An Automatic Segmentation Method for Lung Tumor Based on Improved Region Growing Algorithm
Abstract
:1. Introduction
- (1)
- Combining the prior information of lung tumor with the maximum inter-class variance method (OTSU) algorithm, the method of lung tumor location is constructed, and then the automatic selection of initial seed points of region growing algorithm is realized;
- (2)
- After the seed point expansion, the growth restriction conditions and the automatic updating mechanism of the threshold are established. Finally, the combination result of multi-point growth is taken as the final segmentation result;
- (3)
- The segmentation performance of our proposed method is better than the current popular segmentation algorithm.
2. Related Work
3. Materials and Methods
3.1. CT Data Preprocess
3.2. Region Growing Algorithm
- (1)
- Import a CT slice of the lung with the tumor and serve as an input image;
- (2)
- The user manually determines a point on the tumor as the initial seed point;
- (3)
- The color intensity of the initial seed point is saved as a base value;
- (4)
- Set the threshold;
- (5)
- Similarity check;
- (6)
- The adjacent pixels meeting the conditions are added according to the growth rules and saved as growth points;
- (7)
- Check the new adjacent pixels again, add the adjacent pixels that meet the conditions, and save them as growth points;
- (8)
- Until there are no new growth points, the array of obtained pixel points is the segmented tumor area, and the outermost pixels are the segmented tumor boundary.
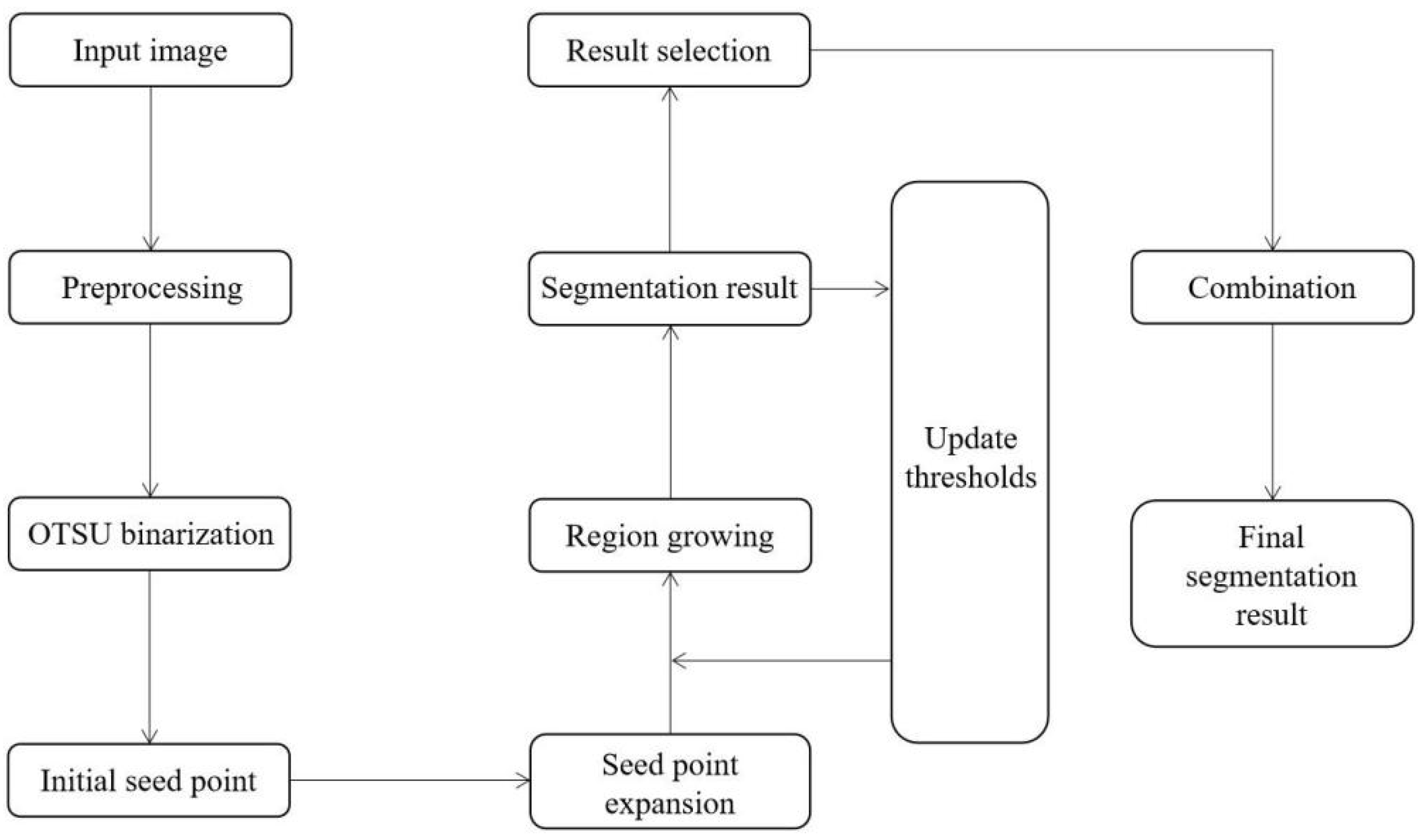
3.3. Improved Region Growing Algorithm
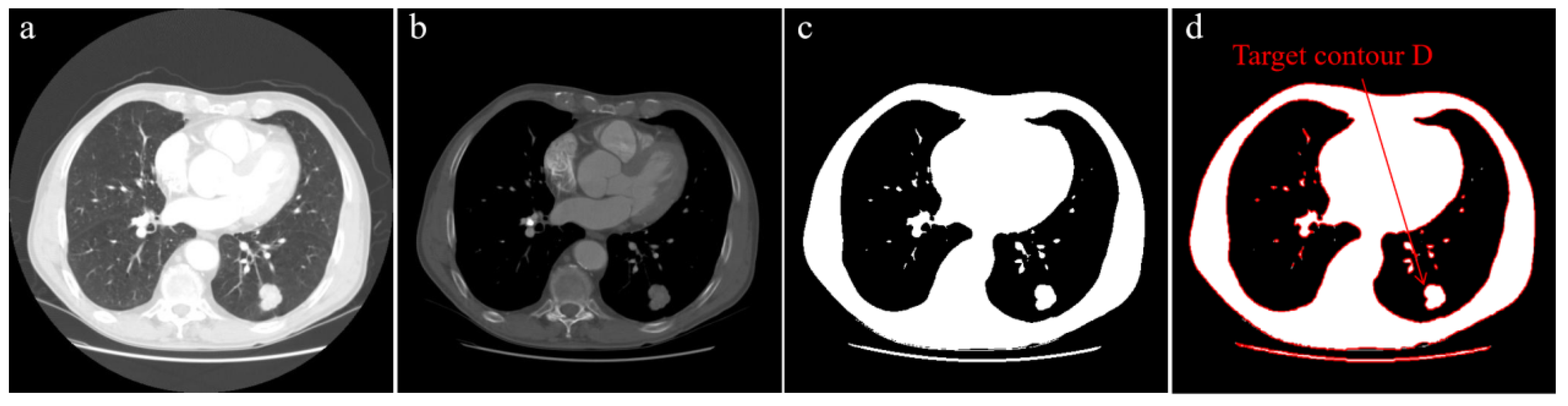
3.3.1. Automatic Selection of Initial Seed Point
- (1)
- (2)
- Extract all contours in the binary image and draw them, as shown in Figure 1d;
- (3)
- Calculate the area and maximum size of all contours, find the contour that conforms to the attribute and set it as target contour D;
- (4)
- Find the centroid C of target contour D by Equation (1), and set the centroid C as the initial seed point.
3.3.2. Initial Seed Point Expansion
3.3.3. Restrictions
3.3.4. Threshold Detection and Update
3.3.5. Automatic Segmentation Steps
- (1)
- Import a CT slice of the lung with the tumor and serve as an input image;
- (2)
- Preprocess the input image;
- (3)
- The binary image is obtained by using the OTSU algorithm;
- (4)
- The initial seed point is automatically obtained;
- (5)
- Initial seed point expansion;
- (6)
- Set the threshold value as the 20% gray threshold of the input image;
- (7)
- The color intensity of the initial seed point is saved as a base value;
- (8)
- Similarity check;
- (9)
- The adjacent pixels meeting the conditions are added according to the growth rules and saved as growth points;
- (10)
- Check the new adjacent pixels again, add the adjacent pixels that meet the conditions, and save them as growth points;
- (11)
- Until there are no new growth points, the array of obtained pixel points is the segmented tumor area, and the outermost pixels are the segmented tumor boundary;
- (12)
- Automatically update thresholds;
- (13)
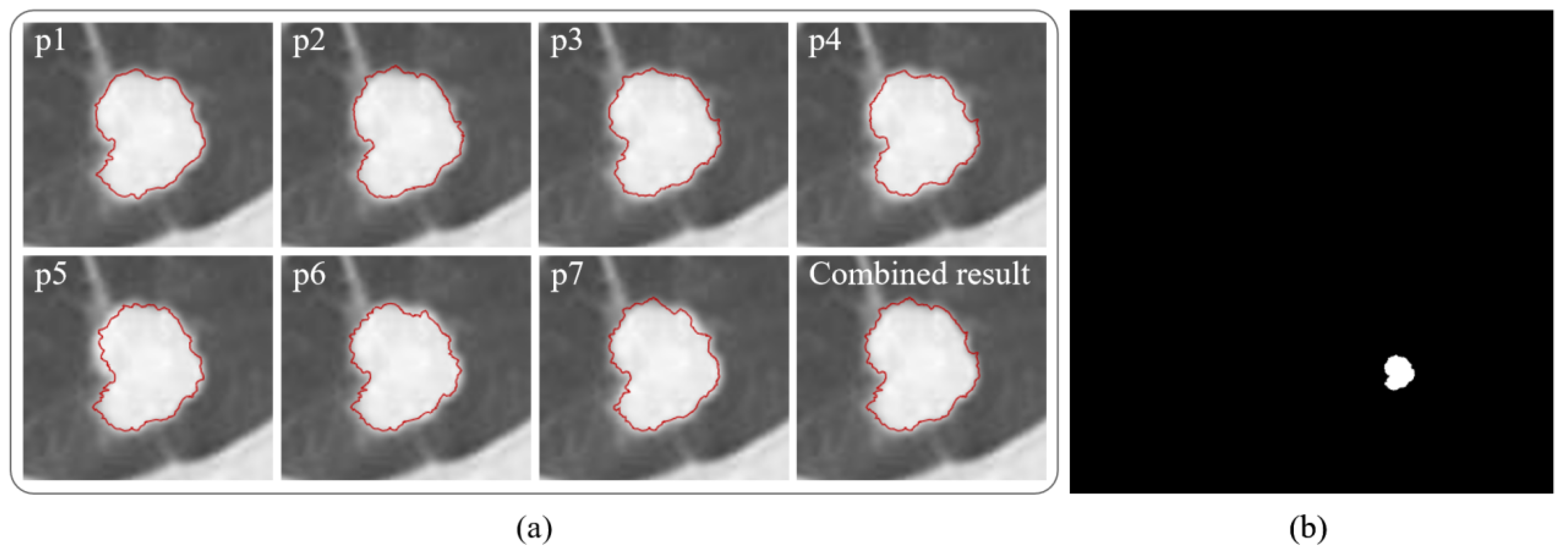
- Repeat steps (7) to (11) with the new threshold obtained, and finally obtain seven tumor boundaries from p1 to p7 of region growth;
- (14)
- According to the growth restriction conditions, the combination of all credible segmentation results obtained from p1–p7 is selected as the final segmentation result.
4. Experiments and Validation
4.1. Performance Evaluation Metrics
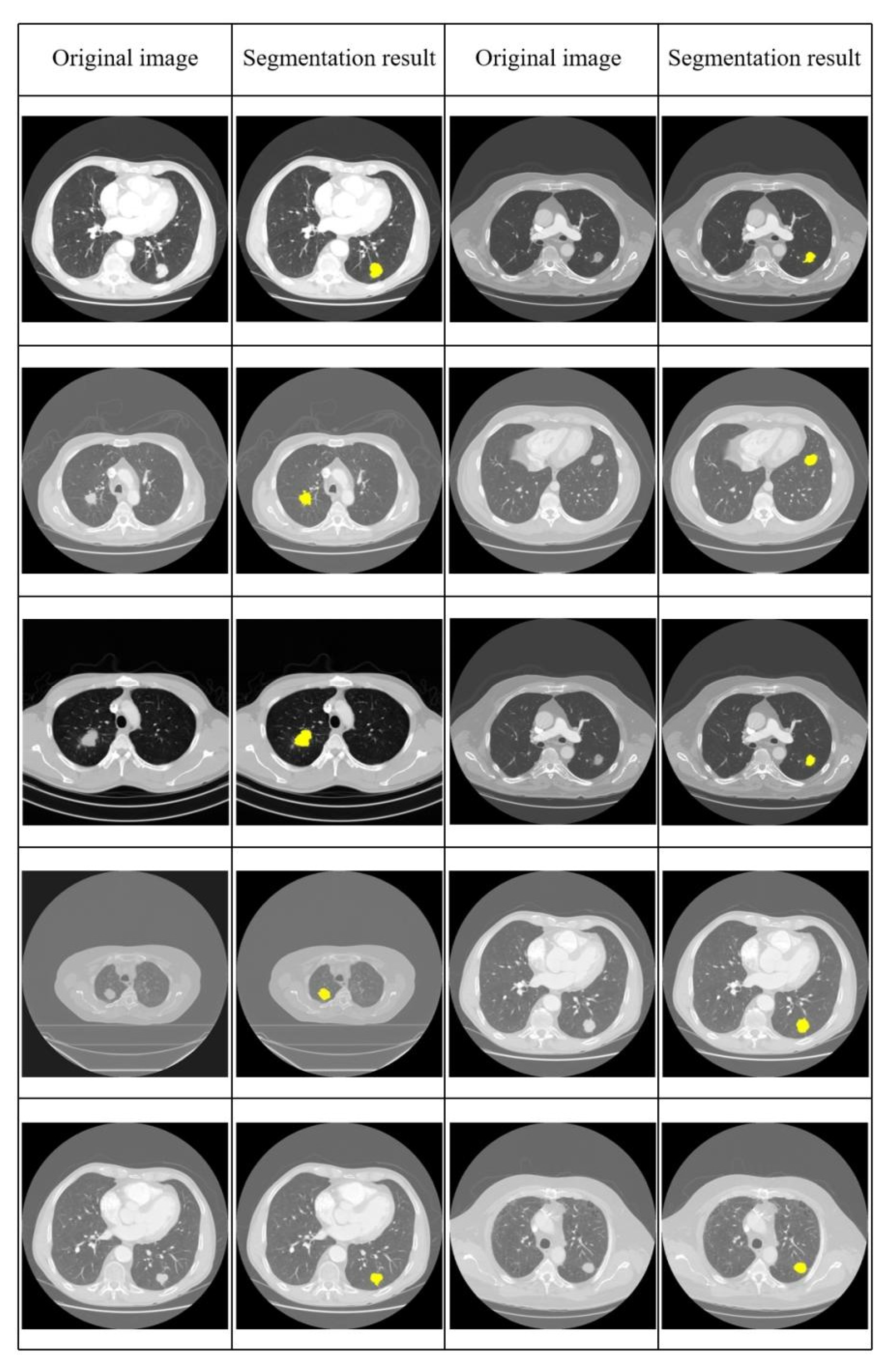
4.2. Results and Performance Comparison
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yu, X.Q.; Luo, Q.; Hughes, S.; Wade, S.; Caruana, M.; Canfell, K.; O’Connell, D.L. Statistical projection methods for lung cancer incidence and mortality: A systematic review. BMJ Open 2019, 9, e028497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLellan, R.; Marshall, H.; Dent, A.; Bowman, R.V.; Yang, I.A.; Fong, K.M. Diagnosis and treatment of early lung cancer. Aust. J. Gen. Pract. 2020, 49, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.T.; Wu, X.; Wen, B.C.; Fowler, J.F.; Fayad, F.; Amendola, B.E.; García, S.; Zerda, A.D.L.; Huang, Z.; Schwade, J.G. Early results of CyberKnife image-guided robotic stereotactic radiosurgery for treatment of lung tumors. Comput. Aided Surg. 2007, 12, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Saboury, B.; Torigian, D.A.; Alavi, A. Current evidence base of FDG-PET/CT imaging in the clinical management of malignant pleural mesothelioma: Emerging significance of image segmentation and global disease assessment. Mol. Imaging Biol. 2011, 13, 801–811. [Google Scholar] [CrossRef]
- Gibbs, J.D.; Graham, M.W.; Higgins, W.E. 3D MDCT-based system for planning peripheral bronchoscopic procedures. Comput. Biol. Med. 2009, 39, 266–279. [Google Scholar] [CrossRef] [Green Version]
- Collins, F.S.; Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 2015, 372, 793–795. [Google Scholar] [CrossRef] [Green Version]
- Reardon, S. Precision-medicine plan raises hopes: US initiative highlights growing focus on targeted therapies. Nature 2015, 517, 540–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, T.; Furukawa, T.; Lami, K.; Pham, H.H.N.; Uegami, W.; Kuroda, K.; Kawai, M.; Sakanashi, H.; Cooper, L.A.D.; Bychkov, A.; et al. A narrative review of digital pathology and artificial intelligence: Focusing on lung cancer. Transl. Lung Cancer Res. 2020, 9, 2255. [Google Scholar] [CrossRef]
- Carlsen, E.A.; Lindholm, K.; Hindsholm, A.; Gæde, M.; Ladefoged, C.N.; Loft, M.; Johnbeck, C.B.; Langer, S.W.; Oturai, P.; Knigge, U.; et al. A convolutional neural network for total tumor segmentation in [64Cu] Cu-DOTATATE PET/CT of patients with neuroendocrine neoplasms. EJNMMI Res. 2022, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Pfaehler, E.; Mesotten, L.; Kramer, G.; Thomeer, M.; Vanhove, K.; Jong, J.D.; Adriaensens, P.; Hoekstra, O.S.; Boellaard, R. Repeatability of two semi-automatic artificial intelligence approaches for tumor segmentation in PET. EJNMMI Res. 2021, 11, 4. [Google Scholar] [CrossRef]
- Kalpathy-Cramer, J.; Zhao, B.; Goldgof, D.; Gu, Y.; Wang, X.; Yang, H.; Tan, Y.; Gillies, R.; Napel, S. A comparison of lung nodule segmentation algorithms: Methods and results from a multi-institutional study. J. Digit. Imaging 2016, 29, 476–487. [Google Scholar] [CrossRef] [Green Version]
- Devi, K.Y.; Sasikala, M. Labeling and clustering-based level set method for automated segmentation of lung tumor stages in CT images. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 2299–2309. [Google Scholar] [CrossRef]
- Comelli, A.; Stefano, A.; Russo, G.; Bignardi, S.; Sabini, M.G.; Petrucci, G.; Ippolito, M.; Yezzi, A. K-nearest neighbor driving active contours to delineate biological tumor volumes. Eng. Appl. Artif. Intell. 2019, 81, 133–144. [Google Scholar] [CrossRef]
- Nithila, E.E.; Kumar, S.S. Segmentation of lung nodule in CT data using active contour model and Fuzzy C-mean clustering. Alex. Eng. J. 2016, 55, 2583–2588. [Google Scholar] [CrossRef] [Green Version]
- Zhou, N.; Yang, T.; Zhang, S. An improved FCM medical image segmentation algorithm based on MMTD. Comput. Math. Method Med. 2014, 2014, 690349. [Google Scholar] [CrossRef] [Green Version]
- Afshar, P.; Ahmadi, A.; Zarandi, M.H.F. Lung tumor area recognition in CT images based on Gustafson-Kessel clustering. In Proceedings of the 2016 IEEE International Conference on Fuzzy Systems (FUZZ-IEEE), IEEE, Vancouver, BC, Canada, 24–29 July 2016; pp. 2302–2308. [Google Scholar]
- Suzuki, K.; Kohlbrenner, R.; Epstein, M.L.; Obajuluwa, A.M.; Xu, J.; Hori, M. Computer-aided measurement of liver volumes in CT by means of geodesic active contour segmentation coupled with level-set algorithms. Med. Phys. 2010, 37, 2159–2166. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Li, C.; Sun, Q.; Xia, D.; Kao, C.Y. Active contours driven by local and global intensity fitting energy with application to brain MR image segmentation. Comput. Med. Imaging Graph. 2009, 33, 520–531. [Google Scholar] [CrossRef]
- Nock, R.; Nielsen, F. Statistical region merging. IEEE Trans. Pattern Anal. Mach. Intell. 2004, 26, 1452–1458. [Google Scholar] [CrossRef]
- Cheng, J.; Li, L.; Luo, B.; Wang, S.; Liu, H. High-resolution remote sensing image segmentation based on improved RIU-LBP and SRM. EURASIP J. Wirel. Commun. Netw. 2013, 2013, 263. [Google Scholar] [CrossRef] [Green Version]
- Dlamini, S.; Chen, Y.H.; Kuo, C.F.J. Complete fully automatic detection, segmentation and 3D reconstruction of tumor volume for non-small cell lung cancer using YOLOv4 and region-based active contour model. Expert Syst. Appl. 2023, 212, 118661. [Google Scholar] [CrossRef]
- Gan, W.; Wang, H.; Gu, H.; Duan, Y.; Shao, Y.; Chen, H.; Feng, A.; Huang, Y.; Fu, X.; Ying, Y.; et al. Automatic segmentation of lung tumors on CT images based on a 2D & 3D hybrid convolutional neural network. Br. J. Radiol. 2021, 94, 20210038. [Google Scholar]
- Wang, B.; Chen, K.; Tian, X.; Yang, Y.; Zhang, X. An effective deep network for automatic segmentation of complex lung tumors in CT images. Med. Phys. 2021, 48, 5004–5016. [Google Scholar] [CrossRef]
- Jiang, J.; Hu, Y.C.; Liu, C.J.; Halpenny, D.; Hellmann, M.D.; Deasy, J.O.; Mageras, G.; Veeraraghavan, H. Multiple resolution residually connected feature streams for automatic lung tumor segmentation from CT images. IEEE Trans. Med. Imaging 2018, 38, 134–144. [Google Scholar] [CrossRef]
- Zhang, G.; Yang, Z.; Jiang, S. Automatic lung tumor segmentation from CT images using improved 3D densely connected UNet. Med. Biol. Eng. Comput. 2022, 60, 3311–3323. [Google Scholar] [CrossRef]
- Cui, H.; Wang, X.; Zhou, J.; Eberl, S.; Yin, Y.; Feng, D.; Fulham, M. Topology polymorphism graph for lung tumor segmentation in PET-CT images. Phys. Med. Biol. 2015, 60, 4893. [Google Scholar] [CrossRef] [PubMed]
- Anshad, P.Y.; Kumar, S.S.; Shahudheen, S. Segmentation of chondroblastoma from medical images using modified region growing algorithm. Clust. Comput. 2019, 22, 13437–13444. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, R.; Zhang, M.; Lin, Y.; Zhang, X.; Wen, Y.; Yang, L.; Huang, Z.; Wang, G.; Zhao, D.; et al. Segmental resection is associated with decreased survival in patients with stage IA non-small cell lung cancer with a tumor size of 21–30 mm. Transl. Lung Cancer Res. 2021, 10, 900. [Google Scholar] [CrossRef] [PubMed]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V. The IASLC lung cancer staging project: Proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goh, T.Y.; Basah, S.N.; Yazid, H.; Safar, M.J.A.; Saad, F.S.A. Performance analysis of image thresholding: Otsu technique. Measurement 2018, 114, 298–307. [Google Scholar] [CrossRef]
- Huo, J.; Okada, K.; Rikxoort, E.M.V.; Kim, H.J.; Alger, J.R.; Pope, W.B.; Goldin, J.G.; Brown, M.S. Ensemble segmentation for GBM brain tumors on MR images using confidence-based averaging. Med. Phys. 2013, 40, 093502. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Kumar, V.; Hall, L.O.; Goldgof, D.B.; Li, C.Y.; Korn, R.; Bendtsen, C.; Velazquez, E.R.; Dekker, A.; Aerts, H.; et al. Automated delineation of lung tumors from CT images using a single click ensemble segmentation approach. Pattern Recognit. 2013, 46, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, M.S.; Shanthini, J.; Karthikeyan, N. Volumetric analysis framework for accurate segmentation and classification (VAF-ASC) of lung tumor from CT images. Soft Comput. 2020, 24, 18489–18497. [Google Scholar] [CrossRef]



| Study | Method | Dataset | Automation |
|---|---|---|---|
| Afshar et al. [16] | Snake algorithm + Gustafson–Kessel clustering | Unpublished | Semi-automatic |
| Dlamini et al. [21] | Region-based active contour | LICD-IDRI | Automatic |
| Gan et al. [22] | Hybrid convolution neural network | Unpublished | Automatic |
| Wang et al. [23] | Deep learning | Unpublished | Automatic |
| Jiang et al. [24] | Deep learning | LICD-IDRI + NSCLC | Automatic |
| Zhang et al. [25] | Deep learning | LICD-IDRI | Automatic |
| Cui et al. [26] | Topo-poly graph model | NSCLC | Semi-automatic |
| Anshad et al. [27] | Modified region growing algorithm | Unpublished | Semi-automatic |
| Dice Coefficients | Jaccard Distance | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Images | FCM | K-Means | SRM | Active Contour | Ours | Images | FCM | K-Means | SRM | Active Contour | Ours |
| 1 | 0.86 | 0.86 | 0.87 | 0.84 | 0.95 | 1 | 0.24 | 0.24 | 0.23 | 0.27 | 0.09 |
| 2 | 0.61 | 0.85 | 0.91 | 0.61 | 0.92 | 2 | 0.56 | 0.26 | 0.16 | 0.56 | 0.14 |
| 3 | 0.78 | 0.79 | 0.85 | 0.75 | 0.93 | 3 | 0.36 | 0.34 | 0.26 | 0.40 | 0.13 |
| 4 | 0.92 | 0.92 | 0.91 | 0.82 | 0.91 | 4 | 0.14 | 0.14 | 0.16 | 0.30 | 0.16 |
| 5 | 0.85 | 0.95 | 0.95 | 0.81 | 0.95 | 5 | 0.26 | 0.09 | 0.09 | 0.31 | 0.09 |
| 6 | 0.87 | 0.88 | 0.91 | 0.87 | 0.97 | 6 | 0.23 | 0.21 | 0.16 | 0.23 | 0.05 |
| 7 | 0.77 | 0.78 | 0.83 | 0.73 | 0.93 | 7 | 0.37 | 0.36 | 0.29 | 0.42 | 0.13 |
| 8 | 0.85 | 0.89 | 0.90 | 0.71 | 0.88 | 8 | 0.26 | 0.19 | 0.18 | 0.44 | 0.21 |
| 9 | 0.92 | 0.96 | 0.92 | 0.84 | 0.97 | 9 | 0.14 | 0.07 | 0.14 | 0.27 | 0.05 |
| 10 | 0.86 | 0.95 | 0.91 | 0.82 | 0.95 | 10 | 0.24 | 0.09 | 0.16 | 0.30 | 0.09 |
| Study | Dice Coefficient (Average) | Jaccard Distance (Average) |
|---|---|---|
| FCM | 0.829 | 0.280 |
| K-means | 0.883 | 0.199 |
| SRM | 0.896 | 0.183 |
| Active contour | 0.780 | 0.350 |
| Dlamini et al. [21] | 0.921 | - |
| Gan et al. [22] | 0.720 | 0.420 |
| Wang et al. [23] | 0.750 | - |
| Jiang et al. [24] | 0.710 | - |
| Zhang et al. [25] | 0.831 | - |
| Cui et al. [26] | 0.881 | - |
| Anshad et al. [27] | 0.912 | 0.161 |
| Ours | 0.936 | 0.114 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Li, D. An Automatic Segmentation Method for Lung Tumor Based on Improved Region Growing Algorithm. Diagnostics 2022, 12, 2971. https://doi.org/10.3390/diagnostics12122971
Wang M, Li D. An Automatic Segmentation Method for Lung Tumor Based on Improved Region Growing Algorithm. Diagnostics. 2022; 12(12):2971. https://doi.org/10.3390/diagnostics12122971
Chicago/Turabian StyleWang, Monan, and Donghui Li. 2022. "An Automatic Segmentation Method for Lung Tumor Based on Improved Region Growing Algorithm" Diagnostics 12, no. 12: 2971. https://doi.org/10.3390/diagnostics12122971
APA StyleWang, M., & Li, D. (2022). An Automatic Segmentation Method for Lung Tumor Based on Improved Region Growing Algorithm. Diagnostics, 12(12), 2971. https://doi.org/10.3390/diagnostics12122971






