Classification of White Blood Cells: A Comprehensive Study Using Transfer Learning Based on Convolutional Neural Networks
Abstract
1. Introduction
- We have applied advanced image processing and data augmentation techniques, i.e., random resizing and cropping, which randomly select different parts of an image, enabling the model to focus on and perceive various features. This improves the model’s generalization capabilities and prevents overfitting.
- We applied advanced fine-tuning techniques such as normalization, mixup augmentation, and label smoothing to train the CNN model and obtain preferable results in comparison with other similar research.
- We investigated and compared the efficiency and complexity of multiple deep neural network (DNN) architectures initialized with pre-trained weights for WBC classification.
2. Materials and Methods
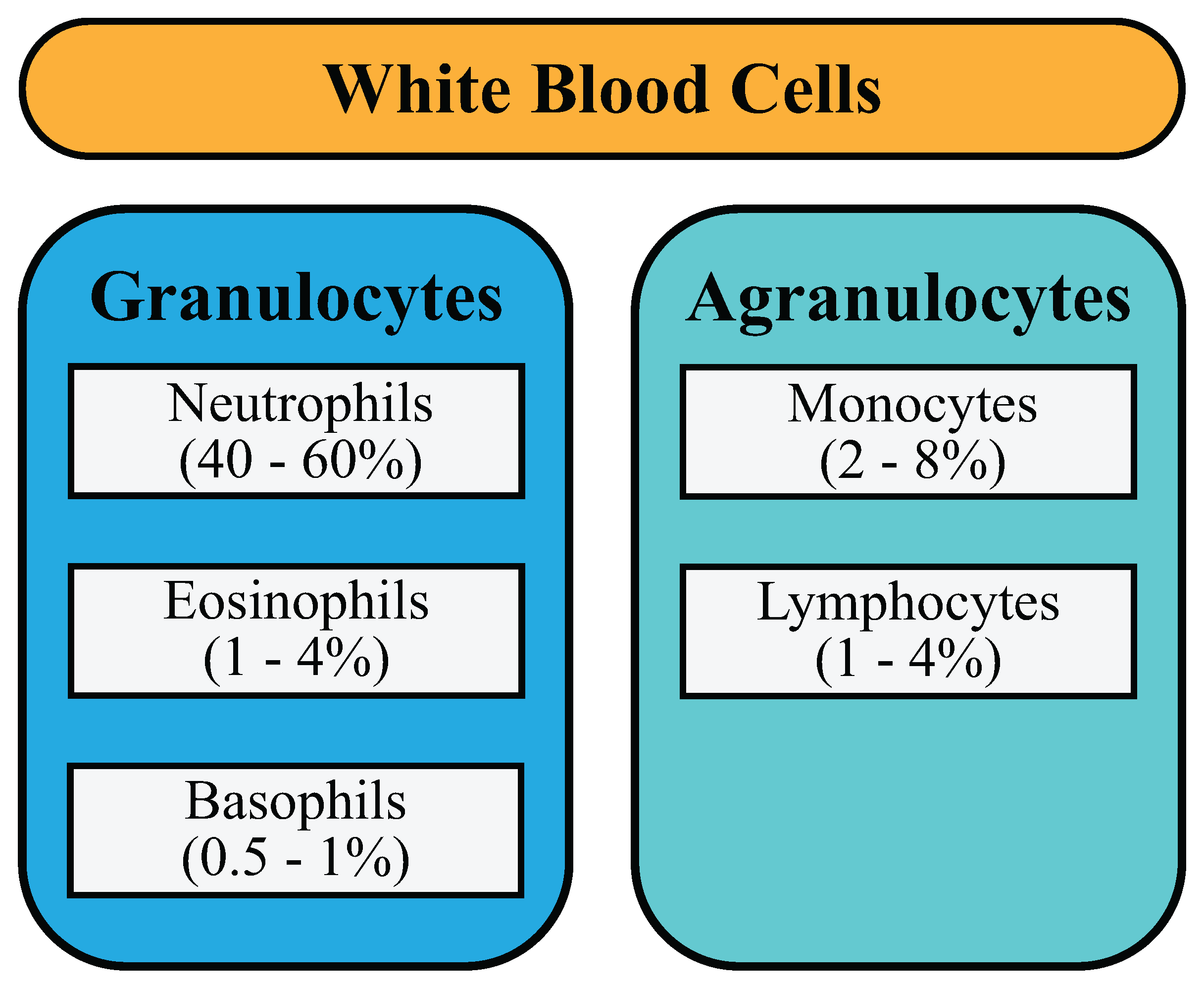
2.1. Dataset
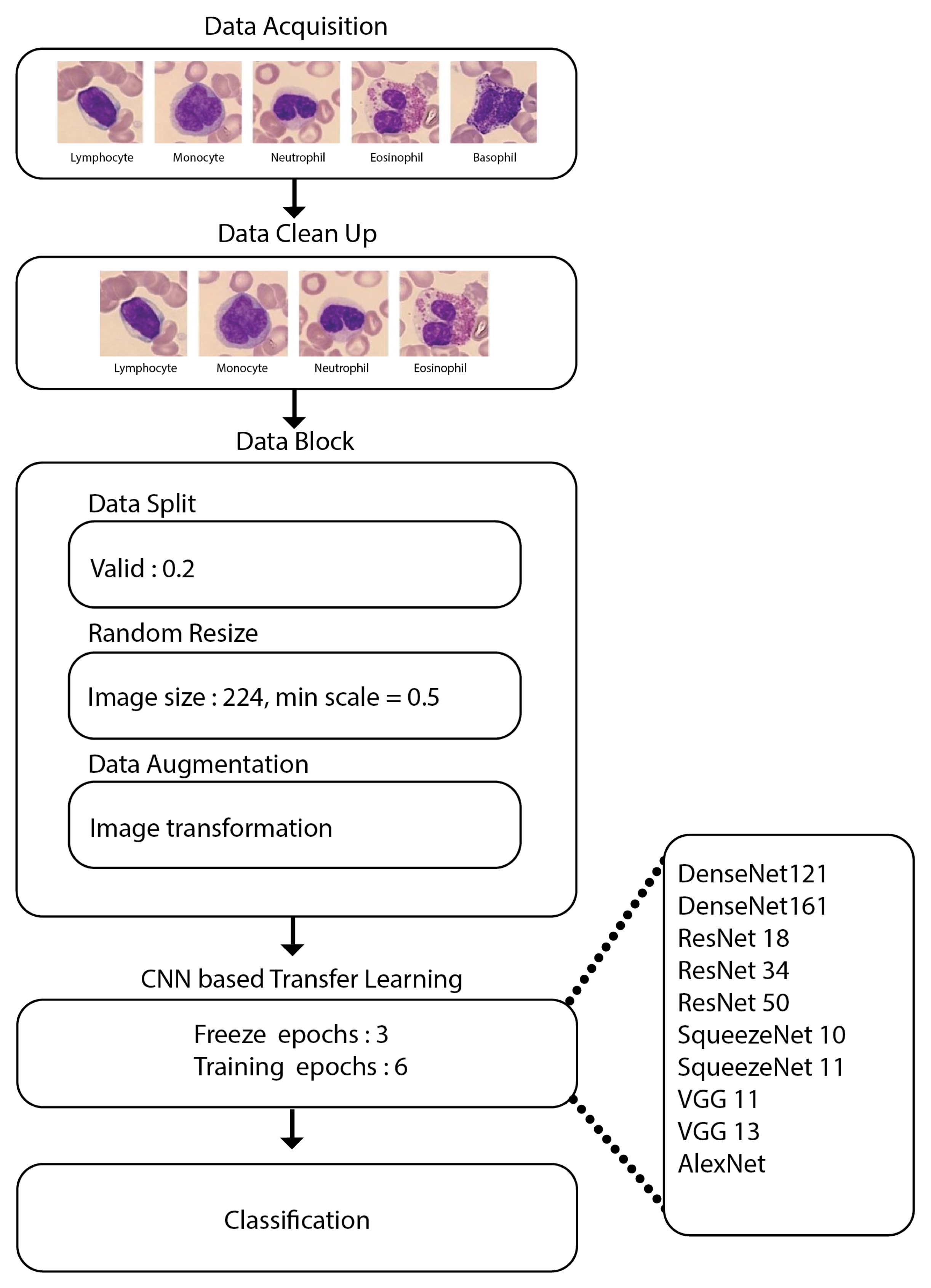
2.2. Data Pre-Processing
- Data split: This helps in folding the dataset into train, test and validation sets.
- Random resize: This block resizes our data to create uniformity in the image size.
- Data augmentation: Image transformations are performed during the run time using this block.
2.3. Convolutional Neural Network
2.4. Transfer Learning
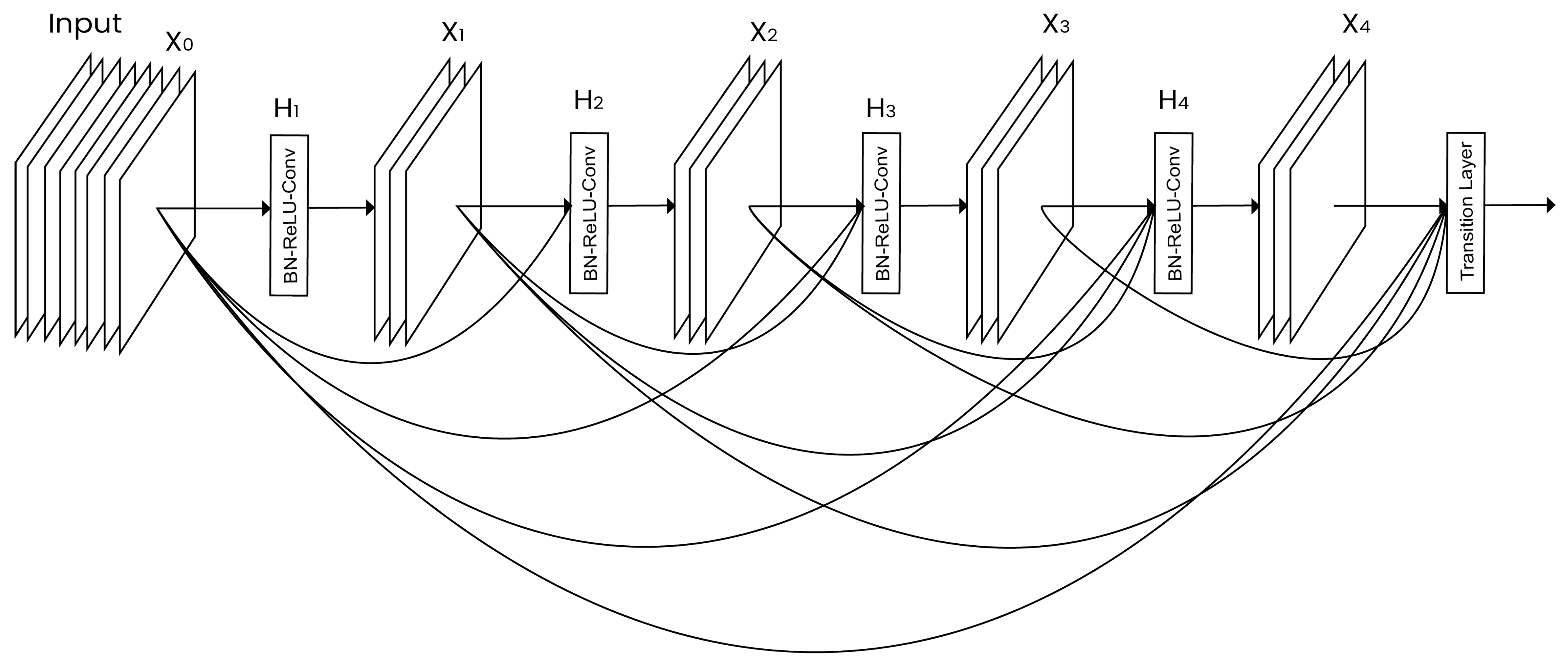
- DenseNet: Introduced by Huang et al. [31], this network contains direct connections between any two layers having the same size as the feature maps. DenseNet, as shown in Figure 3, reduces the vanishing gradient problem, reinforces the feature propagation, vitalizes the reuse of features, and significantly decreases the number of parameters.
- AlexNet: Introduced by Krizhevsky et al. [30]. The architecture is built using eight layers, where the first five are used for the convolution, and the remaining layers are fully connected layers with a softmax function in the last layer.
- SqueezeNet: Introduced by Iandola et al. in 2016 [33]. SqueezeNet begins with a convolutional layer, followed by nine fire modules, and ends with a convolutional layer. The fire modules in the architecture contain a max pooling operation, where the max pooling has a stride of 2.
- VGGNet: This architecture was introduced by Simonyan and Zisserman in 2014 [20]. Only 3 × 3 convolutional layers are used in the VGG network, and they are stacked on top of one another in increasing depths. In addition, max pooling handles reduce volume size. Next comes a softmax classifier, which is composed of two completely connected layers.
2.5. Model Building
2.6. Performance Evaluation Metrics
2.7. Hardware
3. Results and Discussion
3.1. Experimental Details
- Data wrangling: The dataset contains only three images of the basophil class, since it may show little significance when compared to other forms of WBCs. As a result, this form of WBC was removed, and the CNN models were trained with the four different types of WBCs.
- Constructing a datablock: In Fastai [39], a datablock is a high-level application programming interface. It is a method for carefully defining all of the stages involved in preparing data for deep learning systems. The fundamental components of the datablock are listed below:
- (a)
- Train-test division: The splitting ratio was 8:2 for dividing the dataset into training and testing.
- (b)
- Resizing of images: Every image from the dataset was converted to 224 × 224 size, and then the entire batch of images was converted to 128 × 128.
- (c)
- Data augmentation: The resized images underwent further data augmentation operations, i.e., rotation, zoom, perspective warping, and lighting (change in brightness and contrast). This process of generating random variations of the input data so that they appear unique but do not alter the data’s underlying significance is referred to as data augmentation.Data augmentation is the process of generating random variations of the input data so that they appear unique but do not alter the data’s underlying significance.
- (d)
- Model hyperparameters: The settings used for model building were as follows:
- Learning rate: 0.001;
- Optimizer: Adam;
- Epochs: 9;
- Batch size: 64;
- Loss function: cross-entropy loss function.
- Dataloaders: They store multiple dataloader objects (i.e., a training dataloader and a validation dataloader).
3.2. Model Benchmarking
3.3. Model Behavior
3.3.1. Normalization
3.3.2. Mixup Augmentation
- Pick another data at random from your dataset;
- Randomly choose a weight;
- Take a weighted average of your image and the image you choose in step 2; this will serve as your independent variable;
- Take a weighted average of the labels on this image and the labels on your image; the result will be your dependent variable.
3.3.3. Label Smoothing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brundha, M.; Pathmashri, V.; Sundari, S. Quantitative Changes of Red Blood cells in Cancer Patients under Palliative Radiotherapy-A Retrospective Study. Res. J. Pharm. Technol. 2019, 12, 687–692. [Google Scholar] [CrossRef]
- Aliko, V.; Qirjo, M.; Sula, E.; Morina, V.; Faggio, C. Antioxidant defense system, immune response and erythron profile modulation in gold fish, Carassius auratus, after acute manganese treatment. Fish Shellfish. Immunol. 2018, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Seckin, B.; Ates, M.C.; Kirbas, A.; Yesilyurt, H. Usefulness of hematological parameters for differential diagnosis of endometriomas in adolescents/young adults and older women. Int. J. Adolesc. Med. Health 2018, 33. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.C.; Zhang, H.W.; Yu, J.; Xu, H.J.; Chen, H.; Luo, S.P.; Zhang, H.; Liang, L.H.; Wu, X.L.; Lei, Y.; et al. CT imaging and differential diagnosis of COVID-19. Can. Assoc. Radiol. J. 2020, 71, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Vaitkeviciene, G.; Heyman, M.; Jonsson, O.; Lausen, B.; Harila-Saari, A.; Stenmarker, M.; Taskinen, M.; Zvirblis, T.; Åsberg, A.; Groth-Pedersen, L.; et al. Early morbidity and mortality in childhood acute lymphoblastic leukemia with very high white blood cell count. Leukemia 2013, 27, 2259–2262. [Google Scholar] [CrossRef]
- Hu, Z.; Tang, J.; Wang, Z.; Zhang, K.; Zhang, L.; Sun, Q. Deep learning for image-based cancer detection and diagnosis-A survey. Pattern Recognit. 2018, 83, 134–149. [Google Scholar] [CrossRef]
- Shen, L.; Margolies, L.R.; Rothstein, J.H.; Fluder, E.; McBride, R.; Sieh, W. Deep learning to improve breast cancer detection on screening mammography. Sci. Rep. 2019, 9, 12495. [Google Scholar] [CrossRef]
- Mambou, S.J.; Maresova, P.; Krejcar, O.; Selamat, A.; Kuca, K. Breast cancer detection using infrared thermal imaging and a deep learning model. Sensors 2018, 18, 2799. [Google Scholar] [CrossRef]
- Saraswat, M.; Arya, K. Automated microscopic image analysis for leukocytes identification: A survey. Micron 2014, 65, 20–33. [Google Scholar] [CrossRef]
- Anilkumar, K.; Manoj, V.; Sagi, T. A survey on image segmentation of blood and bone marrow smear images with emphasis to automated detection of Leukemia. Biocybern. Biomed. Eng. 2020, 40, 1406–1420. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, Y.; Wang, G.; Liu, J. Fast and robust segmentation of white blood cell images by self-supervised learning. Micron 2018, 107, 55–71. [Google Scholar] [CrossRef]
- Sabino, D.M.U.; da Fontoura Costa, L.; Rizzatti, E.G.; Zago, M.A. A texture approach to leukocyte recognition. Real-Time Imaging 2004, 10, 205–216. [Google Scholar] [CrossRef]
- Hegde, R.B.; Prasad, K.; Hebbar, H.; Singh, B.M.K. Comparison of traditional image processing and deep learning approaches for classification of white blood cells in peripheral blood smear images. Biocybern. Biomed. Eng. 2019, 39, 382–392. [Google Scholar] [CrossRef]
- Singh, I.; Singh, N.P.; Singh, H.; Bawankar, S.; Ngom, A. Blood cell types classification using CNN. In Proceedings of the International Work-Conference on Bioinformatics and Biomedical Engineering, Granada, Spain, 6–8 May 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 727–738. [Google Scholar]
- Ma, L.; Shuai, R.; Ran, X.; Liu, W.; Ye, C. Combining DC-GAN with ResNet for blood cell image classification. Med. Biol. Eng. Comput. 2020, 58, 1251–1264. [Google Scholar] [CrossRef]
- Şengür, A.; Akbulut, Y.; Budak, Ü.; Cömert, Z. White blood cell classification based on shape and deep features. In Proceedings of the 2019 International Artificial Intelligence and Data Processing Symposium (IDAP), Malatya, Turkey, 21–22 September 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–4. [Google Scholar]
- Patil, A.; Patil, M.; Birajdar, G. White blood cells image classification using deep learning with canonical correlation analysis. IRBM 2021, 42, 378–389. [Google Scholar] [CrossRef]
- Hotelling, H. Relations between two sets of variates. In Breakthroughs in Statistics; Springer: Berlin/Heidelberg, Germany, 1992; pp. 162–190. [Google Scholar]
- Wijesinghe, C.B.; Wickramarachchi, D.N.; Kalupahana, I.N.; Lokesha, R.; Silva, I.D.; Nanayakkara, N.D. Fully Automated Detection and Classification of White Blood Cells. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1816–1819. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Jung, C.; Abuhamad, M.; Alikhanov, J.; Mohaisen, A.; Han, K.; Nyang, D. W-net: A CNN-based architecture for white blood cells image classification. arXiv 2019, arXiv:1910.01091. [Google Scholar]
- Rezatofighi, S.H.; Soltanian-Zadeh, H. Automatic recognition of five types of white blood cells in peripheral blood. Comput. Med. Imaging Graph. 2011, 35, 333–343. [Google Scholar] [CrossRef]
- Ucar, F. Deep Learning Approach to Cell Classification in Human Peripheral Blood. In Proceedings of the 2020 5th International Conference on Computer Science and Engineering (UBMK), Diyarbakir, Turkey, 9–11 September 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 383–387. [Google Scholar]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6848–6856. [Google Scholar]
- Karthikeyan, M.; Venkatesan, R.; Vijayakumar, V.; Ravi, L.; Subramaniyaswamy, V. White blood cell detection and classification using Euler’s Jenks optimized multinomial logistic neural networks. J. Intell. Fuzzy Syst. 2020, 39, 8333–8343. [Google Scholar] [CrossRef]
- Ghosh, P.; Bhattacharjee, D.; Nasipuri, M. Blood smear analyzer for white blood cell counting: A hybrid microscopic image analyzing technique. Appl. Soft Comput. 2016, 46, 629–638. [Google Scholar] [CrossRef]
- Chan, T.F.; Vese, L.A. Active contours without edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Rawat, J.; Singh, A.; Bhadauria, H.; Virmani, J.; Devgun, J. Leukocyte classification using adaptive neuro-fuzzy inference system in microscopic blood images. Arab. J. Sci. Eng. 2018, 43, 7041–7058. [Google Scholar] [CrossRef]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a convolutional neural network. In Proceedings of the 2017 International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–6. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Mooney, P. Blood Cell Images. 2018. Available online: www.kaggle.com/datasets/paultimothymooney/blood-cells (accessed on 5 August 2022).
- Valueva, M.V.; Nagornov, N.; Lyakhov, P.A.; Valuev, G.V.; Chervyakov, N.I. Application of the residue number system to reduce hardware costs of the convolutional neural network implementation. Math. Comput. Simul. 2020, 177, 232–243. [Google Scholar] [CrossRef]
- Sabour, S.; Frosst, N.; Hinton, G.E. Dynamic routing between capsules. arXiv 2017, arXiv:1710.09829. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 630–645. [Google Scholar]
- Olson, D.L.; Delen, D. Performance evaluation for predictive modeling. In Advanced Data Mining Techniques; Springer: Berlin/Heidelberg, Germany, 2008; pp. 137–147. [Google Scholar]
- Howard, J.; Gugger, S. Fastai: A layered API for deep learning. Information 2020, 11, 108. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, PMLR, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. mixup: Beyond empirical risk minimization. arXiv 2017, arXiv:1710.09412. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]

| Model | Average Time | Trainable Parameters | Accuracy |
|---|---|---|---|
| AlexNet | 0:52 | 2,735,936 | 0.9702 |
| DenseNet 121 | 2:89 | 8,009,600 | 0.9967 |
| DenseNet 161 | 4:24 | 28,744,896 | 1.0 |
| ResNet 18 | 1:10 | 11,705,920 | 0.9939 |
| ResNet 34 | 1:44 | 21,814,080 | 0.9979 |
| ResNet 50 | 2:52 | 25,616,448 | 0.9991 |
| SqueezeNet 10 | 1:01 | 1,264,832 | 0.9851 |
| SqueezeNet 11 | 0:57 | 1,251,904 | 0.9754 |
| VGG Net 11 | 2:24 | 9,755,392 | 0.9939 |
| VGG Net 13 | 3:40 | 9,940,288 | 0.9967 |
| Epoch | Training Loss | Validation Loss | Accuracy | Error Rate | F1 | Precision | Recall | ROCAUC | Time |
|---|---|---|---|---|---|---|---|---|---|
| Normalization | |||||||||
| 0 | 0.140218 | 0.057371 | 0.978689 | 0.021311 | 0.978645 | 0.978585 | 0.979020 | 0.999439 | 04:24 |
| 1 | 0.100621 | 0.014288 | 0.996783 | 0.003217 | 0.996785 | 0.996747 | 0.996825 | 0.999965 | 04:23 |
| 2 | 0.055214 | 0.010322 | 0.996381 | 0.003619 | 0.996385 | 0.996256 | 0.996565 | 1.000000 | 04:24 |
| 3 | 0.040718 | 0.002779 | 0.999196 | 0.000804 | 0.999196 | 0.999239 | 0.999155 | 1.000000 | 04:23 |
| 4 | 0.022699 | 0.001568 | 1.000000 | 0.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 04:23 |
| 5 | 0.017631 | 0.001189 | 1.000000 | 0.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 04:23 |
| Mixup Augmentation | |||||||||
| 0 | 0.632420 | 0.089475 | 0.978287 | 0.021713 | 0.978325 | 0.978680 | 0.978054 | 0.999194 | 04:29 |
| 1 | 0.555610 | 0.072873 | 0.983514 | 0.016486 | 0.983525 | 0.983634 | 0.984351 | 0.999898 | 04:32 |
| 2 | 0.503748 | 0.042065 | 0.995979 | 0.004021 | 0.995983 | 0.995864 | 0.996143 | 0.999975 | 04:32 |
| 3 | 0.477047 | 0.033872 | 0.999196 | 0.000804 | 0.999196 | 0.999158 | 0.999237 | 0.999999 | 04:29 |
| 4 | 0.452092 | 0.028028 | 0.999196 | 0.000804 | 0.999196 | 0.999196 | 0.999196 | 0.999999 | 04:30 |
| 5 | 0.444563 | 0.027773 | 0.999196 | 0.000804 | 0.999196 | 0.999196 | 0.999196 | 1.000000 | 04:29 |
| Label Smoothing | |||||||||
| 0 | 0.520124 | 0.432285 | 0.979091 | 0.020909 | 0.979164 | 0.979401 | 0.979894 | 0.999494 | 04:29 |
| 1 | 0.442535 | 0.383923 | 0.999598 | 0.000402 | 0.999598 | 0.999619 | 0.999578 | 0.999997 | 04:31 |
| 2 | 0.411049 | 0.366136 | 0.999598 | 0.000402 | 0.999598 | 0.999578 | 0.999618 | 1.000000 | 04:30 |
| 3 | 0.389663 | 0.360205 | 1.000000 | 0.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 04:29 |
| 4 | 0.379095 | 0.357764 | 1.000000 | 0.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 04:28 |
| 5 | 0.374234 | 0.356941 | 1.000000 | 0.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 04:29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamang, T.; Baral, S.; Paing, M.P. Classification of White Blood Cells: A Comprehensive Study Using Transfer Learning Based on Convolutional Neural Networks. Diagnostics 2022, 12, 2903. https://doi.org/10.3390/diagnostics12122903
Tamang T, Baral S, Paing MP. Classification of White Blood Cells: A Comprehensive Study Using Transfer Learning Based on Convolutional Neural Networks. Diagnostics. 2022; 12(12):2903. https://doi.org/10.3390/diagnostics12122903
Chicago/Turabian StyleTamang, Thinam, Sushish Baral, and May Phu Paing. 2022. "Classification of White Blood Cells: A Comprehensive Study Using Transfer Learning Based on Convolutional Neural Networks" Diagnostics 12, no. 12: 2903. https://doi.org/10.3390/diagnostics12122903
APA StyleTamang, T., Baral, S., & Paing, M. P. (2022). Classification of White Blood Cells: A Comprehensive Study Using Transfer Learning Based on Convolutional Neural Networks. Diagnostics, 12(12), 2903. https://doi.org/10.3390/diagnostics12122903





