | FW | Filter Wrapper method |
| N-CK | N-number (combination of multi-kernels) | DMTFS | Discriminative Multi-Task Feature Selection |
| FA | Fisher Algorithm | WA | Wrapper Algorithm |
| MK-SVM | Multi kernel- SVM | PM-Kernel | Probabilistic multi-kernel |
| p-MCI | Progressive-MCI | L-SVM | Lagrangian-SVM |
| s-MCI | Static-MCI | LR, RF | Logistic regression, Random Forest |
References
- Hussain, R.; Zubair, H.; Pursell, S.; Shahab, M. Neurodegenerative Diseases: Regenerative Mechanisms and Novel Therapeutic Approaches. Brain Sci. 2018, 8, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Liu, Y.; Shen, D. Graph-guided joint prediction of class label and clinical scores for the Alzheimer’s disease. Brain Struct. Funct. 2016, 221, 3787–3801. [Google Scholar] [CrossRef] [Green Version]
- Montie, H.L.; Durcan, T.M. The Cell and Molecular Biology of Neurodegenerative Diseases: An Overview. Front. Neurol. 2013, 4, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alzheimer’s Association. 2014 Alzheimer’s Disease Facts and Figures. Alzheimer’s Dement. 2014, 10, e47–e92. [Google Scholar]
- Alzheimer Association. Available online: https://www.alz.org/ (accessed on 5 June 2022).
- Alzheimer’s and Related Disorder Society of India. Available online: https://ardsi.org/ (accessed on 7 January 2022).
- Und Halbach, O.V.B.; Schober, A.; Krieglstein, K. Genes, proteins, and neurotoxins involved in Parkinson’s disease. Prog. Neurobiol. 2004, 73, 151–177. [Google Scholar] [CrossRef]
- Jones, D.T.; Townley, R.A.; Graff-Radford, J.; Botha, H.; Knopman, D.S.; Petersen, R.C.; Jack, C.R.; Lowe, V.J.; Boeve, B.F. Amyloid- and tau-PET imaging in a familial prion kindred. Neurol. Genet. 2018, 4, e290. [Google Scholar] [CrossRef] [Green Version]
- Nelson, T.J.; Sen, A. Apolipoprotein E particle size is increased in Alzheimer’s disease. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2019, 11, 10–18. [Google Scholar] [CrossRef]
- Saar, G.; Koretsky, A.P. Manganese Enhanced MRI for Use in Studying Neurodegenerative Diseases. Front. Neural Circuits 2019, 12, 114. [Google Scholar] [CrossRef] [Green Version]
- Tenreiro, S.; Eckermann, K.; Outeiro, T.F. Protein phosphorylation in neurodegeneration: Friend or foe? Front. Mol. Neurosci. 2014, 7, 42. [Google Scholar] [CrossRef] [Green Version]
- Gorman, A.M. Neuronal cell death in neurodegenerative diseases: Recurring themes around protein handling. J. Cell. Mol. Med. 2008, 12, 2263–2280. [Google Scholar] [CrossRef] [Green Version]
- Davatzikos, C.; Bhatt, P.; Shaw, L.M.; Batmanghelich, K.N.; Trojanowski, J.Q. Prediction of MCI to AD conversion via MRI, CSF biomarkers, and pattern classification. Neurobiol. Aging 2011, 32, 2322.e19–2322.e27. [Google Scholar] [CrossRef] [PubMed]
- Noble, W.; Hanger, D.P.; Miller, C.C.J.; Lovestone, S. The Importance of Tau Phosphorylation for Neurodegenerative Diseases. Front. Neurol. 2013, 4, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emard, J.-F.; Thouez, J.-P.; Gauvreau, D. Neurodegenerative diseases and risk factors: A literature review. Soc. Sci. Med. 1995, 40, 847–858. [Google Scholar] [CrossRef]
- Rathore, S.; Habes, M.; Iftikhar, M.A.; Shacklett, A.; Davatzikos, C. A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer’s disease and its prodromal stages. NeuroImage 2017, 155, 530–548. [Google Scholar] [CrossRef]
- Shimizu, S.; Hirose, D.; Hatanaka, H.; Takenoshita, N.; Kaneko, Y.; Ogawa, Y.; Sakurai, H.; Hanyu, H. Role of Neuroimaging as a Biomarker for Neurodegenerative Diseases. Front. Neurol. 2018, 9, 265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, C.; Song, W.; Chen, Q.; Zheng, Z.; Chen, K.; Cao, B.; Jang, J.; Li, J.; Huang, X.; Gong, Q.; et al. Reduced functional connectivity in early-stage drug-naive Parkinson’s disease: A resting-state fMRI study. Neurobiol. Aging 2014, 35, 431–441. [Google Scholar] [CrossRef]
- Yin, W.; Li, T.; Mucha, P.J.; Cohen, J.R.; Zhu, H.; Zhu, Z.; Lin, W. Altered neural flexibility in children with attention-deficit/hyperactivity disorder. Mol. Psychiatry 2022, 91, 945–955. [Google Scholar] [CrossRef]
- Xie, Y.; Xu, Z.; Xia, M.; Liu, J.; Shou, X.; Cui, Z.; Liao, X.; He, Y. Alterations in Connectome Dynamics in Autism Spectrum Disorder: A Harmonized Mega- and Meta-analysis Study Using the Autism Brain Imaging Data Exchange Dataset. Biol. Psychiatry 2021, 91, 945–955. [Google Scholar] [CrossRef]
- Casanova, R.; Whitlow, C.T.; Wagner, B.; Williamson, J.D.; Shumaker, S.A.; Maldjian, J.A.; Espeland, M.A. High Dimensional Classification of Structural MRI Alzheimer’s Disease Data Based on Large Scale Regularization. Front. Neuroinform. 2011, 5, 22. [Google Scholar] [CrossRef] [Green Version]
- Costafreda, S.G.; Dinov, I.; Tu, Z.; Shi, Y.; Liu, C.-Y.; Kloszewska, I.; Mecocci, P.; Soininen, H.; Tsolaki, M.; Vellas, B.; et al. Automated hippocampal shape analysis predicts the onset of dementia in mild cognitive impairment. NeuroImage 2011, 56, 212–219. [Google Scholar] [CrossRef] [Green Version]
- Battineni, G.; Chintalapudi, N.; Amenta, F.; Traini, E. A comprehensive machine-learning model applied to magnetic resonance imaging (mri) to predict alzheimer’s disease (ad) in older subjects. J. Clin. Med. 2020, 9, 2146. [Google Scholar] [CrossRef] [PubMed]
- Padilla, P.; López, M.; Górriz, J.M.; Ramirez, J.; Salas-Gonzalez, D.; Alvarez, I. NMF-SVM based CAD tool applied to functional brain images for the diagnosis of Alzheimer’s disease. IEEE Trans. Med. Imaging 2011, 31, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tosun, D.; Weiner, M.W.; Schuff, N. Alzheimer’s Disease Neuroimaging Initiative, Locally linear embedding (LLE) for MRI based Alzheimer’s disease classification. Neuroimage 2013, 83, 148–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, K.R.; Aljabar, P.; Heckemann, R.A.; Hammers, A.; Rueckert, D. Alzheimer’s Disease Neuroimaging Initiative, Random forest-based similarity measures for multi-modal classification of Alzheimer’s disease. NeuroImage 2013, 65, 167–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suk, H.I.; Shen, D. Deep Learning-Based Feature Representation for AD/MCI Classification. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Nagoya, Japan, 22–26 September 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 583–590. [Google Scholar]
- Dukart, J.; Mueller, K.; Barthel, H.; Villringer, A.; Sabri, O.; Schroeter, M.L. Alzheimer’s Disease Neuroimaging Initiative. Meta-analysis based SVM classification enables accurate detection of Alzheimer’s disease across different clinical centers using FDG-PET and MRI. Psychiatry Res. Neuroimaging 2013, 212, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, M.; Lan, W.; Wu, F.X.; Pan, Y.; Wang, J. Classification of Alzheimer’s disease using whole brain hierarchical network. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016, 15, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.; Zu, C.; Jie, B.; Shen, D.; Zhang, D. Discriminative multi-task feature selection for multi-modality classification of Alzheimer’s disease. Brain Imaging Behav. 2016, 10, 739–749. [Google Scholar] [CrossRef]
- Zu, C.; Initiative, T.A.D.N.; Jie, B.; Liu, M.; Chen, S.; Shen, D.; Zhang, D. Label-aligned multi-task feature learning for multimodal classification of Alzheimer’s disease and mild cognitive impairment. Brain Imaging Behav. 2015, 10, 1148–1159. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, L.; Igel, C.; Pai, A.; Balas, I.; Anker, C.; Lillholm, M.; Nielsen, M.; Alzheimer’s Disease Neuroimaging Initiative. Differential diagnosis of mild cognitive impairment and Alzheimer’s disease using structural MRI cortical thickness, hippocampal shape, hippocampal texture, and volumetry. NeuroImage Clin. 2017, 13, 470–482. [Google Scholar] [CrossRef]
- Liu, J.; Wang, J.; Tang, Z.; Hu, B.; Wu, F.X.; Pan, Y. Improving Alzheimer’s disease classification by combining multiple measures. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 15, 1649–1659. [Google Scholar] [CrossRef]
- Wang, S.-H.; Phillips, P.; Sui, Y.; Liu, B.; Yang, M.; Cheng, H. Classification of Alzheimer’s Disease Based on Eight-Layer Convolutional Neural Network with Leaky Rectified Linear Unit and Max Pooling. J. Med. Syst. 2018, 42, 85. [Google Scholar] [CrossRef]
- Purushotham, S.; Tripathy, B.K. Evaluation of Classifier Models Using Stratified Tenfold Cross Validation Techniques. In Proceedings of the International Conference on Computing and Communication Systems, Vellore, India, 9–11 September 2011; Springer: Berlin/Heidelberg, Germany, 2011; pp. 680–690. [Google Scholar]
- Ahmed, O.B.; Mizotin, M.; Benois-Pineau, J.; Allard, M.; Catheline, G.; Amar, C.B.; Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s disease diagnosis on structural MR images using circular harmonic functions descriptors on hippocampus and posterior cingulate cortex. Comput. Med. Imaging Graph. 2015, 44, 13–25. [Google Scholar] [CrossRef]
- Chincarini, A.; Bosco, P.; Calvini, P.; Gemme, G.; Esposito, M.; Olivieri, C.; Rei, L.; Squarcia, S.; Rodriguez, G.; Belotti, R.; et al. Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer’s disease. Neuroimage 2011, 58, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Salini, A.; Jeyapriya, U. A majority vote based ensemble classifier for predicting students academic performance. Int. J. Pure Appl. Math. 2018, 118, 24. [Google Scholar]
- Heidari, E.; Sobati, M.A.; Movahedirad, S. Accurate prediction of nanofluid viscosity using a multilayer perceptron artificial neural network (MLP-ANN). Chemom. Intell. Lab. Syst. 2016, 155, 73–85. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, D.; Shen, D.; Alzheimer’s Disease Neuroimaging Initiative. Ensemble sparse classification of Alzheimer’s disease. NeuroImage 2012, 60, 1106–1116. [Google Scholar] [CrossRef] [Green Version]
- Elish, M.O. Assessment of Voting Ensemble for Estimating Software Development Effort. In Proceedings of the 2013 IEEE Symposium on Computational Intelligence and Data Mining (CIDM), Singapore, 16–19 April 2013; pp. 316–321. [Google Scholar]
- Yang, S.; Berdine, G. The receiver operating characteristic (ROC) curve. Southwest Respir. Crit. Care Chron. 2017, 5, 34–36. [Google Scholar] [CrossRef]
- Rezende, E.; Ruppert, G.; Carvalho, T.; Theophilo, A.; Ramos, F.; Geus, P.D. Malicious Software Classification Using VGG16 Deep Neural Network’s Bottleneck Features. In Information Technology-New Generations; Springer: Cham, Switzerland, 2018; pp. 51–59. [Google Scholar]
- Berrar, D. Bayes’ Theorem and Naive Bayes Classifier. In Encyclopedia of Bioinformatics and Computational Biology: ABC of Bioinformatics; Elsevier Science Publisher: Amsterdam, The Netherlands, 2015; pp. 403–412. [Google Scholar]
- Xu, S.; Li, Y.; Wang, Z. Bayesian multinomial Naïve Bayes classifier to text classification. In Advanced Multimedia and Ubiquitous Engineering; Springer: Singapore, 2017; pp. 347–352. [Google Scholar]
- Brijain, M.; Patel, R.; Kushik, M.R.; Rana, K. A survey on decision tree algorithm for classification. Int. J. Eng. Dev. Res. 2014, 2, 1–5. [Google Scholar]
- Jaiswal, J.K.; Samikannu, R. Application of Random Forest Algorithm on Feature Subset Selection and Classification and Regression. In Proceedings of the 2017 World Congress on Computing and Communication Technologies (WCCCT), Tiruchirappalli, India, 2–4 February 2017; pp. 65–68. [Google Scholar]
- Zhang, S.; Li, X.; Zong, M.; Zhu, X.; Cheng, D. Learning k for knn classification. ACM Trans. Intell. Syst. Technol. 2017, 8, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Hamiane, M.; Saeed, F. SVM Classification of MRI Brain Images for Computer-Assisted Diagnosis. Int. J. Electr. Comput. Eng. 2017, 7, 2555. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Y.; Chen, K.; Wu, X.; Zhang, D.; Zhang, J.; Yao, L.; Guo, X. Identification of Conversion from Normal Elderly Cognition to Alzheimer’s Disease using Multimodal Support Vector Machine. J. Alzheimers Dis. 2015, 47, 1057–1067. [Google Scholar] [CrossRef]
- Suthaharan, S. Support Vector Machine. In Machine Learning Models and Algorithms for Big Data Classification; Springer: Boston, MA, USA, 2016; pp. 207–235. [Google Scholar]
- Lin, H.T.; Lin, C.J. A study on sigmoid kernels for SVM and the training of non-PSD kernels by SMO-type methods. Neural Comput. 2003, 3, 16. [Google Scholar]
- Natekin, A.; Knoll, A. Gradient boosting machines, a tutorial. Front. Neurorobotics 2013, 7, 21. [Google Scholar] [CrossRef] [Green Version]
- Chakravarthy, D.G.; Kannimuthu, S. Extreme Gradient Boost Classification Based Interesting User Patterns Discovery for Web Service Composition. Mob. Netw. Appl. 2019, 24, 1883–1895. [Google Scholar] [CrossRef]
- Jiang, J.; Trundle, P.; Ren, J. Medical image analysis with artificial neural networks. Comput. Med. Imaging Graph. 2010, 34, 617–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leon, F.; Floria, S.A.; Bădică, C. Evaluating the Effect of Voting Methods on Ensemble-Based Classification. In Proceedings of the 2017 IEEE International Conference on Innovations in Intelligent Systems and Applications (INISTA), Gdynia, Poland, 3–5 July 2017; pp. 1–6. [Google Scholar]
- Zhou, Z.-H. Ensemble Methods: Foundations and Algorithms; Chapman and Hall/CRC: Boca Raton, FL, USA, 2019. [Google Scholar]
- Zhang, Y.; Zhang, H.; Cai, J.; Yang, B. A Weighted Voting Classifier Based on Differential Evolution. Abstr. Appl. Anal. 2014, 2014, 376950. [Google Scholar] [CrossRef] [Green Version]
- Vovk, V. The Fundamental Nature of the Log Loss Function. In Fields of Logic and Computation II; Springer: Cham, Switzerland, 2015; pp. 307–318. [Google Scholar]
- Sharma, R.; Kaushik, B. Handwritten Indic scripts recognition using neuro-evolutionary adaptive PSO based convolutional neural networks. Sādhanā 2022, 47, 30. [Google Scholar] [CrossRef]
- Madsen, H.; Pinson, P.; Kariniotakis, G.; Nielsen, H.A.; Nielsen, T.S. Standardizing the performance evaluation of short-term wind power prediction models. Wind. Eng. 2005, 29, 475–489. [Google Scholar] [CrossRef] [Green Version]
- Hajian-Tilaki, K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Casp. J. Intern. Med. 2013, 4, 627–635. [Google Scholar]
- Ahmed, U.; Mukhiya, S.K.; Srivastava, G.; Lamo, Y.; Lin, J.C.-W. Attention-Based Deep Entropy Active Learning Using Lexical Algorithm for Mental Health Treatment. Front. Psychol. 2021, 12, 642347. [Google Scholar] [CrossRef]
- Shah, N.; Srivastava, G.; Savage, D.W.; Mago, V. Assessing Canadians Health Activity and Nutritional Habits Through Social Media. Front. Public Health 2020, 7, 400. [Google Scholar] [CrossRef] [PubMed]
- Priya, R.M.S.; Maddikunta, P.K.R.; Parimala, M.; Koppu, S.; Gadekallu, T.R.; Chowdhary, C.L.; Alazab, M. An effective feature engineering for DNN using hybrid PCA-GWO for intrusion detection in IoMT architecture. Comput. Commun. 2020, 160, 139–149. [Google Scholar] [CrossRef]
- Wang, W.; Chen, Q.; Yin, Z.; Srivastava, G.; Gadekallu, T.R.; Alsolami, F.; Su, C. Blockchain and PUF-Based Lightweight Authentication Protocol for Wireless Medical Sensor Networks. IEEE Internet Things J. 2021, 9, 8883–8891. [Google Scholar] [CrossRef]
- Hu, K.; Wang, Y.; Chen, K.; Hou, L.; Zhang, X. Multi-scale features extraction from baseline structure MRI for MCI patient classification and AD early diagnosis. Neurocomputing 2016, 175, 132–145. [Google Scholar] [CrossRef] [Green Version]
- Kavitha, C.; Mani, V.; Srividhya, S.R.; Khalaf, O.I.; Andrés Tavera Romero, C. Early-Stage Alzheimer’s Disease Prediction Using Machine Learning Models. Front. Public Health 2022, 10, 853294. [Google Scholar] [CrossRef]
- Shahbaz, M.; Ali, S.; Guergachi, A.; Niazi, A.; Umer, A. Classification of Alzheimer’s Disease Using Machine Learning Techniques. In Proceedings of the 8th International Conference on Data Science, Technology and Applications (DATA 2019), Prague, Czech Republic, 26–28 July 2019. [Google Scholar] [CrossRef]








| Author | Data Base | Imaging Modality | Total Subjects | Dataset Division | Feature Extraction and Classification Techniques | Classification Accuracy |
|---|---|---|---|---|---|---|
| [25] | ADNI | s-MRI | 259 | sMCI = 139; pMCI = 120 | FW LLE | sMCI/pMCI = 80.0% |
| [29] | ADNI | MRI | 710 | AD = 200; MCI = 280; pMCI = 120; sMCI = 160; CN = 230 | MK-Boost | AD/CN = 94.65% MCI/CN = 85.79% sMCI/pMCI = 72.08% |
| [31] | ADNI | MRI, PET | 726 | AD = 101; CI = 223; sMCI = 93; pMCI = 140; CN= 169 | SVM | AD/CN = 91.20% MCI/CN = 76.40% sMCI/pMCI = 74.20% |
| [32] | ADNI | s-MRI | 303 | AD = 158; CN = 145 | SVM-RFE SVM | AD/CN = 90.76% |
| [35] | ADNI | MRI, PET | 654 | AD = 154; MCI = 346; CN = 154 | RFLD SVM | AD/CN = 88.30% MCI/CN = 79.02% |
| [36] | ADNI | s-MRI, PET | 340 | AD = 113; MCI = 110; CN = 117 | AAL, ROI mSCDDL | AD/CN = 98.5% MCI/CN = 82.8% |
| [37] | ADNI | MRI, PET | 427 | AD = 65; pMCI = 95; sMCI = 132; CN = 135 | MDS, PCA SVM | AD/CN = 96.5% MCI/CN = 91.74% sMCI/pMCI =88.99% |
| [38] | ADNI | MRI, PET | 249 | AD = 70; MCI = 111; CN = 68 | PCA SVM | AD/CN = 92% MCI/CN = 84% |
| [39] | OASIS, LH | MRI | 196 | AD = 98; CN = 98 | 3AF 8L-CNN | AD/CN = 97.65% |
| [40] | ADNI | MRI | 710 | AD = 200; pMCI = 120; sMCI = 160; CN = 230 | ROI, FS MK-BOOST | AD/CN = 95.24% MCI/CN = 86.35% sMCI/pMCI = 74.25% |
| [41] | ADNI1 | MRI | 891 | AD = 199; pMCI = 167; sMCI = 226; CN = 229 | ROI, VBM, CLM LDMIL | AD/CN = 95.86% sMCI/pMCI = 77.64% |
| ADNI2 | MRI | 636 | AD = 159; pMCI = 38; sMCI = 239; CN = 200 | |||
| MIRIAD | MRI | 69 | AD = 23; CN = 46 | AD/CN = 97.16% | ||
| [42] | SELF | rs-MRI | 68 | AD = 34; CN = 34 | 3LHPM-ICA DL Algo. | AD/CN = 95.59% |
| [28] | ADNI | MRI FDG- PET | 1628 | AD = 342; MCI = 418; CN = 866 | RVB, SUVR, ROI L-SVM, LR, RF | AD/CN = 88% MCI/CN = 80% sMCI/pMCI = 73% |
| AIBL | 139 | AD = 72; MCI = 94; pMCI = 21; sMCI = 16; CN = 442 | ||||
| OASIS | 416 | AD = 100; CN = 93 |
| Dataset | Total | Alzheimer’s Disease | Mild Cognitive Impairment | Control Normal |
|---|---|---|---|---|
| Brain Axial MRI T1 and T2 weighted | 2127 | Images = 612 | Images = 538 | Images = 975 |
| Class 1 (AD) | Class 2 (MCI) | Class 3 (CN) | ||
| Label = 0 | Label = 1 | Label = 2 |
| Ground Truth/Actual | |||
|---|---|---|---|
| Predicted | Actual Positive | Actual Negative | |
| Predicted Positive | True positive (TP) | False Negative (FN) | |
| Predicted Negative | False Positive (FP) | True Negative (TN) | |
| Model | Grid Parameters | Optimized Parameters |
|---|---|---|
| Decision Tree | parameters = { ‘max_depth’: [2,4,6,8,10,12,14], ‘min_samples_leaf’: [5,7,9,11,13,15], ‘criterion’: [“gini”, “entropy”], ‘min_samples_split’: [1,2,3,4,5,6,7,8]} | parameters = { ‘max_depth’: [10], ‘min_samples_leaf’: [15], ‘criterion’: [‘gini’], ‘min_samples_split’: [4]} |
| Random Forest | model_params = { ‘n_estimators’: randint(4200), ‘max_features’: truncnorm(a = 0, B = 1, loc = 0.25, scale = 0.1), ‘min_samples_split’:uniform (0.01,0.199)} | model_params = { ‘n_estimators’: [148], ‘max_features’: [0.276163], ‘min_samples_split’: [0.0392]} |
| K-Nearest Neighbor | grid_params = { ‘n_neighbors’: [3,5,11,13,15], ‘weights’: [‘uniform’,’distance’], ‘metric’: [‘minkowski’,’euclidean’,’manhattan’]} | grid_params = { ‘n_neighbors’: [3], ‘weights’: [‘uniform’], ‘metric’: [‘minkowski’]} |
| Naïve Bayes | parameters = { ‘Var_smoothing’:np.logspace (0,9,num = 1000)} | parameters = { ‘Var_smoothing’: [4.328]} |
|
Extreme Gradient Boosting | parameters = { ‘learning_rate’: [0.01,0.1], ‘max_depth’: [3,5,6,10], ‘subsample’: [0.5,0.7], ‘colsample_bytree’: [0.5,0.7], ‘n_estimators’: [100,200,500], ‘objective’: [‘reg:squarederror’]} | parameters = { ‘learning_rate’: [0.1], ‘max_depth’: [6], ‘subsample’: [0.5], ‘colsample_bytree’: [0.5], ‘n_estimators’: [100], ‘objective’: [‘reg:squarederror’]} |
| Gradient Boost | parameters = { “n_estimators”: [50,150], “max_depth”: [1,3,5], “learning_rate”: [0.01,0.1,0.5,1], ‘max_depth’: [3,5,6,10], ‘min_child_weight’: [1,3,5], ‘subsample’: [0.5,0.7], ‘colsample_bytree’: [0.5,0.7], ‘n_estimators’: [100,200,500],} | parameters = { “n_estimators”: [80], “max_depth”: [3], “learning_rate”: [0.5], ‘max_depth’: [6], ‘min_child_weight’: [5], ‘subsample’: [0.5], ‘colsample_bytree’: [0.5], ‘n_estimators’: [100]} |
|
Multi-layer Perceptron Neural Network (MLP-NN) | Parameters = {‘batch_size’: [20,30], ‘nb_epoch’: [50,100,150], ‘optimizer’: [‘adam’,’rmsprop’,’SGD’] ‘units’: [512,1024]} | parameters = {‘batch_size’: [20], ‘nb_epoch’: [50], ‘optimizer’: [‘adam’], ‘units’: [512]} |
|
Proposed Ensemble model XGB + DT + SVM | parameters = { ‘learning_rate’: [0.01,0.1], ‘max_depth’: [3,5,6,10], ‘min_child_weight’: [1,3,5], ‘subsample’: [0.5,0.7], ‘colsample_bytree’: [0.5,0.7], ‘n_estimators’: [100,200,500], ‘objective’: [‘reg:squarederror’], ‘min_samples_leaf’: [5,7,9,11,13,15], ‘criterion’: [“gini”, “entropy”], ‘min_samples_split’: [1,2,3,4,5,6,7,8] ‘kernel’: [‘rbf’, ‘poly’, ‘sigmoid’]} | param_grid = { “n_estimators”: [80], “max_depth”: [3], “learning_rate”: [0.5], ‘max_depth’: [6], ‘min_child_weight’: [5], ‘subsample’: [0.5], ‘colsample_bytree’: [0.5], ‘n_estimators’: [100] ‘kernel’: [‘rbf’]} |
| Model | Accuracy (%) | Sensitivity (%) | Specificity (%) | Log Loss | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AD | CN | MCI | AD | CN | MCI | AD | CN | MCI | ||
| DT | 87.12 | 89.56 | 77.84 | 95.22 | 92.39 | 87.57 | 49.06 | 98.82 | 90.24 | 0.74054 |
| RF | 80.59 | 82.51 | 60.89 | 75.76 | 53.26 | 79.50 | 71.29 | 81.58 | 94.11 | 0.71971 |
| K-NN | 77.42 | 72.09 | 66.30 | 83.61 | 46.63 | 80.12 | 61.16 | 96.39 | 76.96 | 0.38076 |
| NB | 72.22 | 75.86 | 58.75 | 72.01 | 92.39 | 91.30 | 57.74 | 95.46 | 64.40 | 13.6992 |
| GB | 72.25 | 77.98 | 77.58 | 89.76 | 48.91 | 88.19 | 72.27 | 90.00 | 97.51 | 0.53312 |
| XGB | 81.56 | 89.00 | 84.63 | 92.49 | 92.49 | 97.51 | 80.05 | 94.27 | 98.71 | 0.36665 |
| SVM RBF | 71.45 | 88.59 | 86.36 | 98.63 | 54.89 | 99.00 | 79.63 | 99.11 | 96.05 | 0.33290 |
| SVM Poly | 87.84 | 86.21 | 81.48 | 83.95 | 58.15 | 94.40 | 79.20 | 89.23 | 95.40 | 0.29615 |
| SVM Sigmoid | 75.48 | 71.49 | 83.14 | 90.44 | 76.08 | 96.27 | 86.51 | 93.54 | 99.26 | 0.48208 |
| MLP-NN | 89.20 | 81.56 | 84.30 | 95.22 | 64.67 | 98.75 | 82.49 | 96.47 | 98.51 | 0.36665 |
| Proposed XGB + DT + SVM | 87.00 | 90.51 | 90.51 | 91.80 | 77.17 | 98.75 | 88.26 | 94.27 | 99.51 | 0.27795 |
| Model | Accuracy (%) | Sensitivity (%) | Specificity (%) | Log Loss | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AD | CN | MCI | AD | CN | MCI | AD | CN | MCI | ||
| DT | 89.12 | 92.56 | 79.84 | 97.26 | 37.50 | 91.92 | 64.77 | 97.30 | 98.33 | 0.58883 |
| RF | 82.59 | 87.51 | 67.89 | 76.79 | 57.06 | 81.36 | 74.68 | 83.17 | 92.96 | 0.71971 |
| K-NN | 85.84 | 88.21 | 85.48 | 90.44 | 69.02 | 97.51 | 83.77 | 93.56 | 98.74 | 1.31750 |
| NB | 79.22 | 83.86 | 78.75 | 85.32a | 75.43 | 89.44 | 50.00 | 99.87 | 71.42 | 0.27213 |
| GB | 79.25 | 75.98 | 79.58 | 91.46 | 55.43 | 93.78 | 76.43 | 93.11 | 97.88 | 0.39457 |
| XGB | 87.56 | 89.89 | 86.63 | 93.85 | 62.50 | 96.27 | 79.88 | 79.88 | 99.48 | 0.32501 |
| SVM Poly | 88.00 | 86.21 | 85.00 | 92.75 | 77.54 | 87.00 | 86.00 | 81.76 | 84.34 | 0.31123 |
| MLP- NN | 79.45 | 89.79 | 86.36 | 90.78 | 77.17 | 96.27 | 87.09 | 93.76 | 99.27 | 0.42902 |
| Proposed XGB + DT + SVM | 96.12 | 95 | 96.15 | 93.85 | 78.80 | 98.75 | 89.67 | 95.59 | 99.05 | 0.24926 |
| Folds | DT | RF | K-NN | NB | SVM-RBF | SVM- Poly | SVM- Sigmoid | GB | XGB | MLP-NN | Proposed XGB + DT + SVM |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1_fold | 0.71812 | 0.77181 | 0.83892 | 0.83892 | 0.83221 | 0.83221 | 0.65771 | 0.81879 | 0.83892 | 0.84563 | 0.98547 |
| 2_fold | 0.71812 | 0.79865 | 0.90604 | 0.89261 | 0.92617 | 0.92617 | 0.67782 | 0.79867 | 0.87248 | 0.93288 | 0.97564 |
| 3_fold | 0.67114 | 0.73825 | 0.86577 | 0.80536 | 0.87919 | 0.87919 | 0.70469 | 0.83892 | 0.82550 | 0.87248 | 0.97554 |
| 4_fold | 0.67785 | 0.83221 | 0.91275 | 0.89932 | 0.87919 | 0.87919 | 0.77852 | 0.86577 | 0.87919 | 0.87919 | 0.96587 |
| 5_fold | 0.71812 | 0.79865 | 0.87919 | 0.89261 | 0.87248 | 0.87248 | 0.63087 | 0.81208 | 0.83892 | 0.88590 | 0.98658 |
| 6_fold | 0.71812 | 0.75838 | 0.85234 | 0.82550 | 0.88590 | 0.88590 | 0.65771 | 0.80536 | 0.89261 | 0.89261 | 0.97857 |
| 7_fold | 0.67785 | 0.79194 | 0.89932 | 0.87248 | 0.90604 | 0.90604 | 0.66442 | 0.84563 | 0.87919 | 0.91946 | 0.98143 |
| 8_fold | 0.67567 | 0.79054 | 0.88513 | 0.82432 | 0.87837 | 0.87837 | 0.68243 | 0.82432 | 0.87837 | 0.87837 | 0.98543 |
| 9_fold | 0.68243 | 0.79054 | 0.84459 | 0.81756 | 0.89189 | 0.89189 | 0.62162 | 0.84459 | 0.84459 | 0.89189 | 0.99948 |
| 10_fold | 0.75675 | 0.76845 | 0.86486 | 0.85810 | 0.87837 | 0.87837 | 0.64864 | 0.80405 | 0.86486 | 0.88513 | 0.96854 |
| Mean | 0.70141 | 0.78394 | 0.87489 | 0.85268 | 0.87424 | 0.87424 | 0.67043 | 0.82582 | 0.86146 | 0.88835 | 0.98845 |
| Rank | 1st | 2nd | 3rd | 4th | 5th | 6th | 7th | 8th | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| Model | GB | RF | K-NN | NB | DT | XGB | MLP-NN | XGB + DT + SVM | 4.36 × 10−12 |
| Average rank | 2 | 3.05 | 5.25 | 1 | 4.15 | 5.9 | 6.65 | 8 |
| Before Hyperparameter Tuning | After Hyperparameter Tuning | Mean Difference | t-Values | |
|---|---|---|---|---|
| Accuracy (%) | 76.39 | 80.448 | −4.058 ** | −3.225 |
| S. No | Study Reference | Dataset | Technique | Accuracy |
|---|---|---|---|---|
| 1. | Kun Hu et al. [67] | ADNI | SVM | 84.30% |
| 2. | Kavitha et al. [68] | OASIS | Voting classifier | 85.12% |
| 3. | Shahbaz, Muhammad et al. [69] | ADNI | Generalized linear model (GLM) | 88.24% |
| 4. | Classical ML Model | ADNI | XGB | 85.42% |
| 5. | Neural Network | ADNI | MLP-NN | 89.64% |
| 6. | Proposed Voting Ensemble | ADNI | XGB + DT + SVM | 95.75% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, Y.F.; Kaushik, B.; Chowdhary, C.L.; Srivastava, G. Ensemble Model for Diagnostic Classification of Alzheimer’s Disease Based on Brain Anatomical Magnetic Resonance Imaging. Diagnostics 2022, 12, 3193. https://doi.org/10.3390/diagnostics12123193
Khan YF, Kaushik B, Chowdhary CL, Srivastava G. Ensemble Model for Diagnostic Classification of Alzheimer’s Disease Based on Brain Anatomical Magnetic Resonance Imaging. Diagnostics. 2022; 12(12):3193. https://doi.org/10.3390/diagnostics12123193
Chicago/Turabian StyleKhan, Yusera Farooq, Baijnath Kaushik, Chiranji Lal Chowdhary, and Gautam Srivastava. 2022. "Ensemble Model for Diagnostic Classification of Alzheimer’s Disease Based on Brain Anatomical Magnetic Resonance Imaging" Diagnostics 12, no. 12: 3193. https://doi.org/10.3390/diagnostics12123193
APA StyleKhan, Y. F., Kaushik, B., Chowdhary, C. L., & Srivastava, G. (2022). Ensemble Model for Diagnostic Classification of Alzheimer’s Disease Based on Brain Anatomical Magnetic Resonance Imaging. Diagnostics, 12(12), 3193. https://doi.org/10.3390/diagnostics12123193








