1. Introduction
Knee osteoarthritis (OA) is a degenerative disease of the knee joint, which affects three compartments of the knee (lateral, medial, and patella-femoral) and generally develops gradually over 10 to 15 years [
1,
2]. Usually, it results from wear, tear, and progressive loss of articular, followed by infections that damage the joint cavity, causing discomforts such as mobility limitations, joint pain, and swelling [
3]. All joints of the body are somewhat sensitive to alterations and damage to the cartilage tissue, with the knee and hip joints being more susceptible to OA due to their weight-bearing nature. Furthermore, knee OA mostly occurs in people over 55 years old, with a higher prevalence among those over 65 [
4]. By the year 2050, researchers estimate that 130 million individuals worldwide will suffer from knee OA. However, early detection and treatment of knee OA help reduce its progression and improve people’s quality of life [
5].
The cause of OA in the knee is not simple to detect, diagnose, or treat since it is complicated, with a relatively high number of risk variables; that is, advanced age, gender, hormonal state, body mass index (BMI) of individuals, and so on. Besides, there are other medical, environmental, and biological risk factors that are known to have a role in the development and progression of the disease, both modifiable and non-modifiable. In the worst-case scenario, patients with these risk factors undergo a total knee replacement. Currently, the only available therapies for patients suffering from knee OA are behavioral interventions, such as weight loss, physical exercise, and strengthening of joint muscles, which might provide brief pain relief while slowing the course of the disease [
6,
7].
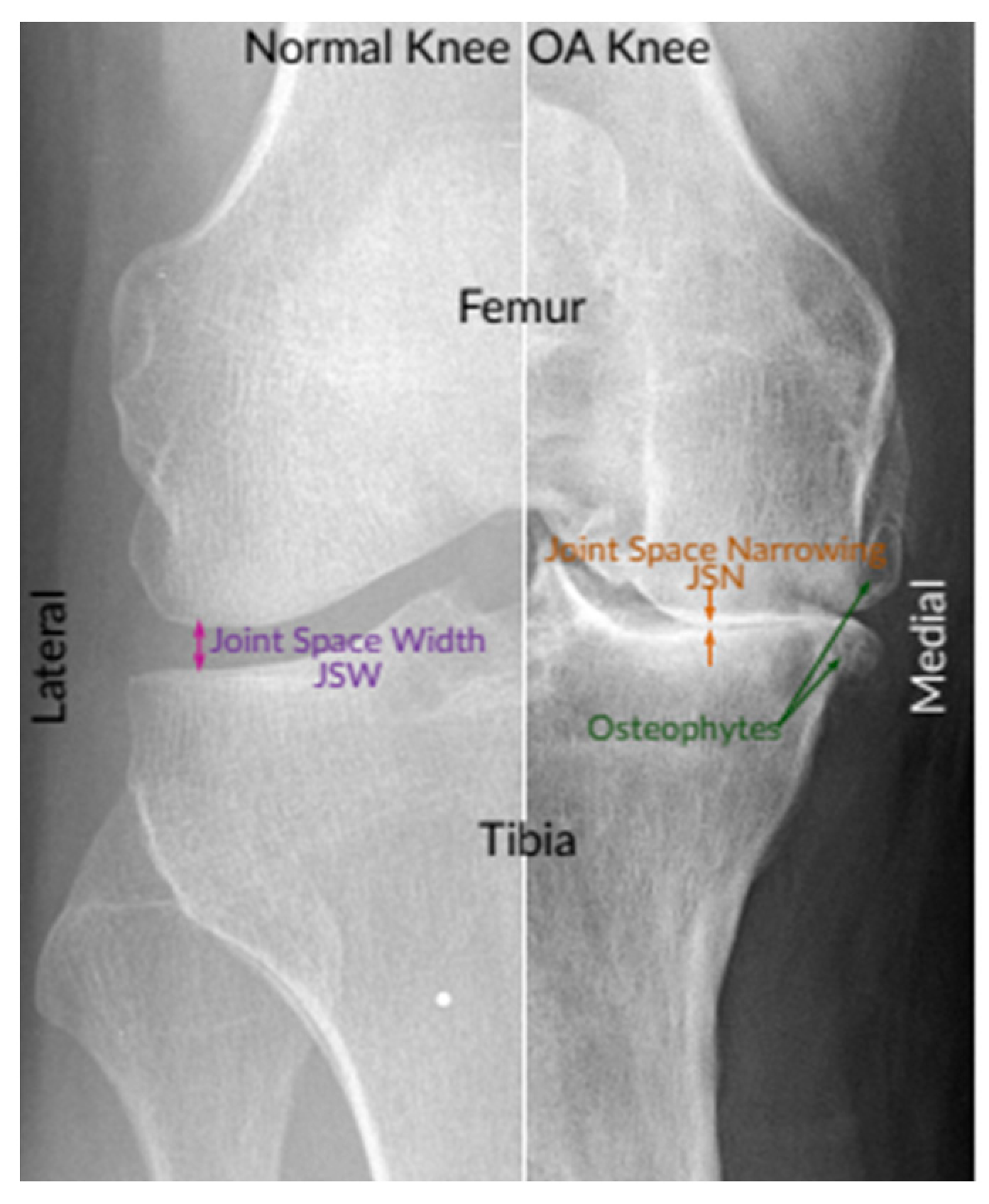
Knee OA is commonly diagnosed and assessed by radiographs (X-rays), which remained the gold benchmark for knee OA screening due to its cost-effectiveness, safety, broad accessibility, and speed. According to radiologists, the most prominent pathological features of the easily observable knee OA are joint space narrowing (JSN) and osteophyte formation, as shown in
Figure 1. These two features can also be used to determine the severity of knee OA using the Kellgren–Lawrence (KL) grading approach. With this approach, knee OA severity is classified depending on the consensus ground truth classification into five grades, namely, grade 0 to grade 4 [
8,
9].
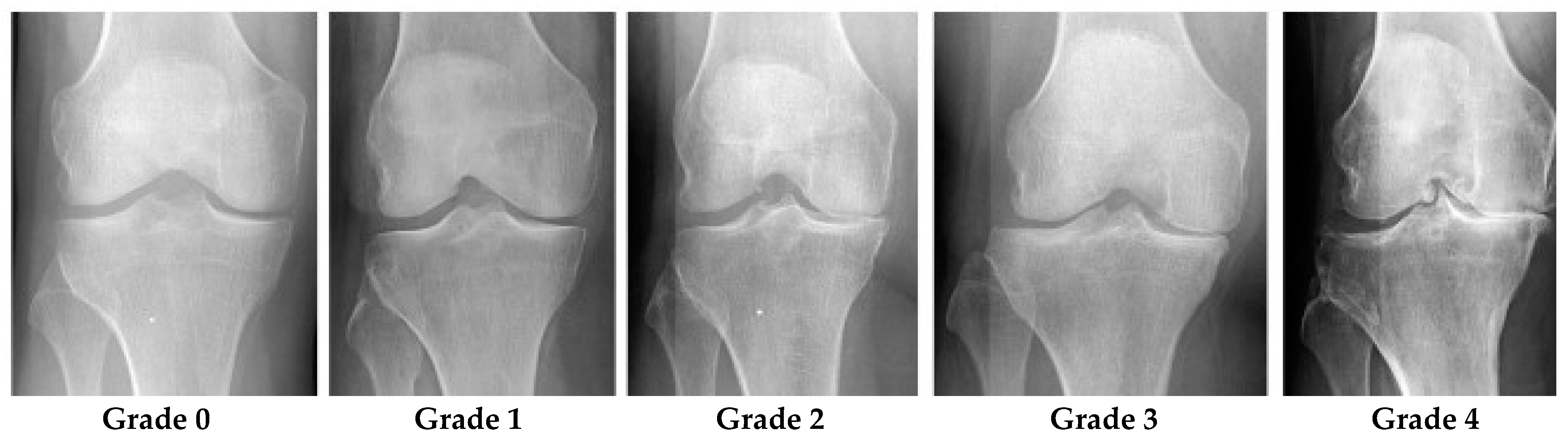
Grade 0 denotes healthy joints in which the radiographic features of knee OA do not exist. Grade 1 denotes doubtful knee OA, which is the possibility of osteophytic lip and questionable JSN. Grade 2 denotes mild OA, which means there are clearly osteophytes as well as the possibility of JSN. Grade 3 denotes moderate OA, which means there are JSN, multiple osteophytes, and sclerosis. The last one, Grade 4, denotes severe OA because of large osteophytes in the joints marked by JSN and severe sclerosis.
Figure 2 shows knee joint samples from all grades of KL [
10].
With the limited number of radiologists, especially in rural areas, as well as the long time required to analyze knee X-ray images, fully automatic classification of knee severity is in great demand since it helps speed up the diagnosis process and increases the rate of early detection. For this reason, many computer-aided diagnosis (CAD)-based medical imaging approaches have been proposed in the literature to detect and analyze knee OA, such as Anifah et al. [
11], Kotti et al. [
12], and Wahyuningrum et al. [
13].
The use of deep learning (DL) and machine learning (ML) techniques in medical imaging has recently increased in order to handle problems of classification [
14,
15], detection [
16,
17], and other associated issues without requiring a radiologist’s expertise [
18].
More specifically, DL-based detection models have been designed and successfully deployed to estimate the severity of knee OA [
14]. Besides, they show staggering performance in the analysis of X-rays in the biomedical domain since it does not require manual feature engineering, which takes place implicitly during the training stage by optimizing its internal parameters to fit the data of interest. Conversely, all standard ML algorithms require the given data to be transformed first using a particular feature engineering or learning algorithm to produce the desired results. Compared to standard ML algorithms, DL algorithms often require inordinate amounts of computational power and resources. Besides, it results in overfitting if fed with too little data. In addition, there are some forms of DL that yield remarkable performance in computer vision, even exceeding that of humans, such as Resnet, Inception, Xception, and CNN-based on TL [
18].
Antony et al. [
19] presented a novel scheme for quantifying the severity of knee OA based on X-ray images. KL grades were used as training input to train FCNN to quantify knee OA severity. Data from the Osteoarthritis Initiative (OAI) and Multicenter Osteoarthritis Study (MOST) were utilized to appraise the effectiveness of this model. Comparing the empirical results of this method to the previously existing methods revealed improvements in classification accuracy, recall, F1 score, and precision.
Norman et al. [
20] proposed a novel approach for the assessment of OA in knee X-rays based on KL grading. Their approach uses state-of-the-art neural networks to implement ensemble learning for precise classification from raw X-ray images. They stated that their approach might be utilized to benefit radiologists in making a quite reliable diagnosis.
Tiulpin et al. [
21] suggested an automated diagnostic technique based on deep Siamese CNNs, which acquire a similarity measure between images. This concept is not limited to simple image pair comparisons but is instead used to compare knee X-rays (with symmetrical joints). Particularly, this network can learn identical weights for both knee joints if the images are split at the central location and fed to a separate CNN branch. Simulation results on the entire OAI dataset demonstrated that their work outperforms previous models, with an accuracy score of 66.71%.
According to Chen et al. [
1], two CNNs were employed to grade knee OA severity based on the KL grading system. A specialized one-stage YOLOv2 network was used to detect X-ray images of knee joints. Using the best-performing CNNs, including versions of YOLO, ResNet, VGG, DenseNet, and InceptionV3, the detected knee joint images were then classified utilizing adjusted ordinal loss analysis. Empirical results revealed that the best classification accuracy and the mean absolute error obtained with their proposed approach are 69.7% and 0.344, respectively.
Moustakidis et al. [
22] developed a deep neural network (DNN)-based technique for knee OA classification, which comprises three processing steps: data preprocessing, data normalization, and a learning procedure for DNN training. Experimental results showed that their presented DNN approach was effective in improving classification accuracy.
Thomas et al. [
23] proposed new deep CNNs for knee OA classification, where this model can take full radiographs as input and expect KL scores with outstanding accuracy. Based on the results reported by this study, an average F1 score of 0.70 and an accuracy of 0.71 was achieved using their proposed model.
Tiulpin et al. [
3] have developed an automatic method to predict OARSI and KL grades from knee radiographs. Based on DeepCNN and leverages an ensemble network of 50 layers, and used TL from ImageNet with a fine-tuning one OAI dataset.
Brahim et al. [
24] presented a computer-aided diagnostic method for early knee osteoarthritis identification utilizing knee X-ray imaging and machine-learning algorithms. Where the proposed approaches have been implemented as follows: first, preprocessing of the X-ray pictures in the Fourier domain has performed using a circular Fourier transform; then MLR (multivariate linear regression) was used to the data to decrease the variability between patients with OA and healthy participants; for feature extraction/selection stage an independent component analysis (ICA) was used for reducing the dimensionality; finally, random forest and Naive Bayes classifier were used for the classification task. Furthermore, the 1024 knee X-ray images from the public database osteoarthritis initiative were used to test this innovative image-based method (OAI).
Wang et al. [
5] suggested a fully automatic scheme based on deep learning to detect knee OA using a pertained YOLO model. Based on the experimental results, their method improves knee OA classification performance compared with the previous state-of-the-art methods.
Yadav et al. [
25] proposed a highly effective and low-cost hybrid SFNet. Due to the lower computation cost and high efficiency attained by training the model at two scales, the hybrid SFNet is a two-scale DL model with fewer neurons. An improved canny edge detection technique is used to locate the fractured bone first. The grey image and its corresponding canny image are then fed into a hybrid SFNet for deep feature extraction. The diagnosis of bone fractures is greatly enhanced by this process.
Lau et al. [
26] developed a method based on ImageNet, the Xception model, and a dataset of X-ray images from total knee arthroplasty (TKA) patients, and the image-based ML model was created. In order to develop a clinical information-based ML model using a random forest classifier was then carried out using a different system built on a dataset with TKA patient clinical parameters. To interpret the prediction choice the model made, class activation maps were also used. The result of the precision rate and recall rate for the ML on the images loosening model reached 0.92 and 0.96, respectively, while a 96.3% accuracy rate for visualization classification was noted. However, the addition of a clinical information-based model, with a precision rate of 0.71 and recall rate of 0.20, did not further demonstrate improvement in the accuracy.
Overall, the approaches reported by the related works and the studies published in Christodoulou et al. [
27], Du et al. [
28], Hirvasniemi et al. [
29], and Sharma et al. [
30] have utilized DL and ML techniques for diagnosis different bone disease. These methods have provided excellent job performance for binary-class classification. In contrast, they are not efficient for multiclass classifying knee OA based on KL grades using X-ray images; it achieved maximum accuracy of 69% [
5]. Thus, it is challenging to propose an effective tool or method for the early classification of knee OA. Therefore, an approach that uses both types of learning is crucial for improving classification performance.
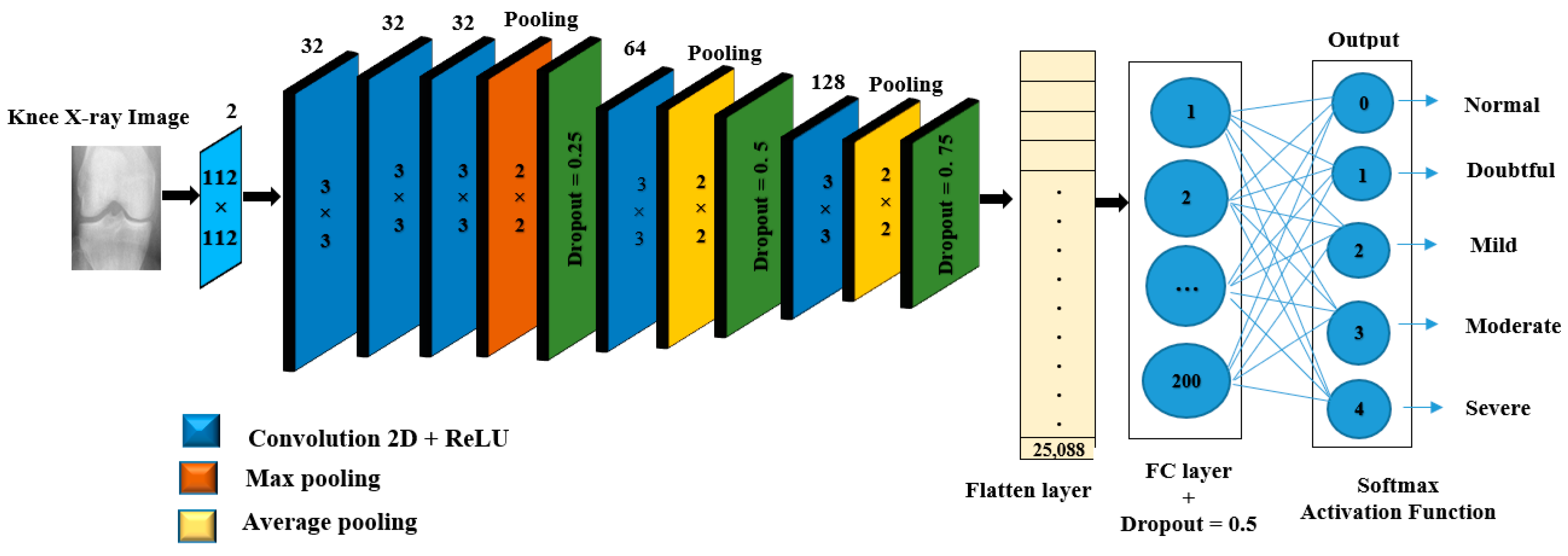
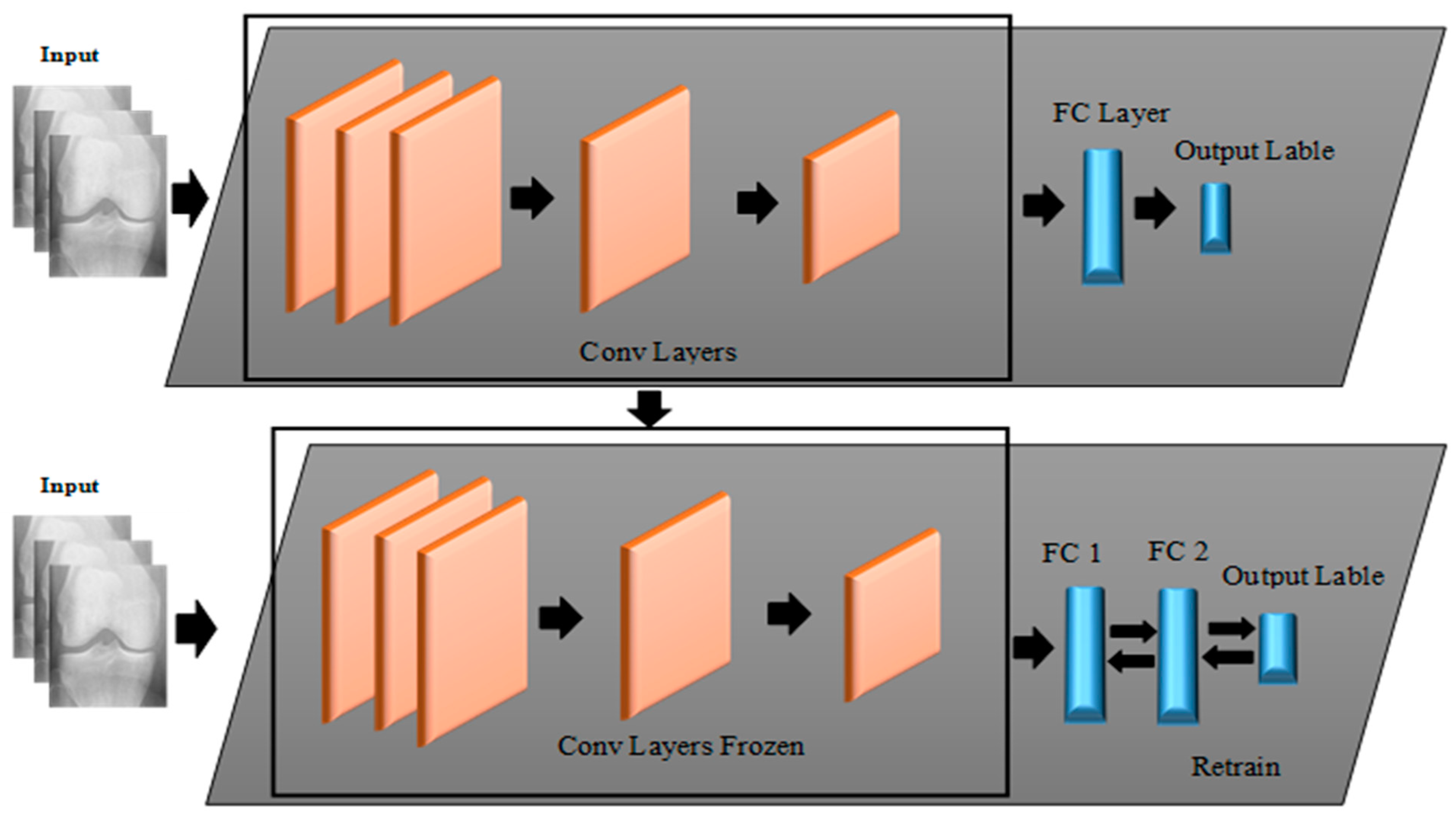
In light of this, this paper aims to propose novel approaches that utilize both DL and ML algorithms to identify a given subject’s class label from a Knee X-ray image according to KL grading criteria. The proposed approaches are mainly based on two forms of learning structures, namely Deep Hybrid Learning-I (DHL-I) and Deep Hybrid Learning-II (DHL-II). The first one, DHL-I, is based on CNN structure, in which a new structure of five classes of prediction was developed to be initially trained on knee X-ray images and then used as feature extraction. A PCA was then applied to reduce these learned features and then fed into SVMs, which can classify knee OA by pattern discrimination. Whereas the second one, DHL-II, is the same as the first one except for the following. The pre-trained CNN developed for the DHL-I was fine-tuned using the TL concept to classify knee OA into four classes, three classes, and two class labels.
The primary motivation of this study is to build up a strategy that mixes ML and DL methods to develop an efficient DHL to classify knee OA severity based on KL grades in different categories. To examine how different class-based classifications (i.e., binary and multiclass) affect prediction performance. As a result of this process, knee OA severity classification is significantly improved on the side of saving time, accuracy, and hardware costs, and these may contribute to early classification in the first stage of the disease to help reduce its progression and improve people’s quality of life.
The following are the main contributions of this paper:
To the best of our knowledge, DL techniques have not yet been used for feature extraction purposes in the literature on knee OA. Thus, the proposed model is the first to show their potential application in this area.
Unlike the existing studies that develop a classification model for specific n-class labels, this paper proposes various classification models for classifying the severity of knee OA, i.e., five classes, four classes, three classes, and two classes model-based.
Despite many articles on TL and fine-tuning, as far as we know, no research has compared or assessed the two approaches in terms of pre-trained deep feature classifications for knee OA.
This model combines deep and hand-crafted features obtained by a proposed pre-trained CNN and the PCA algorithm to generate the most prominent feature set before being sent to the SVM algorithm for classification.
The concept of TL was employed to fine-tune the proposed pre-trained CNN developed on five classes’ labels to fit other class labels, i.e., four classes, three classes, and two classes model-based.
As compared with the existing state-of-the-art methods for predicting knee OA, the proposed DHL models performs significantly better.
The rest of the paper is arranged as follows. The theoretical foundation is presented in
Section 2, and the materials and techniques employed in this work are described in
Section 3.
Section 4 presents the experimental results and their explanations. Finally,
Section 5 brings the paper to a conclusion and discussion.
5. Discussion and Conclusions
In this paper, an efficient method for diagnosing and classifying knee OA severity based on X-ray images has been proposed, which employs pre-trained CNNs for feature extraction as well as fine-tuning the pre-trained CNN using the TL method. In this regard, two DHL (DHL-I and DHL-II) models were created. The first DHL-I approach was based on pre-trained CNNs, PCAs, and SVMs for classifying knee OA severity. Through the pre-trained CNN model, the proposed DHL-I model allows for exploiting the ability of OA to generate diverse features from X-ray images. By incorporating these features into SVMs, we achieved excellent generalization abilities and higher classification accuracy than the previous methods. Experiments were conducted on an OAI dataset to test the performance of the proposed model. Experimental results show that the proposed DHL-I model achieved high accuracy levels both in training and testing data. Further improvements to OA severity diagnostic accuracy were made to classify knee OA into different class labels by testing TL with a pre-trained CNN to see if it handled overfitting and time complexity problems based on the proposed DHL-II model, which showed an improvement in the performance of the model when compared to the related researches. However, the DHL-II shows that the features from the pooling and convolutional layers are more accurate than those from the FC layers. Therefore, fine-tuning networks used to involve replacing the top FC layer, resulting in better classification accuracy. However, there are several limitations faced by our study. Firstly, in the OAI dataset study, skyline view radiographs were not acquired, which would have provided further discriminative information. Therefore, adding lateral view images can be helpful when examining structural features and provide additional information about the patellofemoral joint and femoral osteophytes, which are not visible from PA radiographs alone. Secondly, the proposed models include knee OA radiographs of both tibial condyles and the femoral crista and articulation of both medial and lateral patellofemoral joints, but this requires time. Despite this limitation, this would give the model more information about the relationship between bones in the knee joint. Thirdly, classifying Knee OA images on KL grade 1 is challenging because of the small variations, particularly between grades 0 to grade 2, thus needing powerful feature extraction to address this problem. The final and main limitation of this work is a common problem with ML for medical applications is class imbalance. Therefore, we often have to deal with datasets where one of the classes is significantly under-represented. Consequently, the classification problem becomes harder for the model, and it risks detecting the minority class incorrectly.
Despite the mentioned limitations above, the experimental results of the proposed two DHL models outperformed recent methods compared to the previous work in the literature. To conclude, utilizing the suggested DHL framework, rapid and computer-assisted diagnosis may contribute to early classification in the first stage of the disease to help reduce its progression and improve people’s quality of life.