Breast Cancer Detection in Mammogram Images Using K–Means++ Clustering Based on Cuckoo Search Optimization
Abstract
1. Introduction
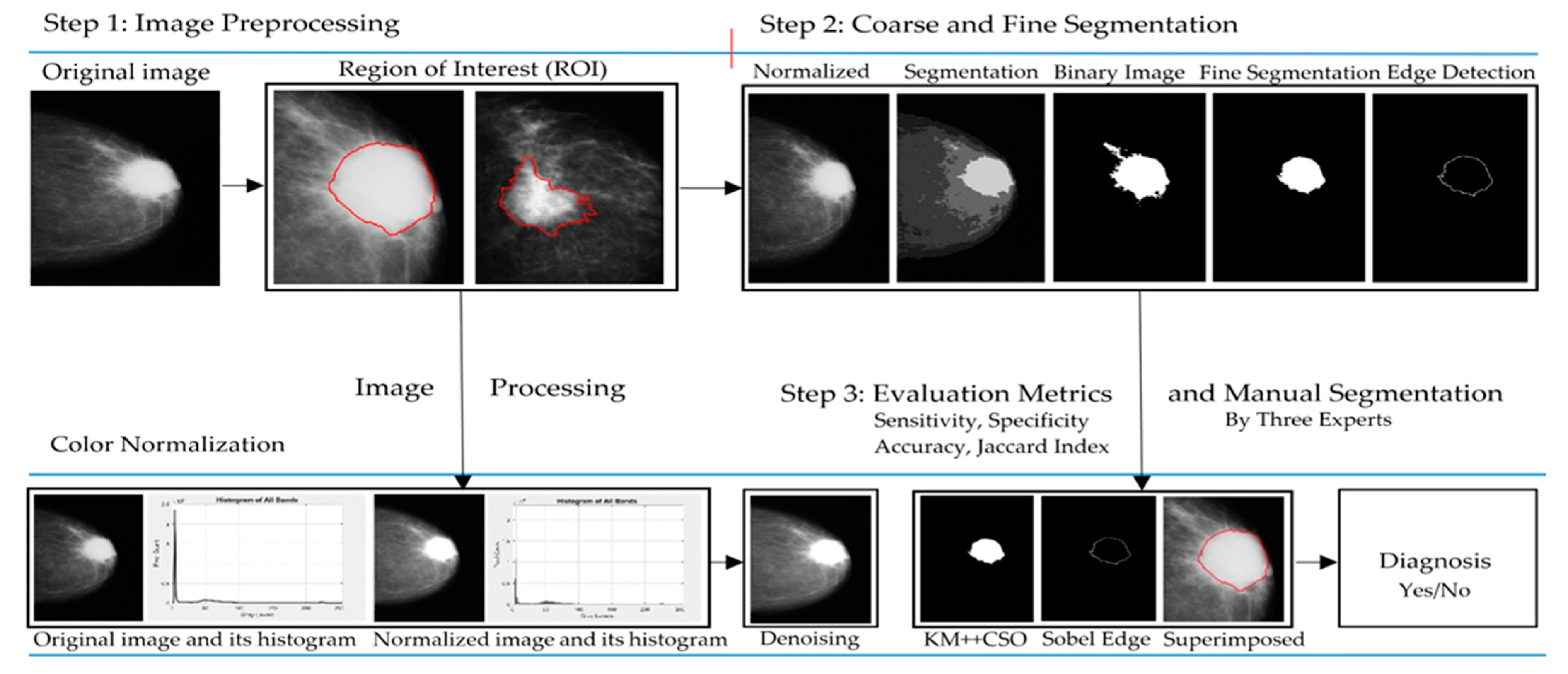
2. Materials and Methods
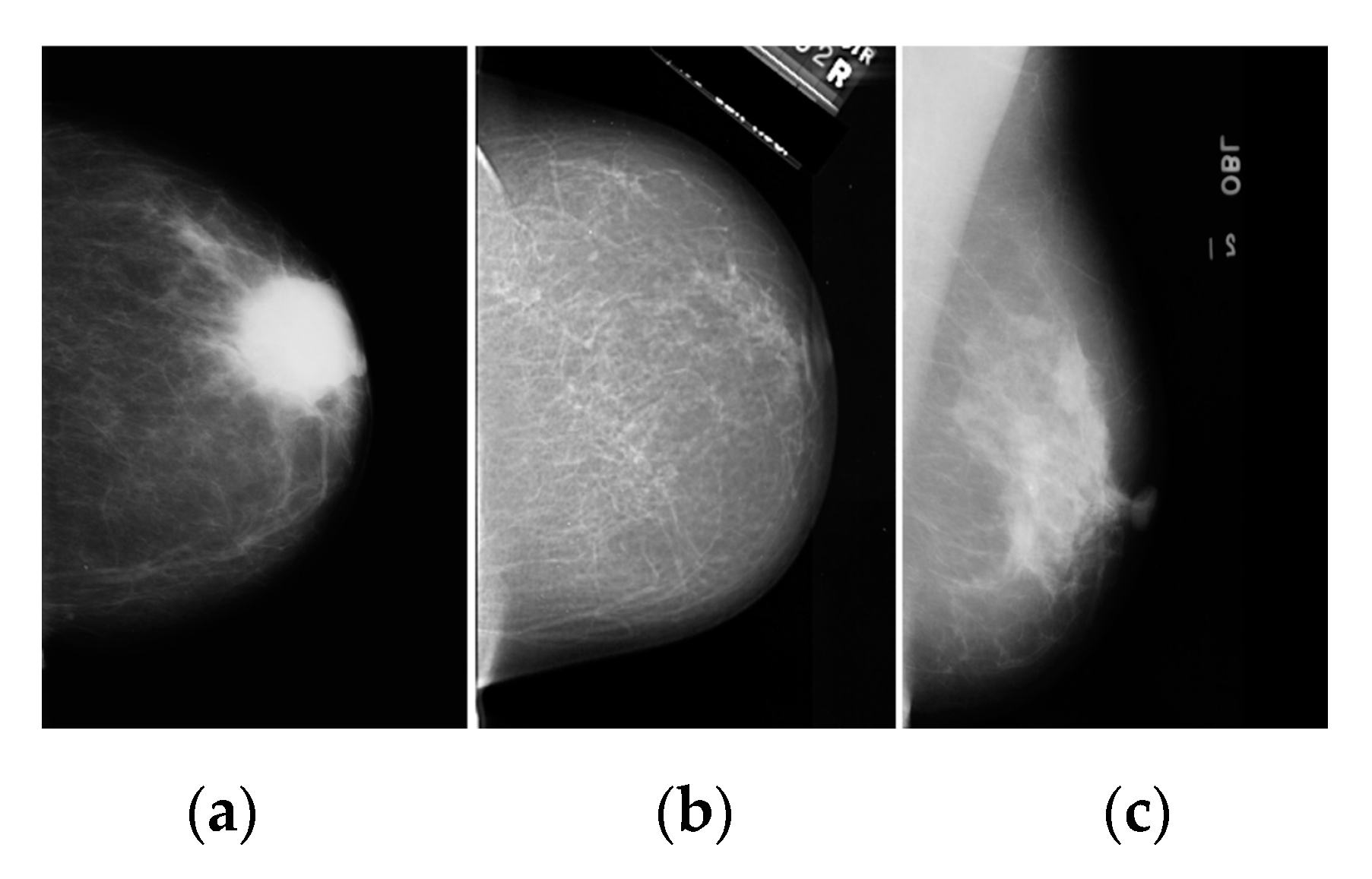
2.1. Dataset and Image Acquisition
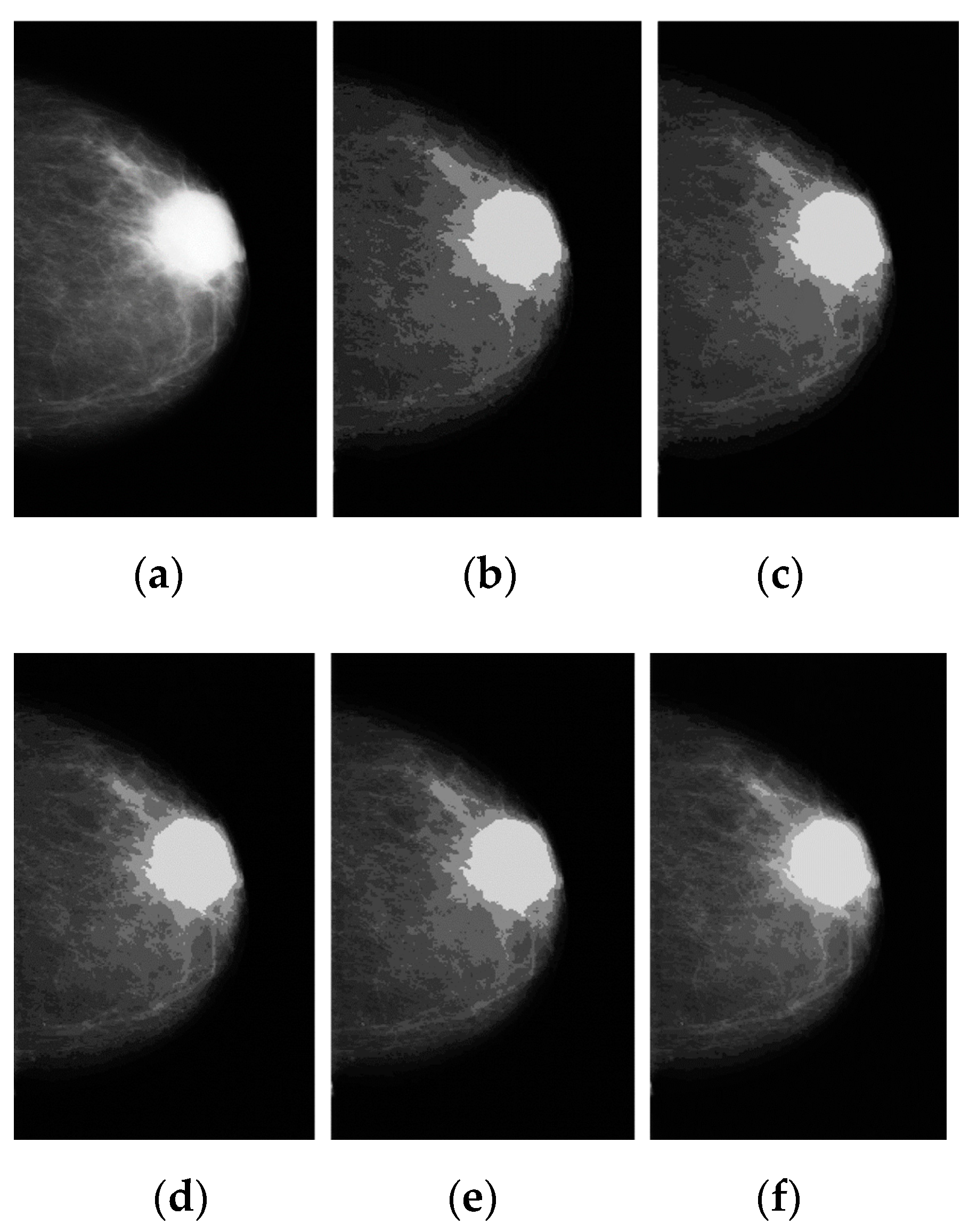
2.2. Image Color Normalization
2.3. Image Denoising
2.4. Breast Cancer Detection
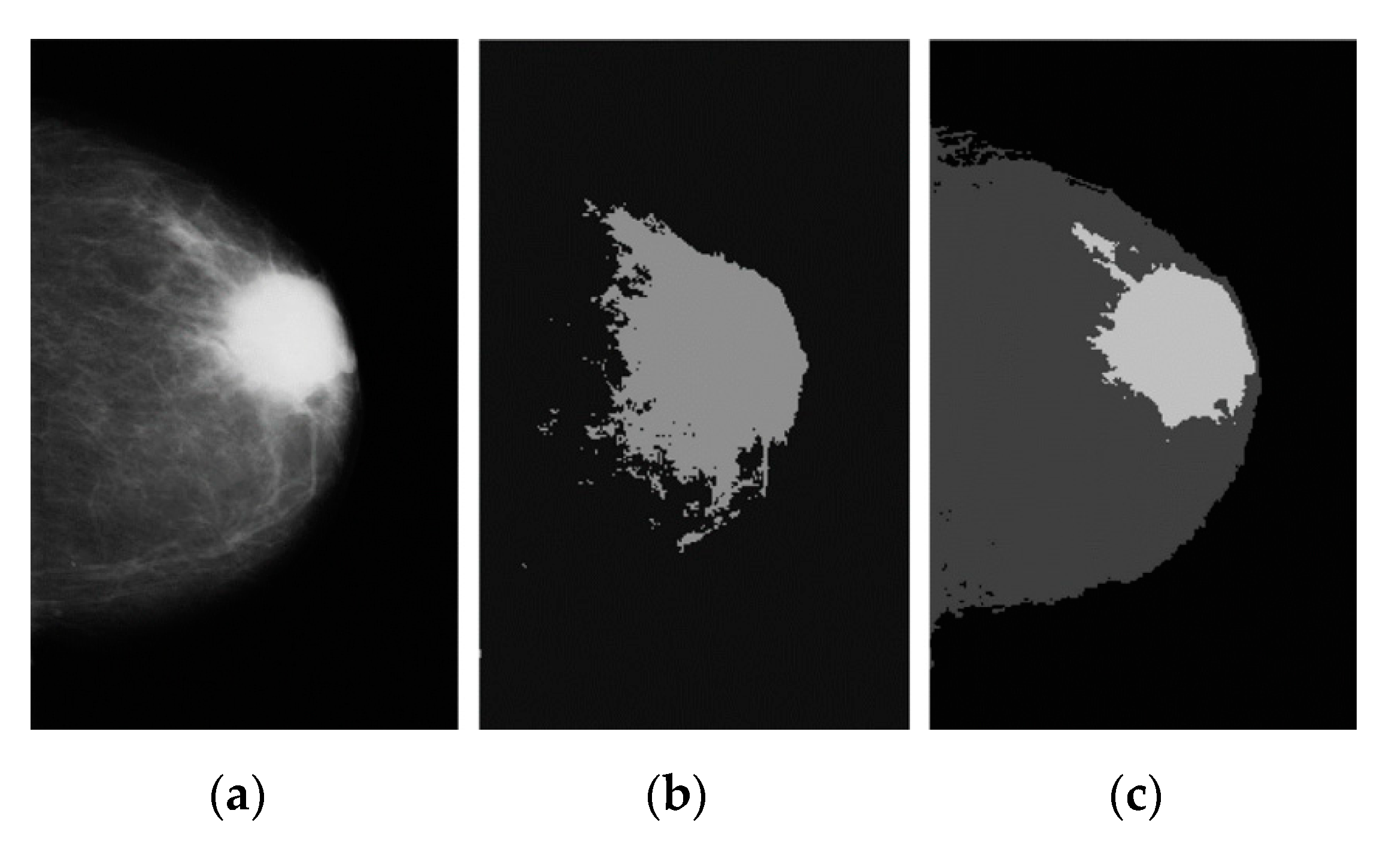
2.4.1. KM Clustering Method
2.4.2. FCM Clustering
2.4.3. FKM Clustering
2.4.4. Optimization Design with KM++CSO
| Algorithm 1: KM++CSO. |
|
2.4.5. Binary Image Detection
2.4.6. Improved Binary Image Using Mathematical Dilation Operators
2.4.7. Edge Detection
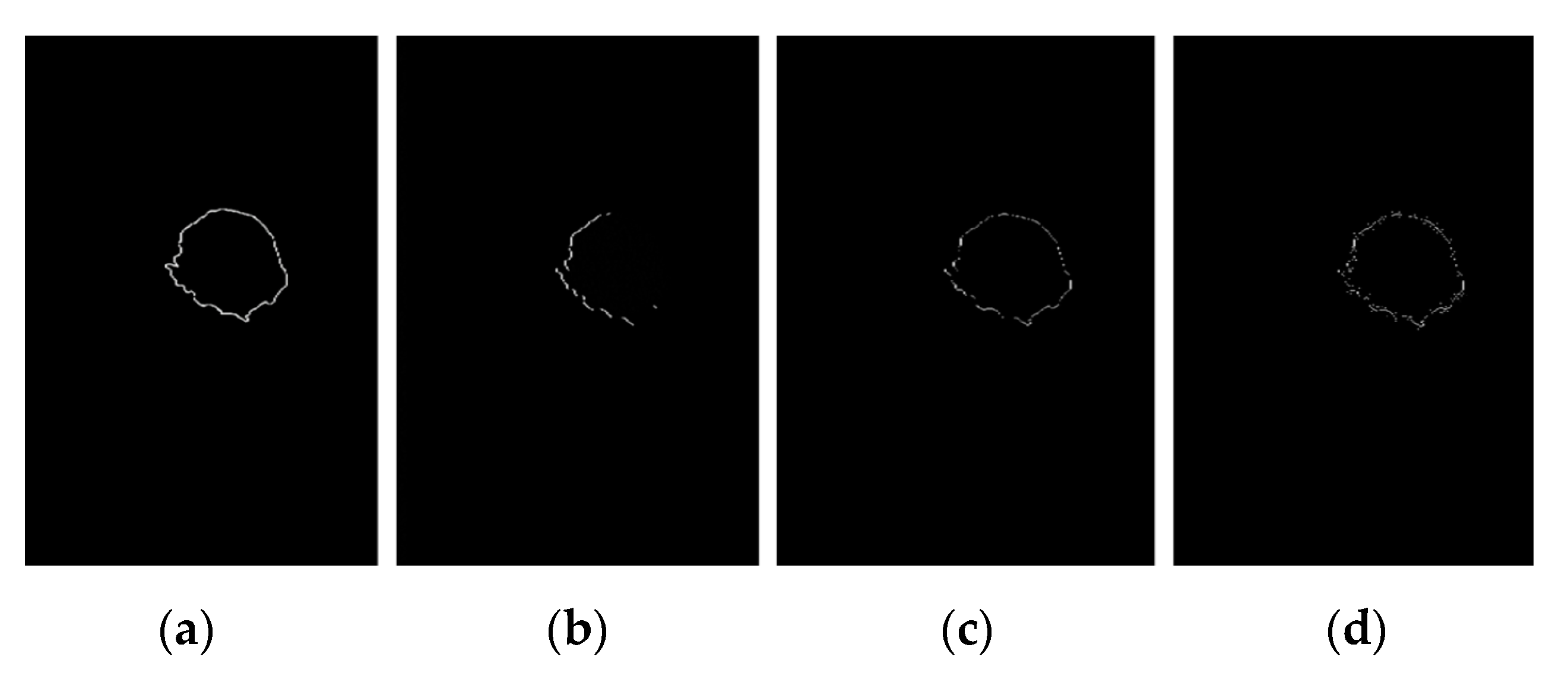
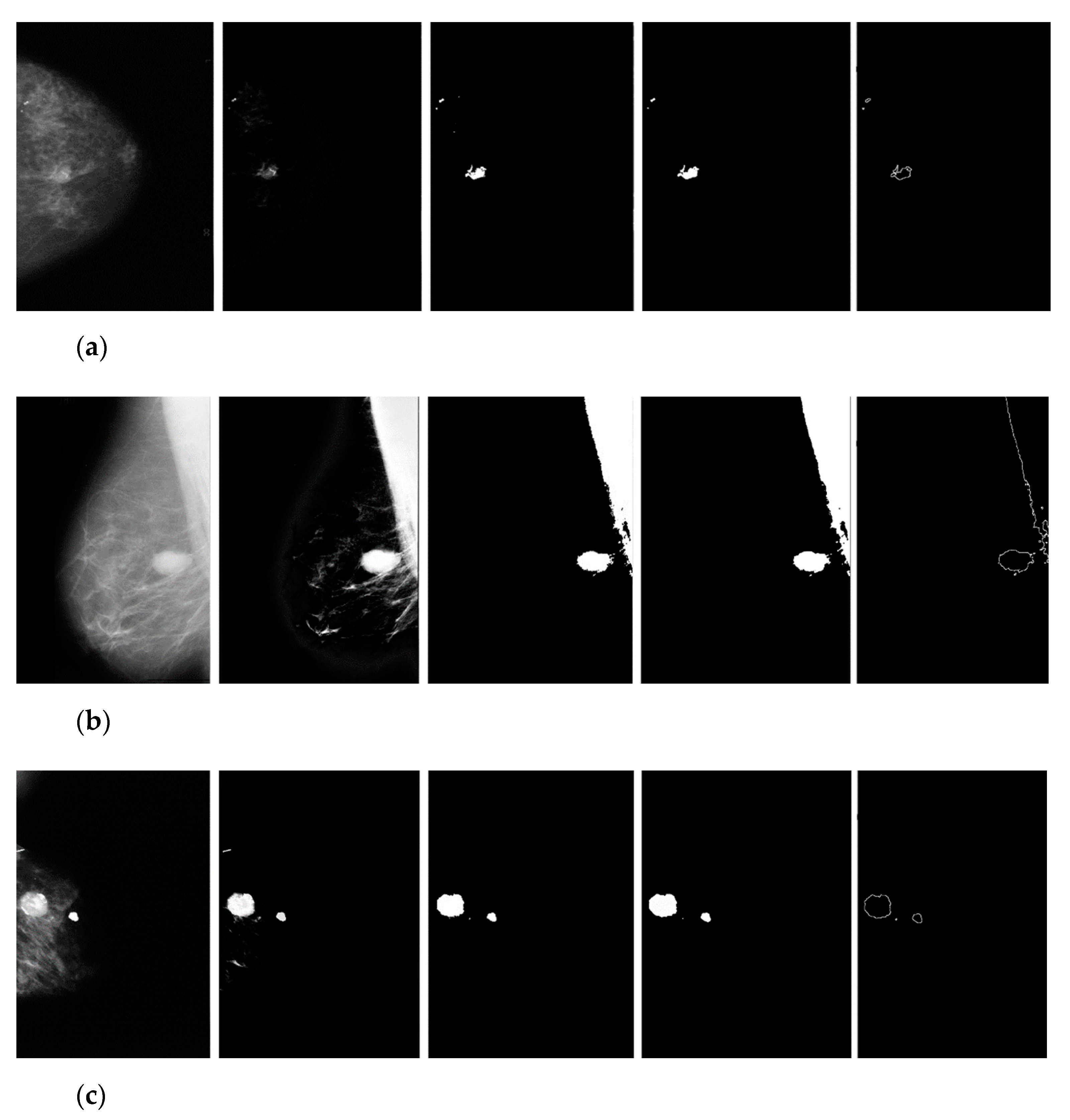
3. Results
3.1. Breast Cancer Detection Results
3.2. Manual Detection Versus Measurement
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, L.R.; Kimberly, M.D.; Hannah, F.E.; Jemal, A. Cancer statistics, CA: A cancer. J. Clin. 2021, 71, 7–33. [Google Scholar]
- Karabatak, M. A new classifier for breast cancer detection based on Naïve Bayesian. Measurement 2015, 72, 32–36. [Google Scholar] [CrossRef]
- Pak, F.; Kanan, H.R.; Alikhassi, A. Breast cancer detection and classification in digital mammography based on Non-Subsampled Contourlet Transform (NSCT) and Super Resolution. Comput. Methods Programs Biomed. 2015, 122, 89–107. [Google Scholar] [CrossRef] [PubMed]
- Sabu, R.S. Analysis of Mammographic Images for Early Detection of Breast Cancer Using Machine Learning Technique. Master’s Thesis, Department of Biotechnology and Medical Engineering, National Institute of Technology, Orissa, India, 2015. [Google Scholar]
- Yang, S.C. A robust approach for subject segmentation of medical Images: Illustration with mammograms and breast magnetic resonance images. Comput. Electr. Eng. 2017, 62, 151–165. [Google Scholar] [CrossRef]
- Punita, S.; Amuthan, A.; Joseph, K.S. Benign and malignant breast cancer segmentation using optimized region growing technique. Future Comput. Inform. J. 2018, 3, 348–358. [Google Scholar] [CrossRef]
- Al-masni, M.A.; Park, J.M.; Gi, G.; Kim, T.Y.; Rivera, P. Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Comput. Methods Programs Biomed. 2018, 157, 85–94. [Google Scholar] [CrossRef]
- Shen, T.; Gou, C.; Wang, J.; Wang, F.Y. Simultaneous segmentation and classification of mass region from mammograms using a mixed-supervision guided deep model. IEEE Signal Process. Lett. 2019, 27, 196–200. [Google Scholar] [CrossRef]
- Punita, M.; Perumal, K. Hybrid segmentation and feature extraction approach to detect tumors based on fuzzy rough-in mammogram images. Procedia Comput. Sci. 2019, 165, 478–484. [Google Scholar] [CrossRef]
- Zebari, D.A.; Zeebaree, D.Q.; Abdulazeez, A.M.; Haron, H.; Hamed, H.N.A. Improved threshold based and trainable fully automated segmentation for breast cancer boundary and pectoral muscle in mammogram images. IEEE Access 2020, 8, 203097–203116. [Google Scholar] [CrossRef]
- Vaka, A.R.; Soni, B.; Reddy, K.S. Breast cancer detection by leveraging machine learning. ICT Express 2020, 6, 320–324. [Google Scholar] [CrossRef]
- Azlan, N.A.N.; Lu, C.K.; Elamvazuthi, I.; Tang, T.B. Automatic detection of masses from mammographic images via artificial intelligence techniques. IEEE Sens. J. 2020, 21, 13094–13102. [Google Scholar] [CrossRef]
- George, K.; Faziludeen, S.; Sankaran, P.; Joseph, K.P. Breast cancer detection from biopsy images using nucleus guided transfer learning and belief-based fusion. Comput. Biol. Med. 2020, 124, 103954. [Google Scholar] [CrossRef] [PubMed]
- Alanazi, S.A.; Kamruzzaman, M.M.; Sarker, M.N.I.; Alruwaili, M.; Alhwaiti, Y. Boosting breast cancer detection using convolutional neural network. J. Healthc. Eng. 2021, 2021, 5528622. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.J.; Abbas, T. Comparison of a classifier performance testing methods: Support vector machine classifier on mammogram images classification. J. Kufa Math. Comput. 2019, 6, 8–12. [Google Scholar] [CrossRef]
- Agnes, S.A.; Anitha, J.; Pandian, S.I.A.; Peter, J.D. Classification of mammogram images using multiscale all convolutional neural network (MA-CNN). J. Med. Syst. 2020, 44, 30. [Google Scholar] [CrossRef] [PubMed]
- Yektaei, H.; Manthouri, M.; Farivar, F. Diagnosis of breast cancer using multi scale convolutional neural network. Biomed. Eng. Appl. Basis Commun. 2019, 31, 1950034. [Google Scholar] [CrossRef]
- Kaur, P.; Singh, G.; Kaur, P. Intellectual detection and validation of automated mammogram breast cancer images by multi-class SVM using deep learning classification. Inform. Med. Unlocked 2019, 16, 100151. [Google Scholar] [CrossRef]
- Viswanath, V.H.; Guachi, L.G.; Thirumuruganandham, S.P. Breast cancer detection using image processing techniques and classification algorithms. EasyChair 2019, 2101, 1–11. [Google Scholar]
- Jin, Y.; Zheng, Y. Medical image processing with deep learning: Mammogram classification and automatic lesion detection. Comput. Sci. Med. 2019, 1–19. Available online: https://nics.utk.edu/wp-content/uploads/sites/275/2021/08/19-Jin-final-report.pdf (accessed on 14 November 2022).
- Kayode, A.A.; Akande, N.O.; Adegun, A.A.; Adebiyi, M.O. An automated mammogram classification system using modified support vector machine. Med. Devices (Auckl.) 2019, 12, 275–284. [Google Scholar] [CrossRef]
- Debelee, T.G.; Gebreselasie, A.; Schwenker, F.; Amirian, M.; Ashenafi, D.Y. Classification of mammograms using texture and CNN based extracted features. J. Biomater. Biomed. Eng. 2019, 42, 79–97. [Google Scholar] [CrossRef]
- Zhang, Y.; Chan, S.; Park, V.Y.; Chang, K.T.; Mehta, S. Automatic detection and segmentation of breast cancer on MRI using mask R-CNN trained on non–fat-sat images and tested on fat-sat images. Acad. Radiol. 2020, 29, S135–S144. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, R.C. Digital Image Processing, 4th ed.; Pearson India: Bangalore, India, 2018. [Google Scholar]
- Bejnordi, B.E.; Litjens, G.; Timofeeva, N. Stain specific standardization of whole-slide histopathological images. IEEE Trans. Med. Imaging 2016, 35, 404–415. [Google Scholar] [CrossRef] [PubMed]
- Janowczyk, A.; Basavanhally, A.; Madabhushi, A. Stain normalization using sparse auto encoders (StaNoSA): Application to digital pathology. Comput. Med. Imaging Graph. 2017, 57, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.; Tapia, D.F.; Pistorius, S. Histogram specification: A fast and flexible method to process digital images. IEEE Trans. Instrum. Meas. 2011, 60, 1565–1578. [Google Scholar] [CrossRef]
- Gao, Z. An adaptive median filtering of salt and pepper noise based on local pixel distribution. In Proceedings of the 2018 International Conference on Transportation & Logistics, Information & Communication, Smart City (TLICSC 2018), Chengdu, China, 30–31 October 2018; Atlantis Press: Paris, France, 2018; Volume 161, pp. 473–483. [Google Scholar]
- Zheng, X.; Lei, Q.; Yao, R.; Gong, Y.; Yin, Q. Image segmentation based on adaptive k-means algorithm. EURASIP J. Image Video Process. 2018, 2018, 68. [Google Scholar] [CrossRef]
- Bezdek, J.C.; Ehrlich, R. FCM-the fuzzy c-means clustering-algorithm. Comput. Geosci. 1984, 10, 191–203. [Google Scholar] [CrossRef]
- Li, M.J.; Ng, M.K.; Cheung, Y.; Huang, J.Z. Agglomerative fuzzy k-means clustering algorithm with selection of number of clusters. IEEE Trans. Knowl. Data Eng. 2008, 20, 1519–1534. [Google Scholar] [CrossRef]
- Arthur, D.; Vassilvitskii, S. K-means++: The advantages of careful seeding. In Proceedings of the Eighteenth Annual ACM-SIAM Symposium on Discrete Algorithms, Society for Industrial and Applied Mathematics Philadelphia, New Orleans, LA, USA, 7–9 January 2007; pp. 1027–1035. [Google Scholar]
- Yang, X.S.; Deb, S. Cuckoo Search via Lévy flights. In Proceedings of the 2009 World Congress on Nature & Biologically Inspired Computing (NaBIC), Coimbatore, India, 9–11 December 2009; pp. 210–214. [Google Scholar]
- Bangare, S.L.; Dubal, A.; Bangare, P.S.; Patil, S. Reviewing Otsu’s method for image thresholding. Int. J. Appl. Eng. Res. 2015, 10, 21777–21783. [Google Scholar] [CrossRef]
- Chudasama, D.; Patel, T.A.; Joshi, S.; Prajapati, G. Image segmentation using morphological operations. Int. J. Comput. Appl. 2015, 117, 16–19. [Google Scholar] [CrossRef]
- Jang, J.; Lee, S.; Hwang, H.J.; Baek, K. Global thresholding algorithm based on boundary selection. In Proceedings of the 2013 13th International Conference on Conference: Control, Automation and Systems (ICCAS), Gwangju, Korea, 20–23 October 2013; pp. 1–3. [Google Scholar]
- Han, L.; Tian, Y.; Qi, Q. Research on edge detection algorithm based on improved Sobel operator. In Proceedings of the 2019 International Conference on Computer Science Communication and Network Security (CSCNS2019), Sanya, China, 22–23 December 2019; Volume 309, pp. 2–6. [Google Scholar]
- Shrivakshan, G.T.; Chandramouli, C. A comparison of various edge detection techniques used in image processing. Int. J. Comput. Sci. 2012, 9, 269–276. [Google Scholar]
- Ferhat, F.A.; Ait, L.; Kerdjidj, O.; Messaoudi, K.; Abdelwahhab, B. Implementation of sobel, prewitt, roberts edge detection on FPGA. In Proceedings of the 2013 World Congress in Computer Science, Computer Engineering, and Applied Computing, Las Vegas, NV, USA, 22–25 July 2013; pp. 1–4. [Google Scholar]
- Wong, H.B.; Lim, G.H. Measures of diagnostic accuracy: Sensitivity, specificity, PPV and NPV. Proc. Singap. Healthc. 2011, 20, 316–318. [Google Scholar] [CrossRef]
- Fletcher, S.; Islam, M.Z. Comparing sets of patterns with the Jaccard index. Australas. J. Inf. Syst. 2018, 22, 1–17. [Google Scholar] [CrossRef]







| Authors | Methods | Dataset | SEN (%) | SPEC (%) | ACC (%) |
|---|---|---|---|---|---|
| Karabatak et al. [2] | Naïve Bayesian | Wisconsin breast cancer | 99.11 | 98.25 | 98.54 |
| Pak et al. [3] | SVM and ANN | Mini–MIAS and DDSM | SVM = 100 | SVM = 80 | SVM = 92.85 |
| ANN = 87.50 | ANN = 93.33 | ANN = 91.31 | |||
| Yang et al. [5] | PSCSM | Mini–MIAS | – | – | 90.9 |
| Punita et al. [6] | RGM, MRF segmentation, Stochastic Relaxation methods, and Fuzzy | DDSM | 98.10 | 97.8 | 98.00 |
| Al-masni et al. [7] | Regional deep learning based on CNN and CLAHE | IN Breast | 97.14 | 92.41 | 95.64 |
| Shen et al. [8] | MSGDM | IN Breast | 97.56 | 88.89 | 94.16 |
| Punita et al. [9] | Fuzzy Rough Segmentation (FR), Rough Segmentation (RS), and Fuzzy Segmentation (FS) | Mini–MIAS | FR = 91.23 | FR = 91.23 | FR = 97.45 |
| RS = 80.25 | RS = 79.45 | RS = 82.45 | |||
| FS = 89.89 | FS = 89.12 | FS = 90.12 | |||
| Zebari et al. [10] | The thresholding method and binary morphological for segmentation | Mini–MIAS, IN BCDR | 98.9 | 98.4 | 98.13 |
| Anji et al. [11] | DNNS | MG Cancer | – | – | 97.21 |
| Azlan et al. [12] | PCA and SVM | DDSM and Mini–MIAS | 93.94 | 96.61 | 95.24 |
| George et al. [13] | The hybrid methods by combined the CNN and SVM | BreaKHis | 97.24 | 96.18 | 96.21 |
| Alanazi et al. [14] | CNN architectures | Kaggle 162 H&E | - | - | 87.00 |
| Mohammed et al. [15] | SVM | Mini–MIAS | - | - | 91.12 |
| Agnes et al. [16] | MA-CNN | Mini–MIAS | 96.00 | - | - |
| Yektaei et al. [17] | Multiscale Convolutional Neural Network (MCNN) | Mini–MIAS | 95.90 | - | 97.03 |
| Kaur et al. [18] | Multiclass Support Vector Machine (MSVM) | Mini–MIAS | - | - | 96.90 |
| Viswanath et al. [19] | SVM | Raw sample images | – | 68 | 84.84 |
| Jin et al. [20] | Binary classifier with CNNI–BCC | Mini–MIAS | – | – | 73.24 |
| Kayode et al. [21] | SVM | Mini–MIAS | 94.4 | 91.3 | – |
| Debelee et al. [22] | SVM and MLP | Mini–MIAS | SVM = 99.48 | SVM = 98.16 | SVM = 99 |
| MLP = 97.40 | MLP = 96.26 | MLP = 97 | |||
| Zhang et al. [23] | Mask R–CNN | DCE–MRI | 80 | 74 | 75 |
| Methods | Datasets | Percentage | |||
|---|---|---|---|---|---|
| SEN (%) | SPEC (%) | ACC (%) | Jaccard Index (%) | ||
| KM | DDSM | 88.38 | 89.10 | 88.89 | 80.45 (2108/2620) |
| FCM | 67.22 | 69.12 | 69.10 | 70.95 (1859/2620) | |
| FKM | 90.15 | 90.18 | 90.16 | 83.55 (2189/2620) | |
| KM++CSO | 95.68 | 95.10 | 95.49 | 87.37 (2289/2620) | |
| KM | Mini–MIAS | 90.10 | 89.78 | 89.92 | 85.13 (275/323) |
| FCM | 60.67 | 60.97 | 60.78 | 64.08 (207/323) | |
| FKM | 91.12 | 91.20 | 91.14 | 86.99 (281/323) | |
| KM++CSO | 96.78 | 97.10 | 96.92 | 89.47 (289/323) | |
| KM | BCDR | 89.96 | 89.89 | 89.92 | 78.86 (97/123) |
| FCM | 61.29 | 61.19 | 61.20 | 65.85 (81/123) | |
| FKM | 91.40 | 91.56 | 91.48 | 88.62 (109/123) | |
| KM++CSO | 96.90 | 96.16 | 96.42 | 91.05 (112/123) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wisaeng, K. Breast Cancer Detection in Mammogram Images Using K–Means++ Clustering Based on Cuckoo Search Optimization. Diagnostics 2022, 12, 3088. https://doi.org/10.3390/diagnostics12123088
Wisaeng K. Breast Cancer Detection in Mammogram Images Using K–Means++ Clustering Based on Cuckoo Search Optimization. Diagnostics. 2022; 12(12):3088. https://doi.org/10.3390/diagnostics12123088
Chicago/Turabian StyleWisaeng, Kittipol. 2022. "Breast Cancer Detection in Mammogram Images Using K–Means++ Clustering Based on Cuckoo Search Optimization" Diagnostics 12, no. 12: 3088. https://doi.org/10.3390/diagnostics12123088
APA StyleWisaeng, K. (2022). Breast Cancer Detection in Mammogram Images Using K–Means++ Clustering Based on Cuckoo Search Optimization. Diagnostics, 12(12), 3088. https://doi.org/10.3390/diagnostics12123088





