A Novel CDH1 Variant Identified in a Chinese Family with Blepharocheilodontic Syndrome
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. WES
2.3. Sanger Sequencing
2.4. Conservation Analysis
2.5. Conformational Analysis
3. Results
3.1. Clinical Findings
3.2. The Identification of the CDH1 Variant
3.3. Conservation and Conformational Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chhabra, N.; Goswami, M.; Chhabra, A. Genetic Basis of Dental Agenesis—Molecular Genetics Patterning Clinical Dentistry. Med. Oral 2014, 19, e112–e119. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Maeda, T. Prevalence and Genetic Basis of Tooth Agenesis. Jpn. Dent. Sci. Rev. 2009, 45, 52–58. [Google Scholar] [CrossRef]
- Rølling, S.; Poulsen, S. Oligodontia in Danish Schoolchildren. Acta Odontol. Scand. 2001, 59, 111–112. [Google Scholar] [CrossRef]
- Polder, B.J.; Van’t Hof, M.A.; Van der Linden, F.P.G.M.; Kuijpers-Jagtman, A.M. A Meta-Analysis of the Prevalence of Dental Agenesis of Permanent Teeth. Community Dent. Oral Epidemiol. 2004, 32, 217–226. [Google Scholar] [CrossRef]
- Gorlin, R.J.; Zellweger, H.; Curtis, M.W.; Wiedemann, H.R.; Warburg, M.; Majewski, F.; Gillessen-Kaesbach, G.; Prahl-Andersen, B.; Zackai, E. Blepharo-Cheilo-Dontic (BCD) Syndrome. Am. J. Med. Genet. 1996, 65, 109–112. [Google Scholar] [CrossRef]
- Weaver, K.N.; Rutledge, K.D.; Grant, J.H.; Robin, N.H. Imperforate Anus Is a Rare Associated Finding in Blepharocheilodontic Syndrome. Am. J. Med. Genet. A 2010, 152A, 438–440. [Google Scholar] [CrossRef]
- Ghoumid, J.; Stichelbout, M.; Jourdain, A.-S.; Frenois, F.; Lejeune-Dumoulin, S.; Alex-Cordier, M.-P.; Lebrun, M.; Guerreschi, P.; Duquennoy-Martinot, V.; Vinchon, M.; et al. Blepharocheilodontic Syndrome Is a CDH1 Pathway-Related Disorder Due to Mutations in CDH1 and CTNND1. Genet. Med. 2017, 19, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Kievit, A.; Tessadori, F.; Douben, H.; Jordens, I.; Maurice, M.; Hoogeboom, J.; Hennekam, R.; Nampoothiri, S.; Kayserili, H.; Castori, M.; et al. Variants in Members of the Cadherin-Catenin Complex, CDH1 and CTNND1, Cause Blepharocheilodontic Syndrome. Eur. J. Hum. Genet. 2018, 26, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Gil da Silva Lopes, V.L.; Guion-Almeida, M.L.; de Oliveira Rodini, E.S. Blepharocheilodontic (BCD) Syndrome: Expanding the Phenotype? Am. J. Med. Genet. A 2003, 121A, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Freitas, E.L.; Martinhago, C.D.; Ramos, E.S.; Murray, J.C.; Gil-da-Silva-Lopes, V.L. Preliminary Molecular Studies on Blepharocheilodontic Syndrome. Am. J. Med. Genet. A 2007, 143A, 2757–2759. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.D.; Chen, C.P.; Bahna, F.; Honig, B.; Shapiro, L. Cadherin-Mediated Cell-Cell Adhesion: Sticking Together as a Family. Curr. Opin. Struct. Biol. 2003, 13, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Gumbiner, B.M. Regulation of Cadherin-Mediated Adhesion in Morphogenesis. Nat. Rev. Mol. Cell Biol. 2005, 6, 622–634. [Google Scholar] [CrossRef]
- Smith, A.L.; Dohn, M.R.; Brown, M.V.; Reynolds, A.B. Association of Rho-Associated Protein Kinase 1 with E-Cadherin Complexes Is Mediated by P120-Catenin. Mol. Biol. Cell 2012, 23, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Thumkeo, D.; Keel, J.; Ishizaki, T.; Oshima, H.; Oshima, M.; Noda, Y.; Matsumura, F.; Taketo, M.M.; Narumiya, S. ROCK-I Regulates Closure of the Eyelids and Ventral Body Wall by Inducing Assembly of Actomyosin Bundles. J. Cell Biol. 2005, 168, 941–953. [Google Scholar] [CrossRef]
- Li, C.-Y.; Cha, W.; Luder, H.-U.; Charles, R.-P.; McMahon, M.; Mitsiadis, T.A.; Klein, O.D. E-Cadherin Regulates the Behavior and Fate of Epithelial Stem Cells and Their Progeny in the Mouse Incisor. Dev. Biol. 2012, 366, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, J.D.; Dobeck, J.M.; Tye, C.E.; Perez-Moreno, M.; Stokes, N.; Reynolds, A.B.; Fuchs, E.; Skobe, Z. Targeted P120-Catenin Ablation Disrupts Dental Enamel Development. PLoS ONE 2010, 5, e12703. [Google Scholar] [CrossRef]
- Benusiglio, P.R.; Caron, O.; Consolino, E.; Duvillard, P.; Coulet, F.; Blayau, M.; Malka, D. Cleft Lip, Cleft Palate, Hereditary Diffuse Gastric Cancer and Germline Mutations in CDH1. Int. J. Cancer 2013, 132, 2470. [Google Scholar] [CrossRef]
- Brito, L.A.; Yamamoto, G.L.; Melo, S.; Malcher, C.; Ferreira, S.G.; Figueiredo, J.; Alvizi, L.; Kobayashi, G.S.; Naslavsky, M.S.; Alonso, N.; et al. Rare Variants in the Epithelial Cadherin Gene Underlying the Genetic Etiology of Nonsyndromic Cleft Lip with or without Cleft Palate. Hum. Mutat. 2015, 36, 1029–1033. [Google Scholar] [CrossRef]
- van der Post, R.S.; Vogelaar, I.P.; Carneiro, F.; Guilford, P.; Huntsman, D.; Hoogerbrugge, N.; Caldas, C.; Schreiber, K.E.C.; Hardwick, R.H.; Ausems, M.G.E.M.; et al. Hereditary Diffuse Gastric Cancer: Updated Clinical Guidelines with an Emphasis on Germline CDH1 Mutation Carriers. J. Med. Genet. 2015, 52, 361–374. [Google Scholar] [CrossRef]
- Kluijt, I.; Siemerink, E.J.M.; Ausems, M.G.E.M.; van Os, T.A.M.; de Jong, D.; Simões-Correia, J.; van Krieken, J.H.; Ligtenberg, M.J.; Figueiredo, J.; van Riel, E.; et al. CDH1-Related Hereditary Diffuse Gastric Cancer Syndrome: Clinical Variations and Implications for Counseling. Int. J. Cancer 2012, 131, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Hansford, S.; Kaurah, P.; Li-Chang, H.; Woo, M.; Senz, J.; Pinheiro, H.; Schrader, K.A.; Schaeffer, D.F.; Shumansky, K.; Zogopoulos, G.; et al. Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond. JAMA Oncol. 2015, 1, 23–32. [Google Scholar] [CrossRef]
- Nishi, E.; Masuda, K.; Arakawa, M.; Kawame, H.; Kosho, T.; Kitahara, M.; Kubota, N.; Hidaka, E.; Katoh, Y.; Shirahige, K.; et al. Exome Sequencing-Based Identification of Mutations in Non-Syndromic Genes among Individuals with Apparently Syndromic Features. Am. J. Med. Genet. A 2016, 170, 2889–2894. [Google Scholar] [CrossRef] [PubMed]
- Prasad, M.K.; Geoffroy, V.; Vicaire, S.; Jost, B.; Dumas, M.; Le Gras, S.; Switala, M.; Gasse, B.; Laugel-Haushalter, V.; Paschaki, M.; et al. A Targeted Next-Generation Sequencing Assay for the Molecular Diagnosis of Genetic Disorders with Orodental Involvement. J. Med. Genet. 2016, 53, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, T.; Zhang, Y.; Zhang, K.; Shi, L.; Chen, Y.; Wang, X.; Sun, Z. Performance Evaluation of Pathogenicity-Computation Methods for Missense Variants. Nucleic Acids Res. 2018, 46, 7793–7804. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, N.M.; Rothstein, J.H.; Pejaver, V.; Middha, S.; McDonnell, S.K.; Baheti, S.; Musolf, A.; Li, Q.; Holzinger, E.; Karyadi, D.; et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016, 99, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A General Framework for Estimating the Relative Pathogenicity of Human Genetic Variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Han, D.; Qu, H.; Gong, Y.; Wu, H.; Zhang, X.; Zhong, N.; Feng, H. EDA Gene Mutations Underlie Non-Syndromic Oligodontia. J. Dent. Res. 2009, 88, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Zhao, R.; He, H.; Zhang, J.; Feng, H.; Lin, L. WNT10A Variants Are Associated with Non-Syndromic Tooth Agenesis in the General Population. Hum. Genet. 2014, 133, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.-W.; Han, D.; Zhang, H.; Liu, Y.; Zhang, X.; Miao, M.Z.; Wang, Y.; Zhao, N.; Zeng, L.; Bai, B.; et al. Nine Novel PAX9 Mutations and a Distinct Tooth Agenesis Genotype-Phenotype. J. Dent. Res. 2018, 97, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Yang, W.; Han, D.; Wang, X.; Guo, S.; Li, J.; Li, F.; Zhang, X.; Wong, S.-W.; Bai, B.; et al. Mutations in WNT10B Are Identified in Individuals with Oligodontia. Am. J. Hum. Genet. 2016, 99, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yu, M.; Liu, H.; Cai, T.; Feng, H.; Liu, Y.; Han, D. Novel MSX1 Variants Identified in Families with Nonsyndromic Oligodontia. Int. J. Oral. Sci. 2021, 13, 2. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, H.; Su, L.; Zheng, J.; Feng, H.; Liu, Y.; Yu, M.; Han, D. Four Novel PAX9 Variants and the PAX9-Related Non-Syndromic Tooth Agenesis Patterns. Int. J. Mol. Sci. 2022, 23, 8142. [Google Scholar] [CrossRef] [PubMed]
- Awadh, W.; Kiukkonen, A.; Nieminen, P.; Arte, S.; Hurmerinta, K.; Rice, D.P. Blepharocheilodontic (BCD) Syndrome: New Insights on Craniofacial and Dental Features. Am. J. Med. Genet. A 2017, 173, 905–913. [Google Scholar] [CrossRef]
- Ababneh, F.K.; Al-Swaid, A.; Elhag, A.; Youssef, T.; Alsaif, S. Blepharo-Cheilo-Dontic (BCD) Syndrome: Expanding the Phenotype, Case Report and Review of Literature. Am. J. Med. Genet. A 2014, 164A, 1525–1529. [Google Scholar] [CrossRef]
- Korula, S.; Wilson, L.; Salomonson, J. Distinct Craniofacial Syndrome of Lagophthalmia and Bilateral Cleft Lip and Palate. Am. J. Med. Genet. 1995, 59, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Petridis, C.; Shinomiya, I.; Kohut, K.; Gorman, P.; Caneppele, M.; Shah, V.; Troy, M.; Pinder, S.E.; Hanby, A.; Tomlinson, I.; et al. Germline CDH1 Mutations in Bilateral Lobular Carcinoma In Situ. Br. J. Cancer 2014, 110, 1053–1057. [Google Scholar] [CrossRef]
- Figueiredo, J.; Söderberg, O.; Simões-Correia, J.; Grannas, K.; Suriano, G.; Seruca, R. The Importance of E-Cadherin Binding Partners to Evaluate the Pathogenicity of E-Cadherin Missense Mutations Associated to HDGC. Eur. J. Hum. Genet. 2013, 21, 301–309. [Google Scholar] [CrossRef]
- Ye, X.; Attaie, A. Genetic Basis of Nonsyndromic and Syndromic Tooth Agenesis. J. Pediatr. Genet. 2016, 5, 198–208. [Google Scholar] [CrossRef]
- Yu, M.; Wong, S.-W.; Han, D.; Cai, T. Genetic Analysis: Wnt and Other Pathways in Nonsyndromic Tooth Agenesis. Oral Dis. 2019, 25, 646–651. [Google Scholar] [CrossRef]
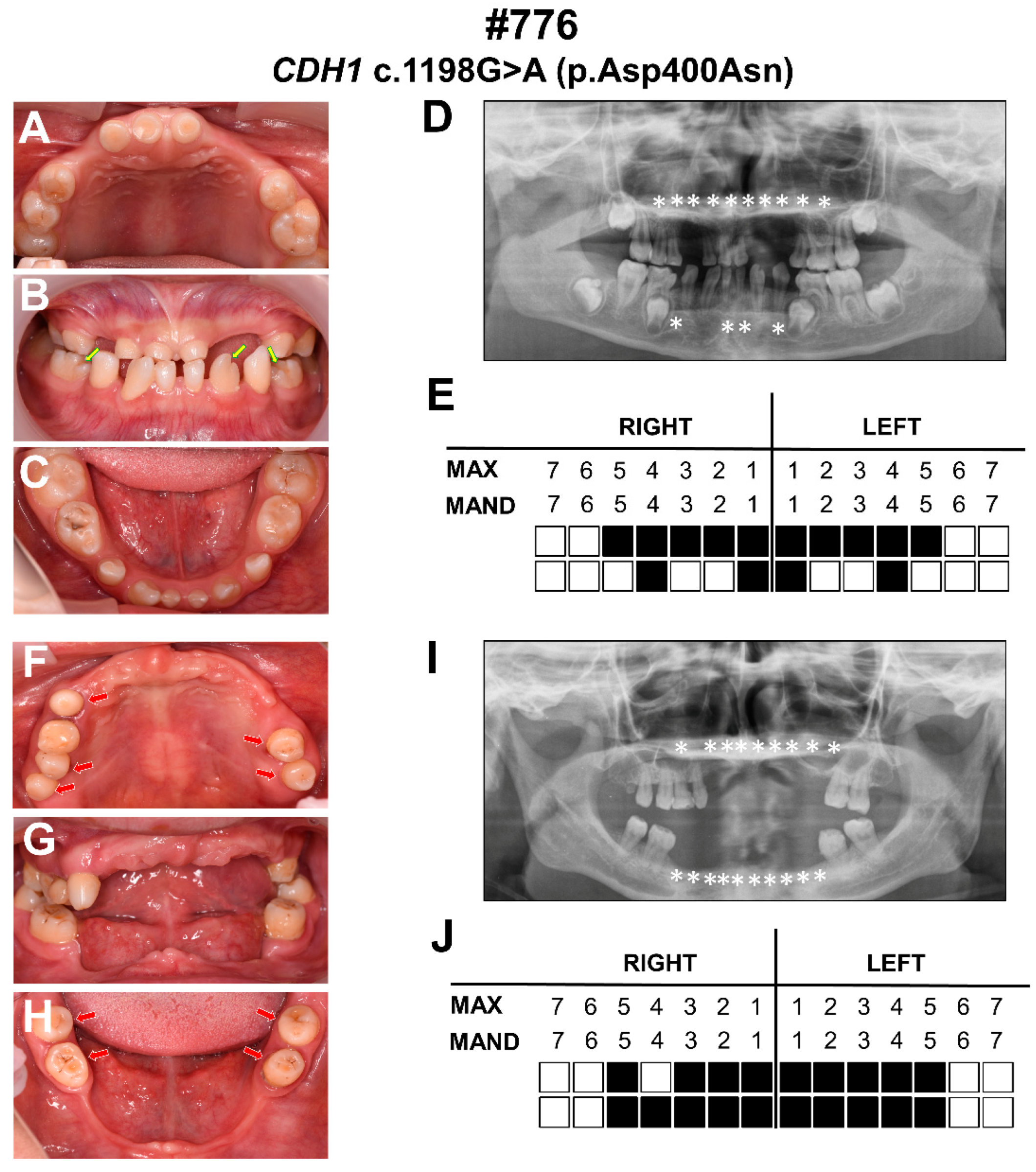
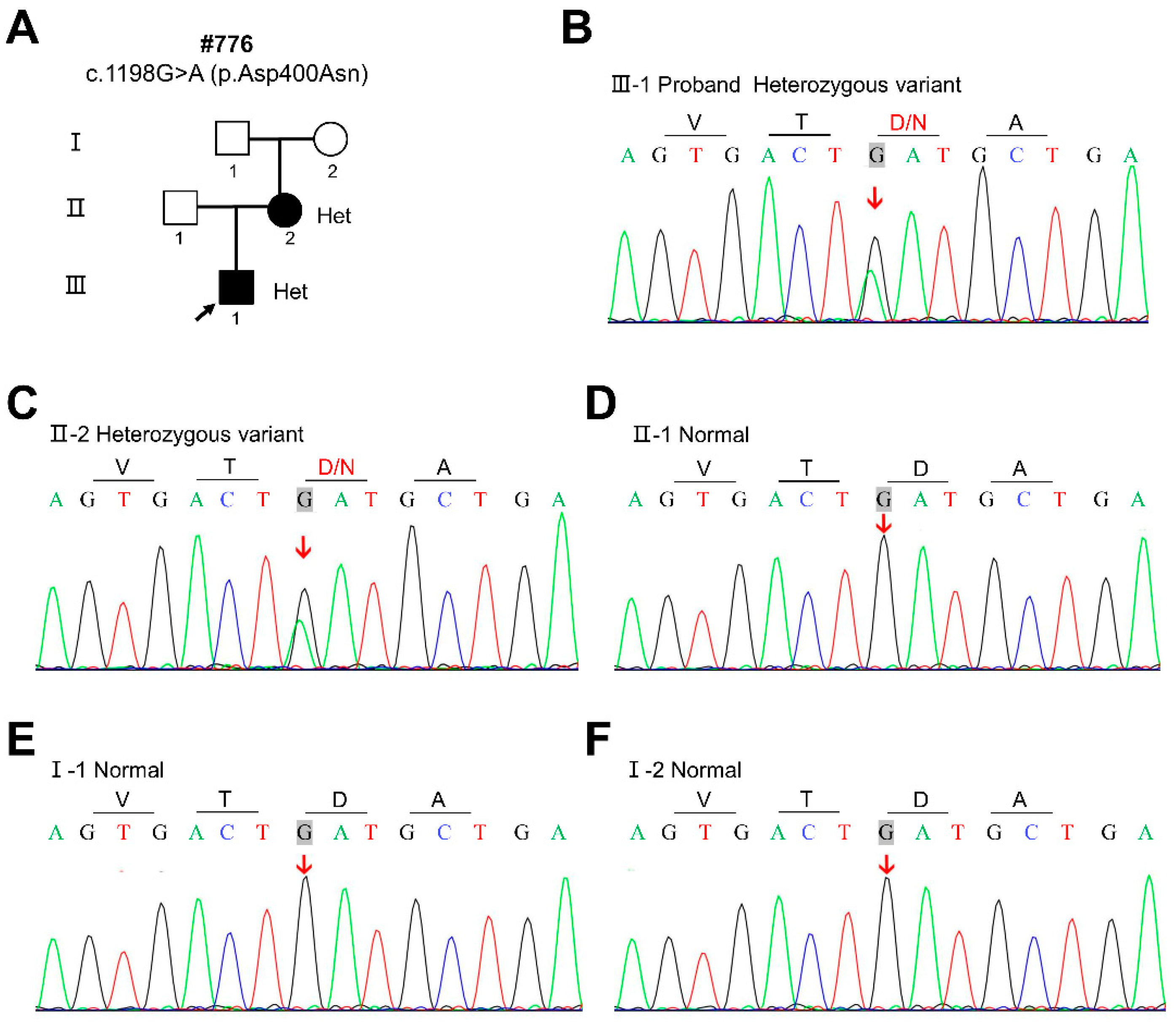
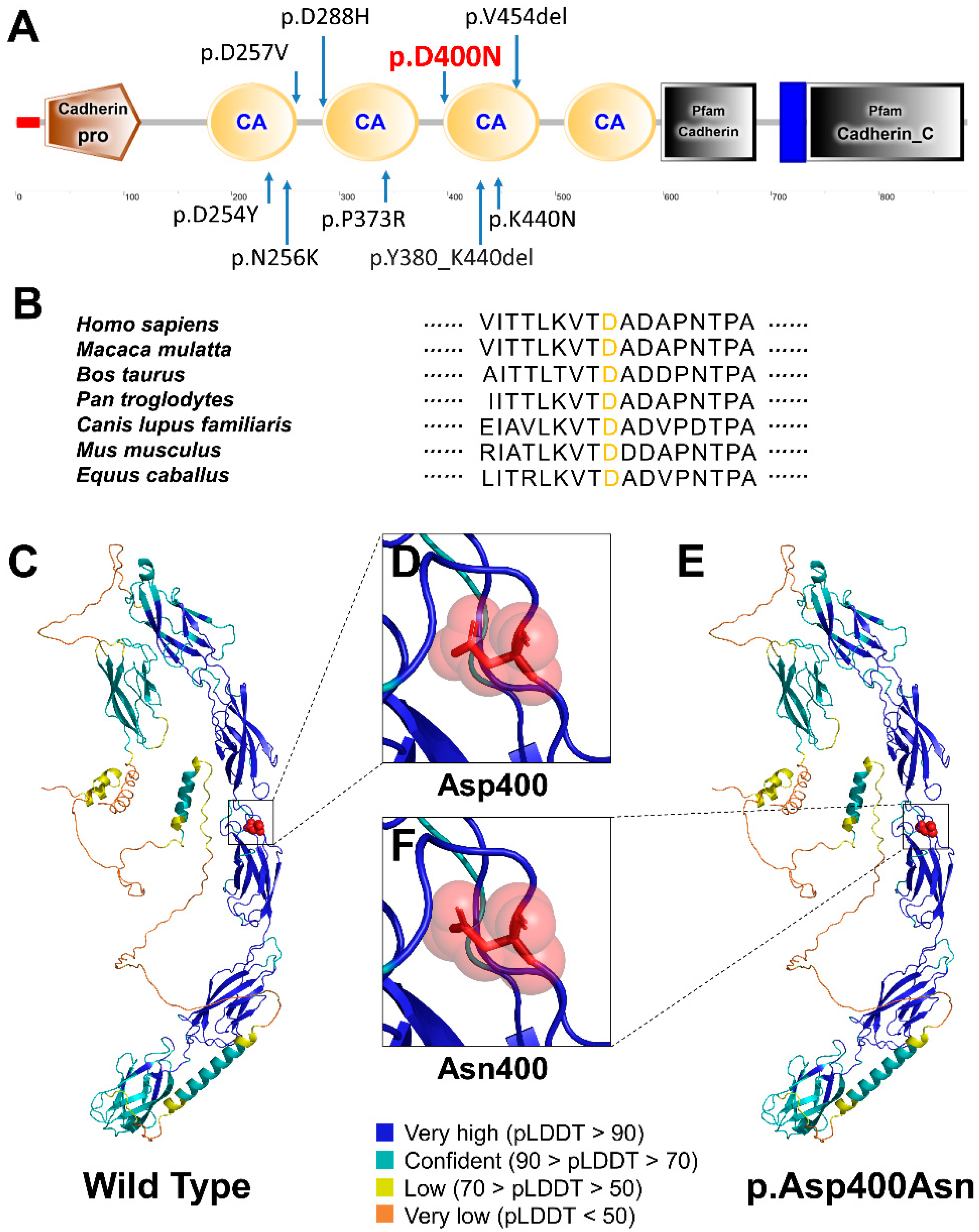
| Clinical Features | The Proband | Ⅱ-2 |
|---|---|---|
| Gender | Male | Female |
| Age | 9 | 35 |
| Number of missing permanent teeth | 14 | 19 |
| Congenital missing of primary teeth | + | + |
| Abnormality in morphology of teeth | + | + |
| CL/P | - | - |
| Euryblepharon | + | + |
| Lagophthalmos | - | - |
| Ectropion | + | + |
| Lacrimal duct abnormalities | - | - |
| Distichiasis | + | + |
| Ankyloblepharon | - | - |
| Sparse hair | + | + |
| High frontal hairline | + | + |
| Broad forehead | + | + |
| Malformed ears | - | - |
| Everted lower lip | - | - |
| Hypothyroidism | - | - |
| Hypertelorism | - | - |
| Imperforate anus | - | - |
| Dermoid cysts | - | - |
| Neural tube defect | - | - |
| Patients | Variant | Variant Type | Domain | ReVe | REVEL | CADD | dbSNP/ gnomAD | ACMG Classification (Evidence of Pathogenicity) |
|---|---|---|---|---|---|---|---|---|
| #776 III-1; #776 II-2 | c.1198G>A/p.Asp400Asn | Hetero-zygous | 3rd CA | 0.680 Disease causing | 0.643 Disease causing | 26.3 Disease causing | - | Likely pathogenic PS2, PM2, PP3, PP4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, B.; Liu, Y.; Su, L.; Liu, H.; Feng, H.; Yu, M.; Liu, H. A Novel CDH1 Variant Identified in a Chinese Family with Blepharocheilodontic Syndrome. Diagnostics 2022, 12, 2936. https://doi.org/10.3390/diagnostics12122936
Lin B, Liu Y, Su L, Liu H, Feng H, Yu M, Liu H. A Novel CDH1 Variant Identified in a Chinese Family with Blepharocheilodontic Syndrome. Diagnostics. 2022; 12(12):2936. https://doi.org/10.3390/diagnostics12122936
Chicago/Turabian StyleLin, Bichen, Yang Liu, Lanxin Su, Hangbo Liu, Hailan Feng, Miao Yu, and Haochen Liu. 2022. "A Novel CDH1 Variant Identified in a Chinese Family with Blepharocheilodontic Syndrome" Diagnostics 12, no. 12: 2936. https://doi.org/10.3390/diagnostics12122936
APA StyleLin, B., Liu, Y., Su, L., Liu, H., Feng, H., Yu, M., & Liu, H. (2022). A Novel CDH1 Variant Identified in a Chinese Family with Blepharocheilodontic Syndrome. Diagnostics, 12(12), 2936. https://doi.org/10.3390/diagnostics12122936







