Computational Evidence for Laboratory Diagnostic Pathways: Extracting Predictive Analytes for Myocardial Ischemia from Routine Hospital Data
Abstract
1. Introduction
1.1. Large but Incomplete Data
1.2. Human Error and Artificial Intelligence
1.3. Our Pilot Study
2. Materials and Methods
2.1. Patient Lab Dataset
2.2. Modelling
2.2.1. Multiple Imputation
2.2.2. Orthogonal Data Augmentation/Bayesian Model Averaging with Regularization (ODA/BMA)
3. Results
4. Discussion
4.1. Interpretations of the Results
4.1.1. TnT, CK, CK-MB-Mass
4.1.2. Potassium, eGFR, Urea
4.1.3. INR, Thrombin Time, Bilirubin
4.1.4. Type of Blood Collection, FiO2
4.1.5. Red Blood Cell Distribution Width (RDW), MCV, MCH, MCHC
4.1.6. HDL Cholesterol and LDL Cholesterol
4.1.7. Total Calcium
4.1.8. Total Glucose
4.1.9. Chloride
4.2. Strengths and Limitations of the Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | artificial intelligence |
| BMA | Bayesian Model Averaging |
| CK | creatine kinase myocardial band |
| CK-MB | creatine kinase MB |
| DOAJ | Directory of Open-Access Journals |
| FiO2 | fraction of inspired oxygen |
| GFR | glomerular Filtration Rate |
| HDL | high-density lipoprotein |
| ICD | International Statistical Classification of Diseases |
| INR | international normalized ratio |
| IQR | interquartile range |
| LD | linear dichroism |
| LDL | low-density lipoprotein |
| MCH | mean corpuscular hemoglobin |
| MCHC | mean corpuscular hemoglobin concentration |
| MCV | mean corpuscular volume |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MI | myocardial ischemia |
| Non-MI | patients without myocardial ischemia |
| NT-proBNP | N-terminal prohormone of brain natriuretic peptide |
| ODA | Orthogonal Data Augmentation |
| RDW | red distribution width |
| TLA | three letter acronym |
| TnT | troponin T |
References
- Knottnerus, J.A.; Van Weel, C.; Muris, J.W. Evaluation of diagnostic procedures. BMJ 2002, 324, 477–480. [Google Scholar] [CrossRef] [PubMed]
- Nakas, C.T.; Schütz, N.; Werners, M.; Leichtle, A.B. Accuracy and Calibration of Computational Approaches for Inpatient Mortality Predictive Modeling. PLoS ONE 2016, 11, e0159046. [Google Scholar] [CrossRef] [PubMed]
- Soffer, S.; Klang, E.; Barash, Y.; Grossman, E.; Zimlichman, E. Predicting In-Hospital Mortality at Admission to the Medical Ward: A Big-Data Machine Learning Model. Am. J. Med. 2021, 134, 227–234.e4. [Google Scholar] [CrossRef] [PubMed]
- Mayer-Schönberger, V. Big Data for cardiology: Novel discovery? Eur. Heart J. 2016, 37, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- Waeschle, R.M.; Bauer, M.; Schmidt, C.E. Fehler in der Medizin: Ursachen, Auswirkungen und Maßnahmen zur Verbesserung der Patientensicherheit. Anaesthesist 2015, 64, 689–704. [Google Scholar] [CrossRef]
- Cadamuro, J. Rise of the machines: The inevitable evolution of medicine and medical laboratories intertwining with artificial intelligence—A narrative review. Diagnostics 2021, 11, 1399. [Google Scholar] [CrossRef]
- Miller, D.D.; Brown, E.W. Artificial Intelligence in Medical Practice: The Question to the Answer? Am. J. Med. 2018, 131, 129–133. [Google Scholar] [CrossRef]
- Johnson, C. The Causes of Human Error in Medicine. Cogn. Technol. Work. 2002, 4, 65–70. [Google Scholar] [CrossRef]
- Shilo, S.; Rossman, H.; Segal, E. Axes of a revolution: Challenges and promises of big data in healthcare. Nat. Med. 2020, 26, 29–38. [Google Scholar] [CrossRef]
- Hoffmann, G.; Bietenbeck, A.; Lichtinghagen, R.; Klawonn, F. Using machine learning techniques to generate laboratory diagnostic pathways—A case study. J. Lab. Precis. Med. 2018, 3, 58. [Google Scholar] [CrossRef]
- Saygitov, R.T.; Glezer, M.G.; Semakina, S.V. Blood urea nitrogen and creatinine levels at admission for mortality risk assessment in patients with acute coronary syndromes. Emerg. Med. J. 2010, 27, 105–109. [Google Scholar] [CrossRef]
- WHO. International Statistical Classification of Diseases and Related Health Problems 10th Revision (ICD-10)-WHO Version for 2019-Covid-Expanded. Available online: https://icd.who.int/browse10/2019/en#/I20-I25 (accessed on 20 October 2022).
- Collet, J.P.; Thiele, H.; Barbato, E.; Bauersachs, J.; Dendale, P.; Edvardsen, T.; Gale, C.P.; Jobs, A.; Lambrinou, E.; Mehilli, J.; et al. 2020 ESC Guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation. Eur. Heart J. 2021, 42, 1289–1367. [Google Scholar] [CrossRef]
- Gelman, A.; Rubin, D.B. Inference from iterative simulation using multiple sequences. Stat. Sci. 1992, 7, 457–472. [Google Scholar] [CrossRef]
- Su, Y.S.; Gelman, A.; Hill, J.; Yajima, M. Multiple imputation with diagnostics (mi) in R: Opening windows into the black box. J. Stat. Softw. 2011, 45, 1–31. [Google Scholar] [CrossRef]
- Yucel, R.M. State of the multiple imputation software. J. Stat. Softw. 2011, 45, 1–7. [Google Scholar] [CrossRef]
- Clyde, M.; Desimone, H.; Parmigiani, G. Prediction via orthogonalized model mixing. J. Am. Stat. Assoc. 1996, 91, 1197–1208. [Google Scholar] [CrossRef]
- Ghosh, J.; Clyde, M.A. Rao-blackwellization for Bayesian variable selection and model averaging in linear and binary regression: A novel data augmentation approach. J. Am. Stat. Assoc. 2011, 106, 1041–1052. [Google Scholar] [CrossRef]
- Herold, G. Innere Medizin; Gerd Herold: Köln, Germany, 2007; pp. 131–155. [Google Scholar] [CrossRef]
- Moen, M.F.; Zhan, M.; Hsu, V.D.; Walker, L.D.; Einhorn, L.M.; Seliger, S.L.; Fink, J.C. Frequency of hypoglycemia and its significance in chronic kidney disease. Clin. J. Am. Soc. Nephrol. 2009, 4, 1121–1127. [Google Scholar] [CrossRef]
- Ronco, C.; McCullough, P.; Anker, S.D.; Anand, I.; Aspromonte, N.; Bagshaw, S.M.; Bellomo, R.; Berl, T.; Bobek, I.; Cruz, D.N.; et al. Cardio-renal syndromes: Report from the consensus conference of the acute dialysis quality initiative. Eur. Heart J. 2010, 31, 703–711. [Google Scholar] [CrossRef]
- Shlipak, M.G.; Massie, B.M. The clinical challenge of cardiorenal syndrome. Circulation 2004, 110, 1514–1517. [Google Scholar] [CrossRef]
- Lim, A.K. Diabetic nephropathy—Complications and treatment. Int. J. Nephrol. Renov. Dis. 2014, 7, 361–381. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, G.; Virk, H.U.H.; Khalid, M.; Lavie, C.J.; Ventura, H.; Mukherjee, D.; Ramu, V.; Bhogal, S.; Kumar, G.; Shanmugasundaram, M.; et al. Diabetic cardiomyopathy—A comprehensive updated review. Prog. Cardiovasc. Dis. 2019, 62, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Dalfó Baqué, A. New equation to estimate glomerular filtration rate? FMC Form. Med. Contin. Aten. Prim. 2009, 16, 614. [Google Scholar] [CrossRef]
- Higgins, C. Urea and the Clinical Value of Measuring Blood Urea Concentration. pp. 1–6. 2016. Available online: https://acutecaretesting.org/en/articles/urea-and-the-clinical-value-of-measuring-blood-urea-concentration (accessed on 20 October 2022).
- Saner, F.H.; Heuer, M.; Meyer, M.; Canbay, A.; Sotiropoulos, G.C.; Radtke, A.; Treckmann, J.; Beckebaum, S.; Dohna-Schwake, C.; Oldedamink, S.W.; et al. When the heart kills the liver: Acute liver failure in congestive heart failure. Eur. J. Med. Res. 2009, 14, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Naschitz, J.E.; Slobodin, G.; Lewis, R.J.; Zuckerman, E.; Yeshurun, D. Heart diseases affecting the liver and liver diseases affecting the heart. Am. Heart J. 2000, 140, 111–120. [Google Scholar] [CrossRef]
- Zalawadiya, S.K.; Veeranna, V.; Niraj, A.; Pradhan, J.; Afonso, L. Red cell distribution width and risk of coronary heart disease events. Am. J. Cardiol. 2010, 106, 988–993. [Google Scholar] [CrossRef]
- Allison, M.A.; Wright, C.M. A comparison of HDL and LDL cholesterol for prevalent coronary calcification. Int. J. Cardiol. 2004, 95, 55–60. [Google Scholar] [CrossRef]
- Cheung, B.M.; Wat, N.M.; Tam, S.; Thomas, G.N.; Leung, G.M.; Cheng, C.H.; Woo, J.; Janus, E.D.; Lau, C.P.; Lam, T.H.; et al. Components of the metabolic syndrome predictive of its development: A 6-year longitudinal study in Hong Kong Chinese. Clin. Endocrinol. 2008, 68, 730–737. [Google Scholar] [CrossRef]
- Després, J.P.; Lemieux, I.; Dagenais, G.R.; Cantin, B.; Lamarche, B. HDL-cholesterol as a marker of coronary heart disease risk: The Québec cardiovascular study. Atherosclerosis 2000, 153, 263–272. [Google Scholar] [CrossRef]
- Grundy, S.M.; Arai, H.; Barter, P.; Bersot, T.P.; Betteridge, D.J.; Carmena, R.; Cuevas, A.; Davidson, M.H.; Genest, J.; Kesäniemi, Y.A.; et al. An International Atherosclerosis Society Position Paper: Global recommendations for the management of dyslipidemia. Executive summary. Atherosclerosis 2014, 232, 410–413. [Google Scholar] [CrossRef]
- Blaha, M.J.; Blumenthal, R.S.; Brinton, E.A.; Jacobson, T.A. The importance of non-HDL cholesterol reporting in lipid management. J. Clin. Lipidol. 2008, 2, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; Chapman, M.J.; De Backer, G.G.; Delgado, V.; Ference, B.A.; et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk. Eur. Heart J. 2020, 41, 111–188. [Google Scholar] [CrossRef] [PubMed]
- Grandi, N.C.; Brenner, H.; Hahmann, H.; Wüsten, B.; März, W.; Rothenbacher, D.; Breitling, L.P. Calcium, phosphate and the risk of cardiovascular events and all-cause mortality in a population with stable coronary heart disease. Heart 2012, 98, 926–933. [Google Scholar] [CrossRef] [PubMed]
- Pentti, K.; Tuppurainen, M.T.; Honkanen, R.; Sandini, L.; Kröger, H.; Alhava, E.; Saarikoski, S. Use of calcium supplements and the risk of coronary heart disease in 52–62-year-old women: The Kuopio Osteoporosis Risk Factor and Prevention Study. Maturitas 2009, 63, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Boniatti, M.M.; Cardoso, P.R.; Castilho, R.K.; Vieira, S.R. Is hyperchloremia associated with mortality in critically ill patients? A prospective cohort study. J. Crit. Care 2011, 26, 175–179. [Google Scholar] [CrossRef]
- Neyra, J.A.; Canepa-Escaro, F.; Li, X.; Manllo, J.; Adams-Huet, B.; Yee, J.; Yessayan, L. Association of Hyperchloremia with Hospital Mortality in Critically Ill Septic Patients. Crit. Care Med. 2015, 43, 1938–1944. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Blackwell, M.; Honaker, J.; King, G. A Unified Approach to Measurement Error and Missing Data: Overview and Applications. Sociol. Methods Res. 2017, 46, 303–341. [Google Scholar] [CrossRef]
- Kush, R.D.; Warzel, D.; Kush, M.A.; Sherman, A.; Navarro, E.A.; Fitzmartin, R.; Pétavy, F.; Galvez, J.; Becnel, L.B.; Zhou, F.L.; et al. FAIR data sharing: The roles of common data elements and harmonization. J. Biomed. Inform. 2020, 107, 103421. [Google Scholar] [CrossRef]
- Eurlings, C.G.M.J.; Bektas, S.; Sanders-van Wijk, S.; Tsirkin, A.; Vasilchenko, V.; Meex, S.J.R.; Failer, M.; Oehri, C.; Ruff, P.; Zellweger, M.J.; et al. Use of artificial intelligence to assess the risk of coronary artery disease without additional (non-invasive) testing: Validation in a low-risk to intermediate-risk outpatient clinic cohort. BMJ Open 2022, 12, e055170. [Google Scholar] [CrossRef]
- Liu, R.; Wang, M.; Zheng, T.; Zhang, R.; Li, N.; Chen, Z.; Yan, H.; Shi, Q. An artificial intelligence-based risk prediction model of myocardial infarction. BMC Bioinform. 2022, 23, 217. [Google Scholar] [CrossRef]
- Wang, H.; Zu, Q.; Chen, J.; Yang, Z.; Ahmed, M.A. Application of Artificial Intelligence in Acute Coronary Syndrome: A Brief Literature Review. Adv. Ther. 2021, 38, 5078–5086. [Google Scholar] [CrossRef]
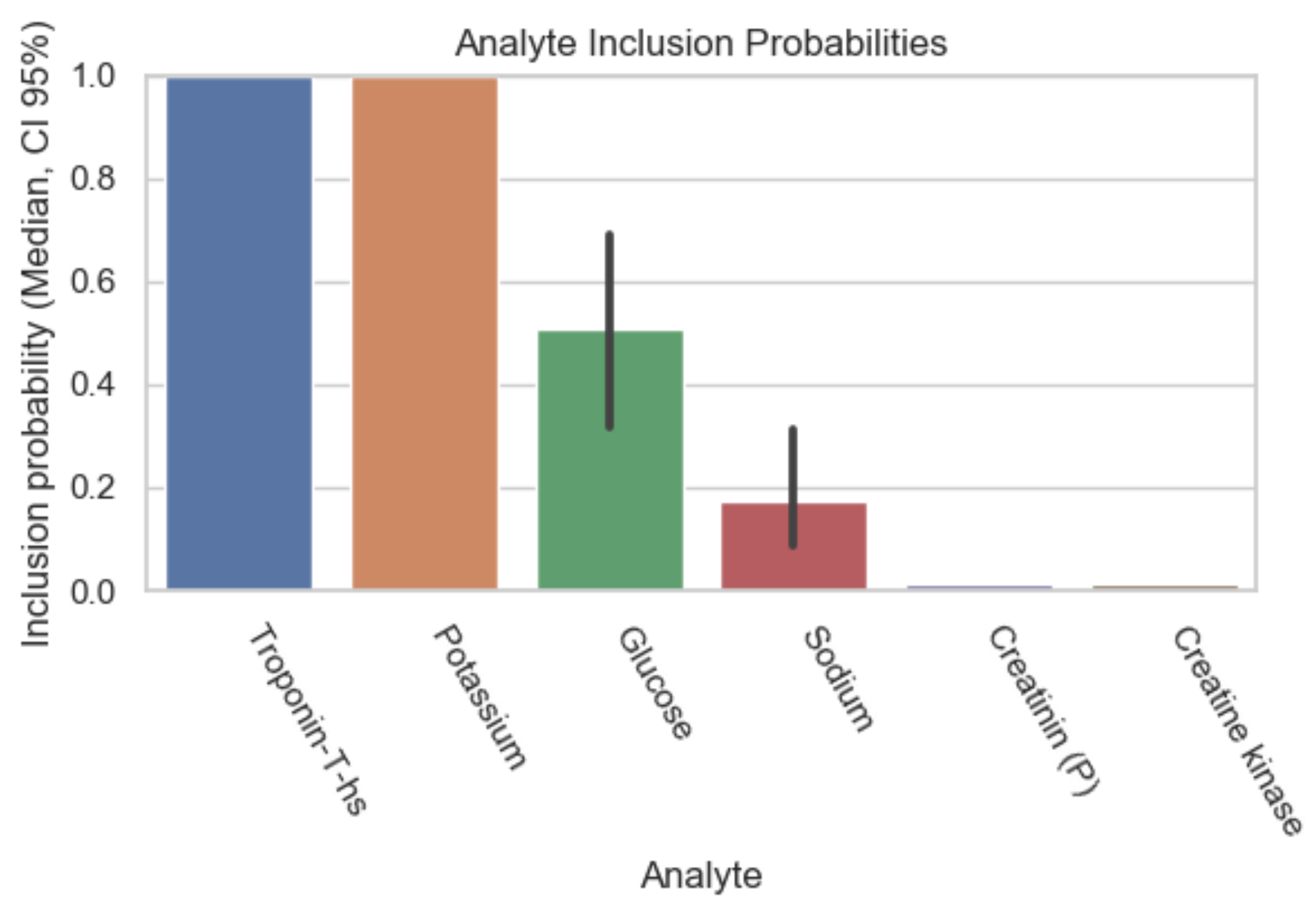
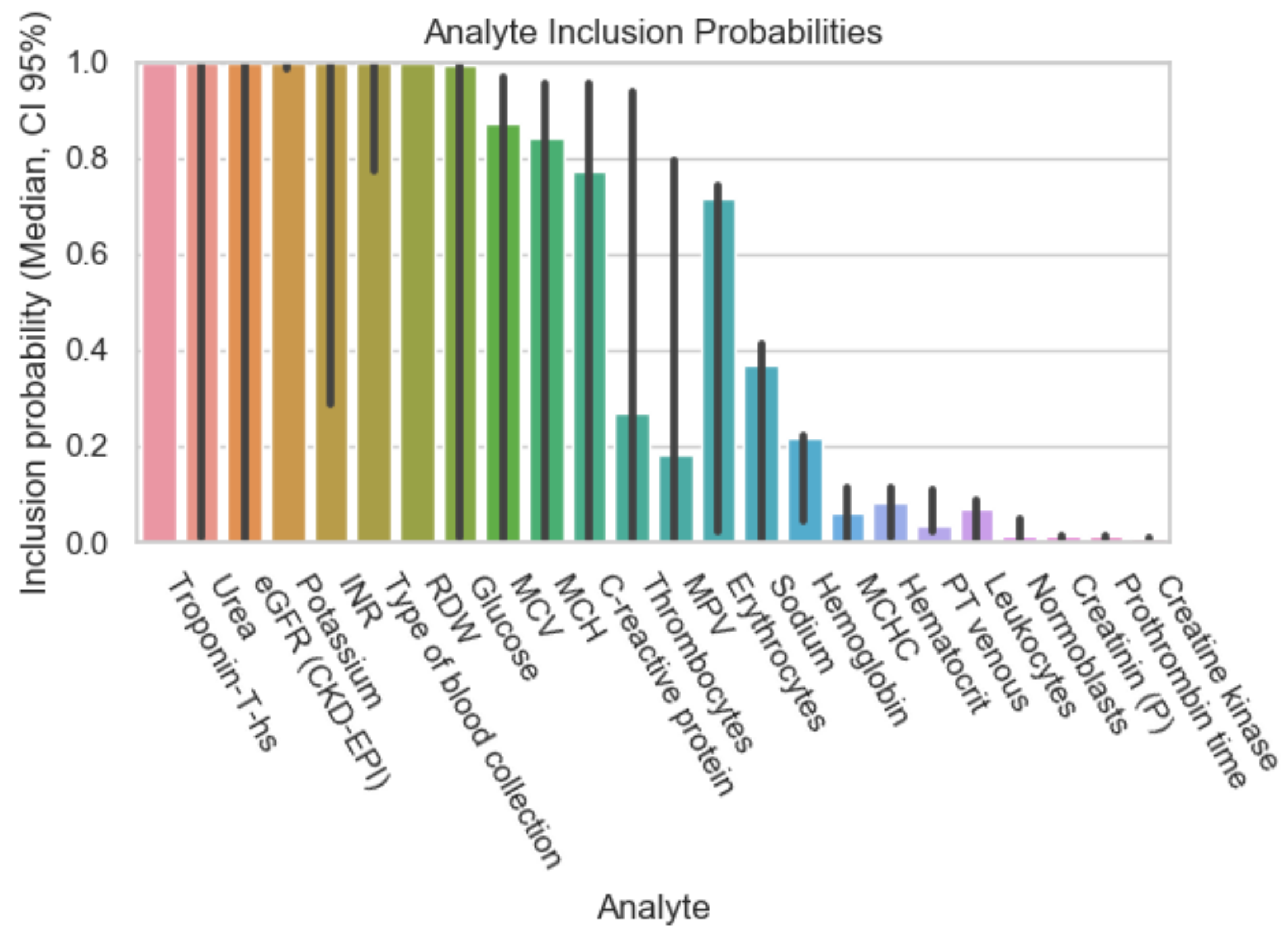

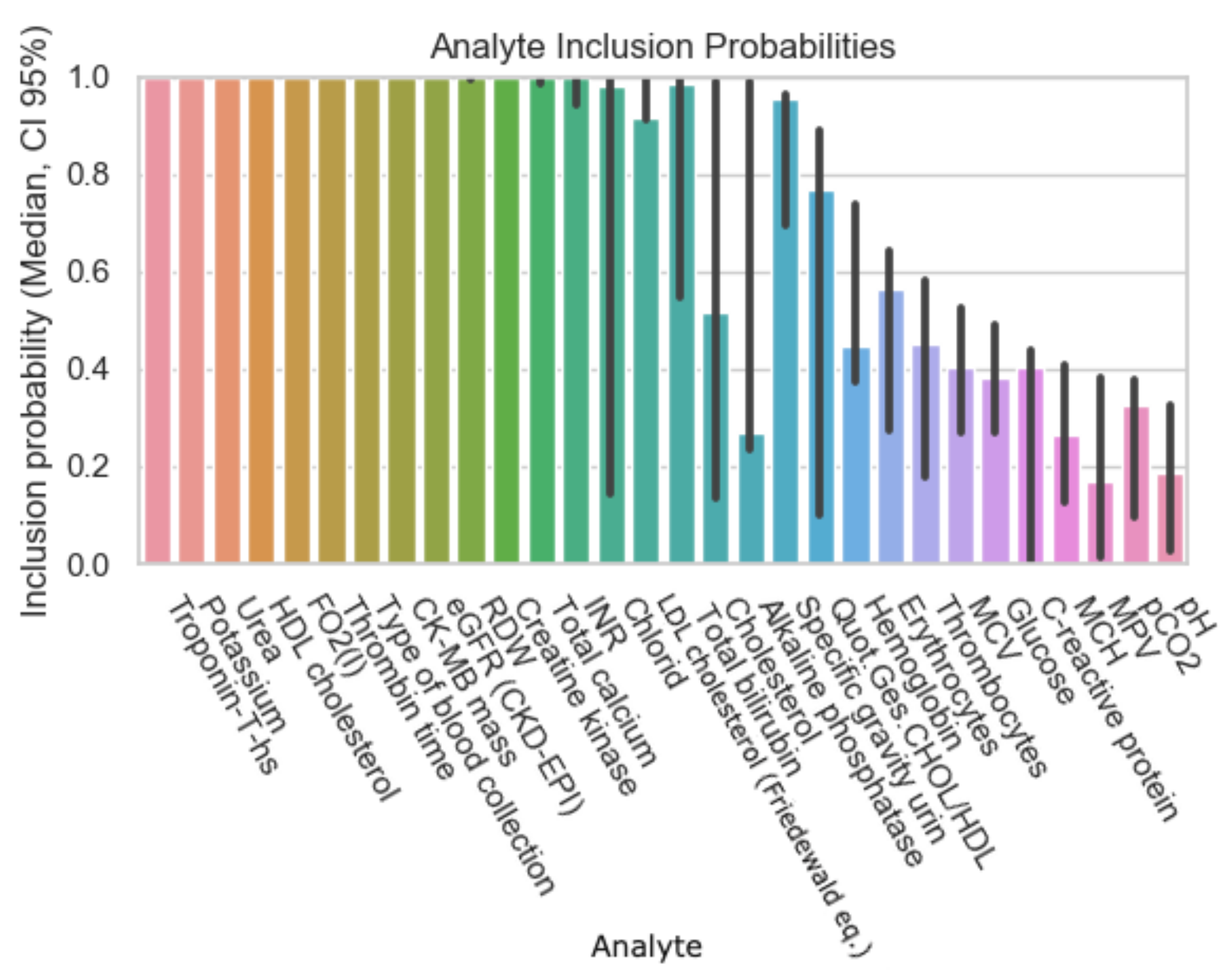
| Dataset ID | Number of Analytes |
|---|---|
| 20% sparsity | 8 |
| 40% sparsity | 26 |
| 60% sparsity | 59 |
| 80% sparsity | 110 |
| Analyte | Inclusion Figure 1 | Inclusion Figure 2 | Inclusion Figure 3 | Inclusion Figure 4 | Total Inclusion |
|---|---|---|---|---|---|
| Troponin | + | + | + | + | 4/4 |
| Potassium | + | + | + | + | 4/4 |
| Type of blood collection | + | + | + | 3/4 | |
| Red Distribution width | + | + | + | 3/4 | |
| eGFR | + | + | + | 3/4 | |
| Urea | + | + | + | 3/4 | |
| INR | + | + | + | 3/4 | |
| CK | + | + | 2/4 | ||
| CK-MB-Masse | + | + | 2/4 | ||
| Thrombin time | + | + | 2/4 | ||
| HDL-Cholesterol | + | + | 2/4 | ||
| Calcium total | + | + | 2/4 | ||
| FO2 | + | 1/4 | |||
| Bilirubin | + | 1/4 | |||
| Chloride | + | 1/4 | |||
| Glucose | + | 1/4 | |||
| LDL-Cholesterol | + | 1/4 | |||
| MCV, MCH, MCHC | + | 1/4 |
| Analyte Name | NON-MI | MI | Total |
|---|---|---|---|
| Troponin T [ng/L] | 20[43.02] | 248.1[1233.44] | 25[78.62] |
| Potassium (mmol/L) | 4 [0.5] | 4.1 [0.5] | 4.1 [0.5] |
| Type of blood collection | 0[0] | 0[0] | 0[0] |
| RDW (%) | 13.5[1.9] | 13.4[1.6] | 13.5[1.9] |
| eGFR (mL/min/1.73 m ) | 85[40] | 76[36] | 84[39] |
| Urea (mmol/L) | 5.9[4.4] | 6.3[4.1] | 5.9[4.4] |
| INR | 1.1[0.13] | 1.03[0.13] | 1.01[0.07] |
| CK (U/L) | 96[126] | 191[500] | 103[154] |
| CK-MB-Masse (%) | 3.9[7.2] | 12[44.3] | 4.6[11.5] |
| Thrombin time (s) | 16[2.5] | 17.6[23.2] | 16.1[2.5] |
| HDL-Cholesterol (mmol/L) | 1.24[0.24] | 1.14[0.47] | 1.23[0.55] |
| Calcium total (mmol/L) | 2.25[0.21] | 2.21[0.17] | 2.24[0.2] |
| FO2 (mmHg) | 21[41] | 59[54] | 32[43] |
| Bilirubin (µmol/L) | 8[10] | 9[8.5] | 8[10] |
| Chloride (mmol/L) | 107[7] | 108[5] | 107[7] |
| Glucose (mmol/L) | 5.96[2.11] | 6.5[2.5] | 6[2.1] |
| LDL-Cholesterol (mmol/L) | 2.23[1.36] | 2.39[1.49] | 2.25[1.38] |
| MCV (fl) | 86[7] | 86[6] | 86[7] |
| MCH (pg) | 30[3] | 30[2] | 30[3] |
| MCHC (g/dL) | 342.5[17] | 344[17] | 343[17] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liniger, Z.; Ellenberger, B.; Leichtle, A.B. Computational Evidence for Laboratory Diagnostic Pathways: Extracting Predictive Analytes for Myocardial Ischemia from Routine Hospital Data. Diagnostics 2022, 12, 3148. https://doi.org/10.3390/diagnostics12123148
Liniger Z, Ellenberger B, Leichtle AB. Computational Evidence for Laboratory Diagnostic Pathways: Extracting Predictive Analytes for Myocardial Ischemia from Routine Hospital Data. Diagnostics. 2022; 12(12):3148. https://doi.org/10.3390/diagnostics12123148
Chicago/Turabian StyleLiniger, Zara, Benjamin Ellenberger, and Alexander Benedikt Leichtle. 2022. "Computational Evidence for Laboratory Diagnostic Pathways: Extracting Predictive Analytes for Myocardial Ischemia from Routine Hospital Data" Diagnostics 12, no. 12: 3148. https://doi.org/10.3390/diagnostics12123148
APA StyleLiniger, Z., Ellenberger, B., & Leichtle, A. B. (2022). Computational Evidence for Laboratory Diagnostic Pathways: Extracting Predictive Analytes for Myocardial Ischemia from Routine Hospital Data. Diagnostics, 12(12), 3148. https://doi.org/10.3390/diagnostics12123148







