A Computerized Analysis with Machine Learning Techniques for the Diagnosis of Parkinson’s Disease: Past Studies and Future Perspectives
Abstract
1. Introduction
1.1. Artificial Intelligence and Machine Learning-Based Detection of Parkinson’s Disease
1.2. Research Problem and Motivation of Current Systematic Review
1.3. Contribution
- In this paper, we reviewed the significant statistics and relevant information collected from 217 articles (from various resources) published from 2015–2022 on the diagnosis and classification of PD.
- The fundamental discussion on AI and ML with their significance in the field of medical healthcare.
- In order to improve the prediction of PD, we also present recommendations for future perspectives to help researchers and scholars in recognizing various plausible paths for them to work in the future.
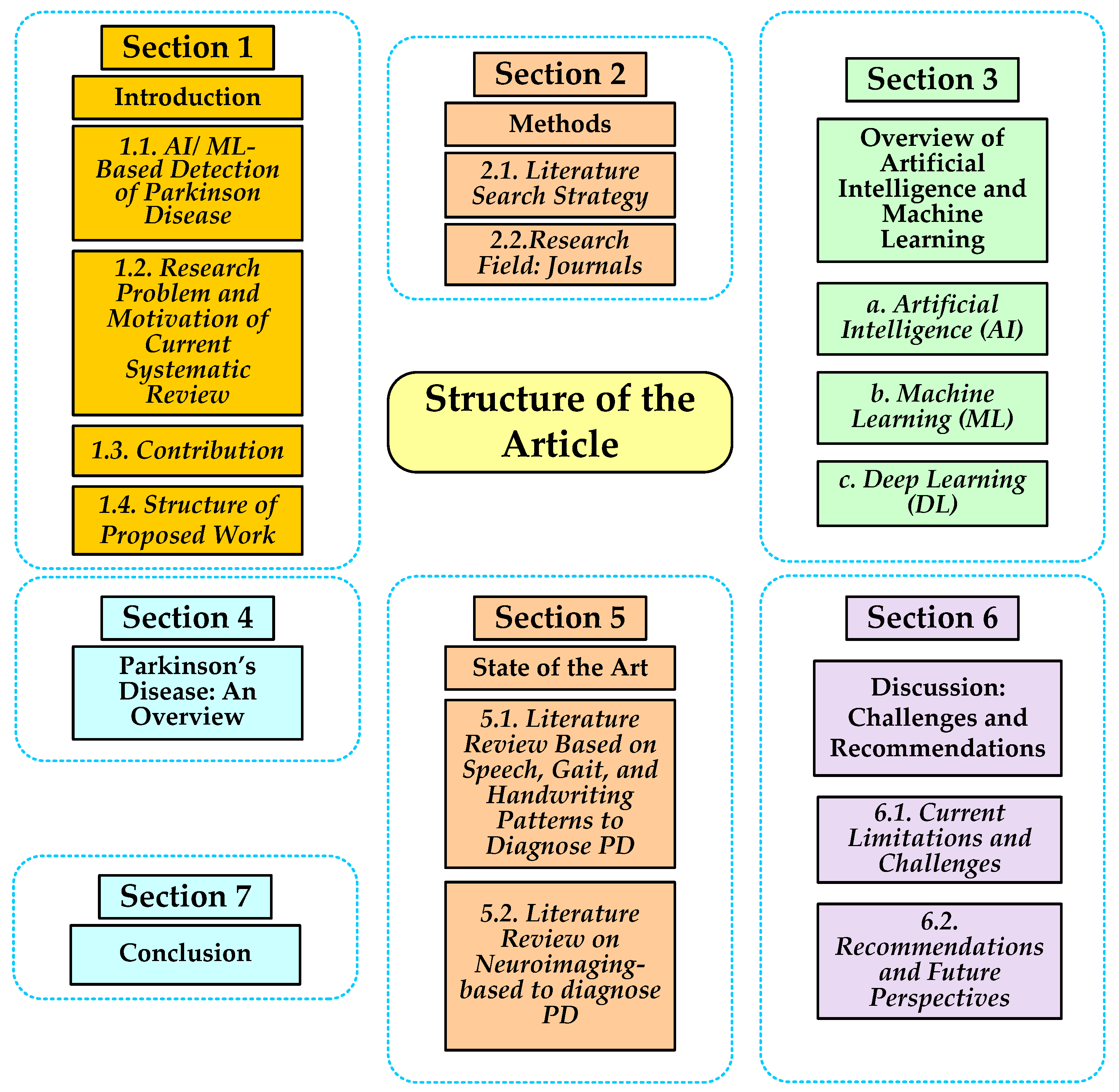
1.4. Structure of Proposed Work
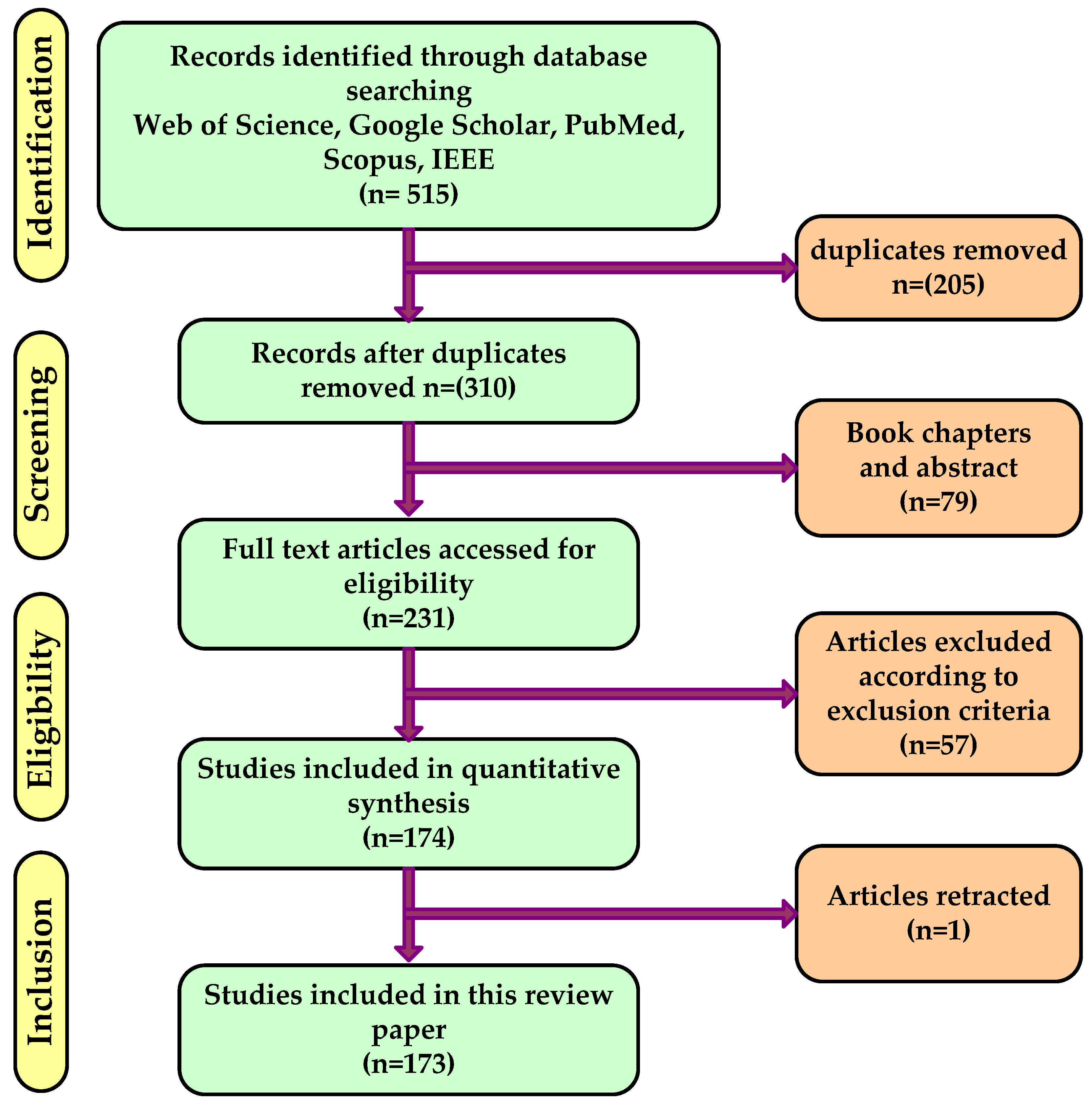
2. Methods
2.1. Literature Search Strategy
2.2. Research Field: Journals
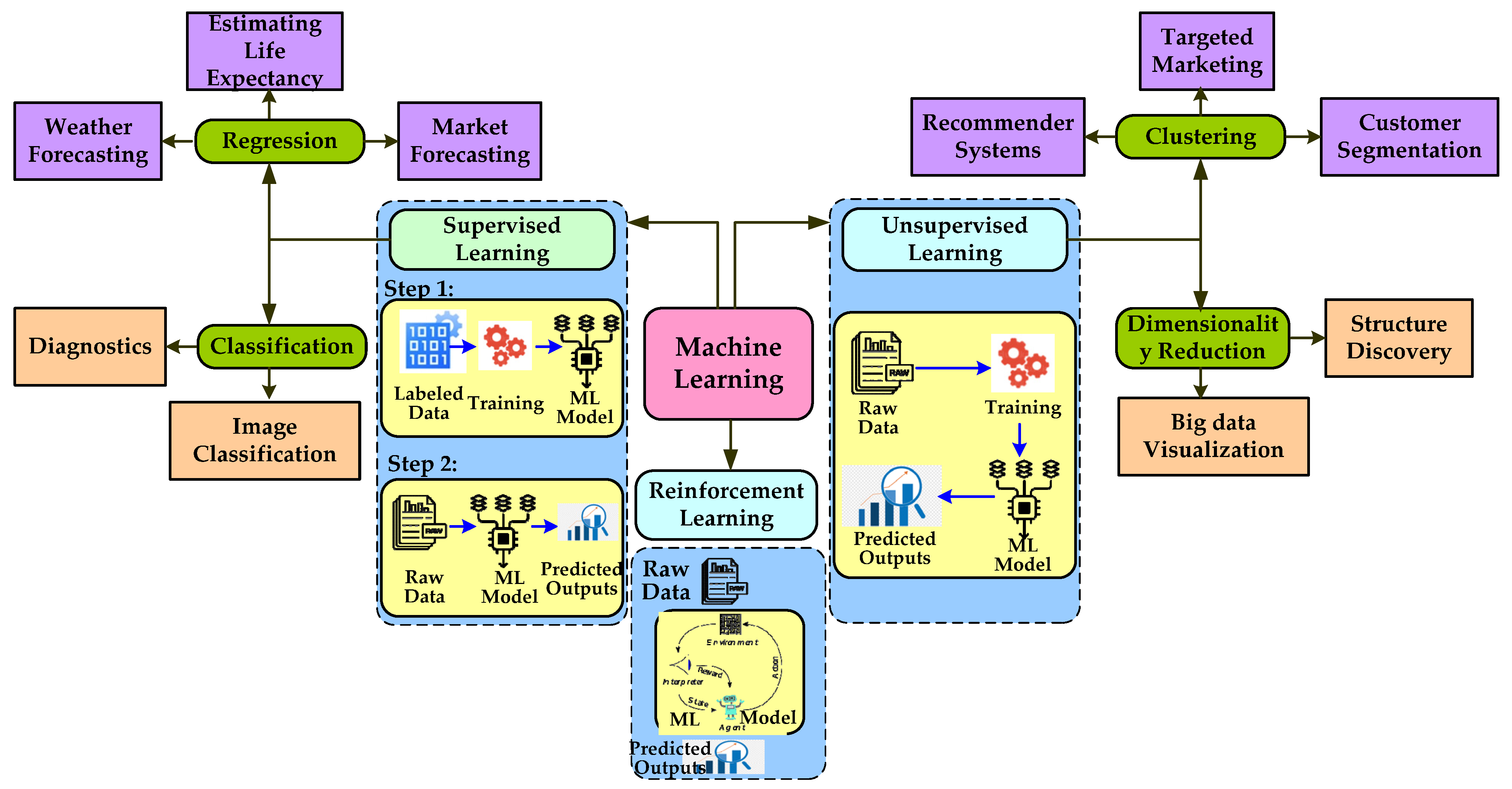
3. Overview of Artificial Intelligence, Machine Learning, and Deep Learning
- (a)
- Artificial Intelligence
- (b)
- Machine Learning
- (c)
- Deep Learning
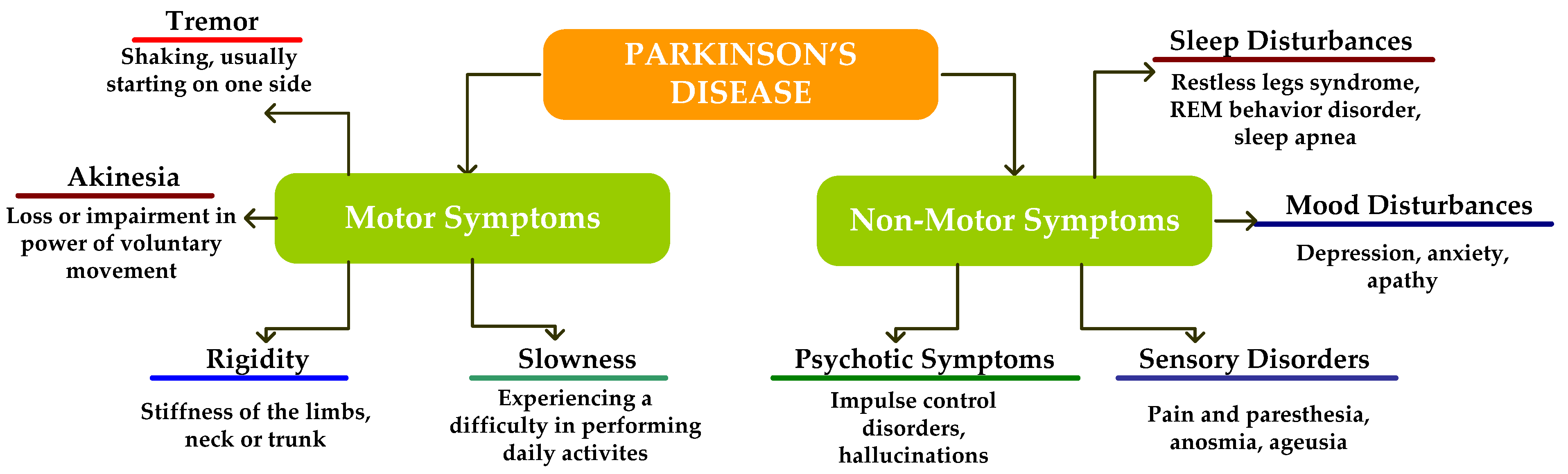
4. Parkinson’s Disease: An Overview
Medical Approaches for Parkinson’s Disease Diagnosis
- a.
- Medical Treatment
- b.
- Dopamine Agonists
- c.
- COMT Inhibitors
- d.
- Selegiline
- e.
- Anticholinergic medications
- f.
- Amantadine
5. State of the Art
5.1. Literature Review Based on Speech, Gait, and Handwriting Patterns to Diagnose PD
5.2. Literature Review on Neuroimagingto Diagnose PD
6. Discussion: Challenges and Recommendations
6.1. Current Limitations and Challenges
- a.
- Issues with Multimodality datasets for PD detection
- b.
- Issues with Machine Learning Techniques
- c.
- Issues with Clinical Validation
6.2. Recommendations and Future Perspectives
- Make the most of all available data sources. It might be difficult to gather all available modalities for each subject. For instance, a PET scan, a costly neuroimaging modality, is not performed on some participants, although practically all of the subjects’ clinical records list an MRI. This is true for publicly accessible data like ADNI. To fill up the gaps in the data, we advise employing certain methods. For instance, utilizing MRI data to complete missing PET data [204], creating CT scans from MRIs [205,206,207], cycle-consistent generative adversarial networks (GAN) [208], and feature-consistent GAN [207]. As an alternative, deep designs that include handling procedures for missing data can be applied [209,210,211,212,213]. Additionally, data augmentation might be useful in this context to increase the dataset and address unbalanced classes. Through image modification operators including rotation [214], scaling and shifting [215], changing intensity, contrast, and saturation [216], as well as noise injection and random translation [217], data augmentation may be accomplished.
- The widespread usage of the CNN algorithm [218,219] on MRI image data is a significant discovery. Comparing these models to other algorithms, they frequently produce favorable outcomes. Researchers might wish to conduct further research and use more CNN-based hybrid algorithms [220,221,222]. Additionally, we found that classifying images has not frequently utilized the learning algorithm. This is an opportunity for future researchers to utilize attention to raise the precision of deep learning models.
- At present, wearable sensors are only useful for using gait parameters to diagnose PD. The wearable device that can detect PD must have the other modules included in it. In addition to one symptom, researchers must concentrate on creating wearable sensor systems that can diagnose additional symptoms also. For instance, a wrist-worn sensor might be created that can track data constantly over a long period of time and recognize various PD symptoms.
- Currently, various ML models have been developed by researchers that can diagnose PD based on a patient’s specific symptoms. The researchers should focus on establishing an ML model for diagnosing PD that takes all the symptoms as input. A lightweight portable device can be used to diagnose the various symptoms of PD by measuring several parameters such as accuracy, precision, sensitivity, recall, etc. This device should be easily wearable and washable, and it should be able to identify the different stages of the disease, along with analyzing the changes due to medication treatment.
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 3-OMD | 3-O-Methyldopa |
| ACC | Accuracy |
| ACh | Acetylcholine |
| AD | Alzheimer’s Disease |
| AI | Artificial Intelligence |
| ANN | Artificial Neural Network |
| ASD | Autism Spectrum Disorders |
| AUC | Area Under the Curve |
| CAD | Computer-Aided Diagnosis |
| CNN | Convolutional Neural Network |
| COMT | Catechol-O-Methyl Transferase |
| CSF | Cerebrospinal Fluid |
| DaTSCAN | Dopamine Transporter Scan |
| DBF | Discrimination-Based Function |
| DBSCAN | Density-Based Spatial |
| DL | Deep Learning |
| DWT | Discrete Wavelet Transform |
| ET | Essential Tremors |
| GAN | Generative Adversarial Networks |
| GD | Gradient Descent |
| GDS | Geriatric Depression Scale |
| HC | Healthy Controls |
| IEEE | Institute of Electrical and Electronics Engineers |
| KNN | K-Nearest Neighbor |
| LDA | Linear Discriminant Analysis |
| LIME | Local Interpretable Model-Agnostic Explainer |
| LSTM | Long Short-Term Memory |
| LSVRC | Large Scale Visual Recognition Competition |
| MAO-B | Monoamine oxidase B |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MFCC | Mel-Frequency Cepstral Coefficients |
| ML | Machine Learning |
| MLP | Multilayer Perceptron |
| MoCA | Montreal Cognitive Assessment |
| MMSE | Mini-Mental Score examination |
| MSA-P | Multiple System Atrophy with Predominant Parkinsonian Features |
| MVDA | Multi-Variate Vocal Data Analysis |
| NB | Naive Bayes |
| NC | Normal Control |
| ND | Normal Development |
| NLP | Natural Language Processing |
| OCT | Optical Coherence Tomography |
| PCA | Principal Component Analysis |
| PD | Parkinson’s Disease |
| PDSS | Panic Disorder Severity Scale |
| PET | Positron Emission Tomography |
| PHT | Pathological Hand Tremor |
| PIGD | Postural Instability Gait Disorder |
| PLOS | Public Library of Science |
| PPMI | Parkinson’s Progression Markers Initiative |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| PSP | Progressive Supranuclear Palsy |
| RBD | Random Eye Movement Sleep Behavior Disorder |
| RL | Reinforcement Learning |
| SCOPA-AUT | Scales for Outcomes in Parkinson’s Disease-Autonomic |
| SDC | Shifted Delta Cepstral |
| SFFCC | Single Frequency Filtering Cepstral Coefficients |
| SL | Supervised Learning |
| sMRI | Structural Magnetic Resonance Imaging |
| SPECT | Single-Photon Emission Computerized Tomography |
| STAI | State-Trait Anxiety Inventory for Adults |
| SVM | Support Vector Machine |
| SZ | Schizophrenia |
| UL | Unsupervised Learning |
| UPDRS | Unified Parkinson’s Disease Rating Scale |
| WHO | World Health Organization |
References
- DeMaagd, G.; Philip, A. Parkinson’s Disease and Its Management: Part 1: Disease Entity, Risk Factors, Pathophysiology, Clinical Presentation, and Diagnosis. Pharm. Ther. 2015, 40, 504–532. [Google Scholar]
- Rizek, P.; Kumar, N.; Jog, M.S. An update on the diagnosis and treatment of Parkinson disease. CMAJ 2016, 188, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Arias-Vergara, T.; Vásquez-Correa, J.C.; Orozco-Arroyave, J.R. Parkinson’s Disease and Aging: Analysis of Their Effect in Phonation and Articulation of Speech. Cogn. Comput. 2017, 9, 731–748. [Google Scholar] [CrossRef]
- De Rijk, M.D.; Launer, L.J.; Berger, K.; Breteler, M.M.; Dartigues, J.F.; Baldereschi, M.; Fratiglioni, L.; Lobo, A.; Martinez-Lage, J.; Trenkwalder, C.; et al. Prevalence of Parkinson’s disease in Europe: A collaborative study of population-based cohorts. Neurologic Diseases in the Elderly Research Group. Neurology 2000, 54 (Suppl. 5), S21–S23. [Google Scholar] [PubMed]
- Cantürk, İ.; Karabiber, F. A machine learning system for the diagnosis of Parkinson’s disease from speech signals and its application to multiple speech signal types. Arab. J. Sci. Eng. 2016, 41, 5049–5059. [Google Scholar] [CrossRef]
- Singh, N.; Pillay, V.; Choonara, Y.E. Advances in the treatment of Parkinson’s disease. Prog. Neurobiol. 2007, 81, 29–44. [Google Scholar] [CrossRef]
- Sakar, B.E.; Isenkul, M.E.; Sakar, C.O.; Sertbas, A.; Gurgen, F.; Delil, S.; Apaydin, H.; Kursun, O. Collection and analysis of a Parkinson speech dataset with multiple types of sound recordings. IEEE J. Biomed. Health Inform. 2013, 17, 828–834. [Google Scholar] [CrossRef]
- Abujrida, H.; Agu, E.; Pahlavan, K. Smartphone-based gait assessment to infer Parkinson’s disease severity using crowdsourced data. In Proceedings of the 2017 IEEE Healthcare Innovations and Point of Care Technologies (HI-POCT), Bethesda, MD, USA, 6–8 November 2017; pp. 208–211. [Google Scholar]
- Adams, W.R. High-accuracy detection of early Parkinson’s Disease using multiple characteristics of finger movement while typing. PLoS ONE 2017, 12, e0188226. [Google Scholar] [CrossRef]
- Adeli, E.; Shi, F.; An, L.; Wee, C.-Y.; Wu, G.; Wang, T.; Shen, D. Joint feature-sample selection and robust diagnosis of Parkinson’s disease from MRI data. Neuroimage 2016, 14, 206–219. [Google Scholar] [CrossRef]
- Adeli, E.; Thung, K.-H.; An, L.; Wu, G.; Shi, F.; Wang, T.; Shen, D. Semi-Supervised Discriminative Classification Robust to Sample-Outliers and Feature-Noises. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 41, 515–522. [Google Scholar] [CrossRef]
- Agarwal, A.; Chandrayan, S.; Sahu, S.S. Prediction of Parkinson’s disease using speech signal with Extreme Learning Machine. In Proceedings of the 2016 International Conference on Electrical, Electronics, and Optimization Techniques (ICEEOT), Chennai, India, 3–5 March 2016; pp. 3776–3779. [Google Scholar]
- Ahmadi, S.-A.; Vivar, G.; Frei, J.; Nowoshilow, S.; Bardins, S.; Brandt, T.; Krafczyk, S. Towards computerized diagnosis of neurological stance disorders: Data mining and machine learning of posturography and sway. J. Neurol. 2019, 266 (Suppl. 1), 108–117. [Google Scholar] [CrossRef] [PubMed]
- Aich, S.; Kim, H.; younga, K.; Hui, K.L.; Al-Absi, A.A.; Sain, M. A Supervised Machine Learning Approach Using Different Feature Selection Techniques on Voice Datasets for Prediction of Parkinson’s Disease. In Proceedings of the 2019 21st International Conference on Advanced Communication Technology (ICACT), Pyeong Chang, Korea, 17–20 February 2019; pp. 1116–1121. [Google Scholar]
- Al-Fatlawi, A.H.; Jabardi, M.H.; Ling, S.H. Efficient diagnosis system for Parkinson’s disease using deep belief network. In Proceedings of the 2016 IEEE Congress on Evolutionary Computation (CEC), Vancouver, BC, Canada, 24–29 July 2016; pp. 1324–1330. [Google Scholar]
- Alam, M.N.; Garg, A.; Munia, T.T.K.; Fazel-Rezai, R.; Tavakolian, K. Vertical ground reaction force marker for Parkinson’s disease. PLoS ONE 2017, 12, e0175951. [Google Scholar] [CrossRef] [PubMed]
- Alaskar, H.; Hussain, A. Prediction of Parkinson Disease Using Gait Signals. In Proceedings of the 2018 11th International Conference on Developments in eSystems Engineering (DeSE), Cambridge, UK, 2–5 September 2018; pp. 23–26. [Google Scholar]
- Alharthi, A.S.; Ozanyan, K.B. Deep Learning for Ground Reaction Force Data Analysis: Application to Wide-Area Floor Sensing. In Proceedings of the 2019 IEEE 28th International Symposium on Industrial Electronics (ISIE), Vancouver, BC, Canada, 12–14 June 2019; pp. 1401–1406. [Google Scholar]
- Ali, L.; Khan, S.U.; Arshad, M.; Ali, S.; Anwar, M. A Multi-model Framework for Evaluating Type of Speech Samples having Complementary Information about Parkinson’s Disease. In Proceedings of the 2019 International Conference on Electrical, Communication, and Computer Engineering (ICECCE), Swat, Pakistan, 24–25 July 2019; pp. 1–5. [Google Scholar]
- Ali, L.; Zhu, C.; Golilarz, N.A.; Javeed, A.; Zhou, M.; Liu, Y. Reliable Parkinson’s Disease Detection by Analyzing Handwritten Drawings: Construction of an Unbiased Cascaded Learning System Based on Feature Selection and Adaptive Boosting Model. IEEE Access 2019, 7, 116480–116489. [Google Scholar] [CrossRef]
- Ali, L.; Zhu, C.; Zhang, Z.; Liu, Y. Automated Detection of Parkinson’s Disease Based on Multiple Types of Sustained Phonations Using Linear Discriminant Analysis and Genetically Optimized Neural Network. IEEE J. Transl. Eng. Health Med. 2019, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, E.J.; Alshamrani, F.H.; Syed, H.F.; Olatunji, S.O. Classification of Parkinson’s Disease Using NNge Classification Algorithm. In Proceedings of the 2018 21st Saudi Computer Society National Computer Conference (NCC), Riyadh, Saudi Arabia, 25–26 April 2018; pp. 1–7. [Google Scholar]
- Amoroso, N.; la Rocca, M.; Monaco, A.; Bellotti, R.; Tangaro, S. Complex networks reveal early MRI markers of Parkinson’s disease. Med. Image Anal. 2018, 48, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; Haque, M.A.; Alex, J.S.R.; Venkatesan, N. Evaluation of Machine learning and Deep learning algorithms combined with dimentionality reduction techniques for classification of Parkinson’s Disease. In Proceedings of the 2018 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT), Louisville, KY, USA, 6–8 December 2018; pp. 342–347. [Google Scholar]
- Khodatars, M.; Shoeibi, A.; Sadeghi, D.; Ghaasemi, N.; Jafari, M.; Moridian, P.; Berk, M. Deep learning for neuroimaging-based diagnosis and rehabilitation of autism spectrum disorder: A review. Comput. Biol. Med. 2021, 139, 104949. [Google Scholar] [CrossRef] [PubMed]
- Baby, M.S.; Saji, A.J.; Kumar, C.S. Parkinsons disease classification using wavelet transform based feature extraction of gait data. In Proceedings of the 2017 International Conference on Circuit, Power and Computing Technologies (ICCPCT), Kollam, India, 20–21 April 2017; pp. 1–6. [Google Scholar]
- Baggio, H.C.; Abos, A.; Segura, B.; Campabadal, A.; Uribe, C.; Giraldo, D.M.; Perez-Soriano, A.; Muñoz, E.; Compta, Y.; Junque, C.; et al. Cerebellar resting-state functional connectivity in Parkinson’s disease and multiple system atrophy: Characterization of abnormalities and potential for differential diagnosis at the single-patient level. Neuroimage Clin. 2019, 22, 101720. [Google Scholar] [CrossRef]
- Bakar, Z.A.; Ispawi, D.I.; Ibrahim, N.F.; Tahir, N.M. Classification of Parkinson’s disease based on Multilayer Perceptrons (MLPs) Neural Network and ANOVA as a feature extraction. In Proceedings of the 2012 IEEE 8th International Colloquium on Signal Processing and Its Applications, Malacca, Malaysia, 23–25 March 2012; pp. 63–67. [Google Scholar]
- Banerjee, M.; Chakraborty, R.; Archer, D.; Vaillancourt, D.; Vemuri, B.C. DMR-CNN: A CNN Tailored for DMR Scans with Applications to PD Classification. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 388–391. [Google Scholar]
- Benba, A.; Jilbab, A.; Hammouch, A. Discriminating between Patients with Parkinson’s and Neurological Diseases Using Cepstral Analysis. IEEE Trans. Neural Syst. Rehabil. Eng. 2016, 24, 1100–1108. [Google Scholar] [CrossRef]
- Benba, A.; Jilbab, A.; Hammouch, A.; Sandabad, S. Using RASTA-PLP for discriminating between different Neurological diseases. In Proceedings of the 2016 International Conference on Electrical and Information Technologies (ICEIT), Tangiers, Morocco, 4–7 May 2016; pp. 406–409. [Google Scholar]
- Bernad-Elazari, H.; Herman, T.; Mirelman, A.; Gazit, E.; Giladi, N.; Hausdorff, J.M. Objective characterization of daily living transitions in patients with Parkinson’s disease using a single body-fixed sensor. J. Neurol. 2016, 263, 1544–1551. [Google Scholar] [CrossRef]
- Bhati, S.; Velazquez, L.M.; Villalba, J.; Dehak, N. LSTM Siamese Network for Parkinson’s Disease Detection from Speech. In Proceedings of the 2019 IEEE Global Conference on Signal and Information Processing (GlobalSIP), Ottawa, ON, Canada, 11–14 November 2019; pp. 1–5. [Google Scholar]
- Buongiorno, D.; Bortone, I.; Cascarano, G.D.; Trotta, G.F.; Brunetti, A.; Bevilacqua, V. A low-cost vision system based on the analysis of motor features for recognition and severity rating of Parkinson’s Disease. BMC Med. Inform. Decis. Mak. 2019, 19 (Suppl. 9), 243. [Google Scholar] [CrossRef]
- Lakany, H. Extracting a diagnostic gait signature. Pattern Recogn. 2008, 41, 1627–1637. [Google Scholar] [CrossRef]
- Figueiredo, J.; Santos, C.P.; Moreno, J.C. Automatic recognition of gait patterns in human motor disorders using machine learning: A review. Med. Eng. Phys. 2018, 53, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hazan, H.; Hilu, D.; Manevitz, L.; Ramig, L.O.; Sapir, S. Early diagnosis of Parkinson’s disease via machine learning on speech data. In Proceedings of the 27th Convention of Electrical and Electronics Engineers in Israel, Eilat, Israel, 14–17 November 2012; pp. 1–4. [Google Scholar]
- Karan, B.; Sahu, S.S.; Mahto, K. Parkinson disease prediction using intrinsic mode function based features from speech signal. Biocybern. Biomed. Eng. 2020, 40, 249–264. [Google Scholar] [CrossRef]
- Frid, A.; Hazan, H.; Hilu, D.; Manevitz, L.; Ramig, L.O.; Sapir, S. Computational diagnosis of Parkinson’s Disease directly from natural speech using machine learning techniques. In Proceedings of the International Conference on Software Science, Technology and Engineering, Ramat Gan, Israel, 11–12 June 2014; pp. 50–53. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Sibley, K.G.; Girges, C.; Hoque, E.; Foltynie, T. Video-based analyses of Parkinson’s disease severity: A brief review. J. Parkinson’s Dis. 2021, 11, S83–S93. [Google Scholar] [CrossRef]
- Belić, M.; Bobić, V.; Badža, M.; Šolaja, N.; Đurić-Jovičić, M.; Kostić, V.S. Artificial intelligence for assisting diagnostics and assessment of Parkinson’s disease—A review. Clin. Neurol. Neurosurg. 2019, 184, 105442. [Google Scholar] [CrossRef]
- Landers, M.; Saria, S.; Espay, A.J. Will Artificial Intelligence Replace the Movement Disorders Specialist for Diagnosing and Managing Parkinson’s Disease? J. Parkinson’s Dis. 2021, 11, S117–S122. [Google Scholar] [CrossRef]
- Palumbo, B.; Bianconi, F.; Nuvoli, S.; Spanu, A.; Fravolini, M.L. Artificial intelligence techniques support nuclear medicine modalities to improve the diagnosis of Parkinson’s disease and Parkinsonian syndromes. Clin. Transl. Imaging 2021, 9, 19–35. [Google Scholar] [CrossRef]
- Saravanan, S.; Ramkumar, K.; Adalarasu, K.; Sivanandam, V.; Kumar, S.R.; Stalin, S.; Amirtharajan, R. A Systematic Review of Artificial Intelligence (AI) Based Approaches for the Diagnosis of Parkinson’s Disease. Arch. Comput. Methods Eng. 2022, 29, 3639–3653. [Google Scholar] [CrossRef]
- Perju-Dumbrava, L.; Barsan, M.; Leucuta, D.C.; Popa, L.C.; Pop, C.; Tohanean, N.; Popa, S.L. Artificial intelligence applications and robotic systems in Parkinson’s disease. Exp. Ther. Med. 2022, 23, 153. [Google Scholar] [CrossRef]
- Giannakopoulou, K.M.; Roussaki, I.; Demestichas, K. Internet of Things Technologies and Machine Learning Methods for Parkinson’s Disease Diagnosis, Monitoring and Management: A Systematic Review. Sensors 2022, 22, 1799. [Google Scholar] [CrossRef]
- Khachnaoui, H.; Mabrouk, R.; Khlifa, N. Machine learning and deep learning for clinical data and PET/SPECT imaging in Parkinson’s disease: A review. IET Image Process. 2020, 14, 4013–4026. [Google Scholar] [CrossRef]
- Narayanan, R.R.; Durga, N.; Nagalakshmi, S. Impact of Artificial Intelligence (AI) on Drug Discovery and Product Development. Indian J. Pharm. Educ. Res. 2022, 56, S387–S397. [Google Scholar] [CrossRef]
- Termine, A.; Fabrizio, C.; Strafella, C.; Caputo, V.; Petrosini, L.; Caltagirone, C.; Cascella, R. Multi-Layer Picture of Neurodegenerative Diseases: Lessons from the Use of Big Data through Artificial Intelligence. J. Personal. Med. 2021, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Lim AC, Y.; Natarajan, P.; Fonseka, R.D.; Maharaj, M.; Mobbs, R.J. The application of artificial intelligence and custom algorithms with inertial wearable devices for gait analysis and detection of gait-altering pathologies in adults: A scoping review of literature. Digit. Health 2022, 8, 20552076221074128. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, M. Use of magnetic resonance imaging and artificial intelligence in studies of diagnosis of Parkinson’s disease. ACS Chem. Neurosci. 2019, 10, 2658–2667. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, G.; Xu, Y.; Tang, X. Application of artificial intelligence in the MRI classification task of human brain neurological and psychiatric diseases: A scoping review. Diagnostics 2021, 11, 1402. [Google Scholar] [CrossRef]
- Yadav, D.; Garg, R.K.; Chhabra, D.; Yadav, R.; Kumar, A.; Shukla, P. Smart diagnostics devices through artificial intelligence and mechanobiological approaches. 3 Biotech 2020, 10, 351. [Google Scholar] [CrossRef]
- Patil, A.D.; Biousse, V.; Newman, N.J. Artificial intelligence in ophthalmology: An insight into neurodegenerative disease. Curr. Opin. Ophthalmol. 2022, 33, 432–439. [Google Scholar] [CrossRef]
- Suri, J.S.; Paul, S.; Maindarkar, M.A.; Puvvula, A.; Saxena, S.; Saba, L.; Turk, M.; Laird, J.R.; Khanna, N.N.; Viskovic, K.; et al. Cardiovascular/Stroke Risk Stratification in Parkinson’s Disease Patients Using Atherosclerosis Pathway and Artificial Intelligence Paradigm: A Systematic Review. Metabolites 2022, 12, 312. [Google Scholar] [CrossRef]
- Vitale, A.; Villa, R.; Ugga, L.; Romeo, V.; Stanzione, A.; Cuocolo, R. Artificial intelligence applied to neuroimaging data in Parkinsonian syndromes: Actuality and expectations. Math. Biosci. Eng. 2021, 18, 1753–1773. [Google Scholar] [CrossRef] [PubMed]
- Cascianelli, S.; Scialpi, M.; Amici, S.; Forini, N.; Minestrini, M.; Fravolini, M.; Sinzinger, H.; Schillaci, O.; Palumbo, B. Role of artificial intelligence techniques (automatic classifiers) in molecular imaging modalities in neurodegenerative diseases. Curr. Alzheimer Res. 2017, 14, 198–207. [Google Scholar] [CrossRef]
- Rana, A.; Dumka, A.; Singh, R.; Panda, M.K.; Priyadarshi, N.; Twala, B. Imperative Role of Machine Learning Algorithm for Detection of Parkinson’s Disease: Review, Challenges and Recommendations. Diagnostics 2022, 12, 2003. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, U.; Acharya, U.R.; Adeli, H. Artificial intelligence techniques for automated diagnosis of neurological disorders. Eur. Neurol. 2019, 82, 41–64. [Google Scholar] [CrossRef]
- Singh, A.V.; Chandrasekar, V.; Janapareddy, P.; Mathews, D.E.; Laux, P.; Luch, A.; Yang, Y.; Garcia-Canibano, B.; Balakrishnan, S.; Abinahed, J.; et al. Emerging application of nanorobotics and artificial intelligence to cross the BBB: Advances in design, controlled maneuvering, and targeting of the barriers. ACS Chem. Neurosci. 2021, 12, 1835–1853. [Google Scholar] [CrossRef] [PubMed]
- Maitin, A.M.; Romero Muñoz, J.P.; García-Tejedor, Á.J. Survey of Machine Learning Techniques in the Analysis of EEG Signals for Parkinson’s Disease: A Systematic Review. Appl. Sci. 2022, 12, 6967. [Google Scholar] [CrossRef]
- Vatansever, S.; Schlessinger, A.; Wacker, D.; Kaniskan, H.Ü.; Jin, J.; Zhou, M.M.; Zhang, B. Artificial intelligence and machine learning-aided drug discovery in central nervous system diseases: State-of-the-arts and future directions. Med. Res. Rev. 2021, 41, 1427–1473. [Google Scholar] [CrossRef]
- Hansen, C.; Sanchez-Ferro, A.; Maetzler, W. How mobile health technology and electronic health records will change care of patients with Parkinson’s disease. J. Parkinson’s Dis. 2018, 8, S41–S45. [Google Scholar] [CrossRef]
- Luis-Martínez, R.; Monje, M.H.; Antonini, A.; Sánchez-Ferro, Á.; Mestre, T.A. Technology-enabled care: Integrating multidisciplinary care in Parkinson’s disease through digital technology. Front. Neurol. 2020, 11, 575975. [Google Scholar] [CrossRef]
- Fiandaca, M.S.; Lonser, R.R.; Elder, J.B.; Ząbek, M.; Bankiewicz, K.S. Advancing gene therapies, methods, and technologies for Parkinson’s disease and other neurological disorders. Neurol. Neurochir. Pol. 2020, 54, 220–231. [Google Scholar] [CrossRef]
- Kubota, K.J.; Chen, J.A.; Little, M.A. Machine learning for large-scale wearable sensor data in Parkinson’s disease: Concepts, promises, pitfalls, and futures. Mov. Disord. 2016, 31, 1314–1326. [Google Scholar] [CrossRef] [PubMed]
- Rana, A.; Rawat, A.S.; Bijalwan, A.; Bahuguna, H. Application of multi-layer (perceptron) artificial neural network in the diagnosis system: A systematic review. In Proceedings of the 2018 International Conference on Research in Intelligent and Computing in Engineering (RICE), San Salvador, El Salvador, 22–24 August 2018; pp. 1–6. [Google Scholar]
- Baduge, S.K.; Thilakarathna, S.; Perera, J.S.; Arashpour, M.; Sharafi, P.; Teodosio, B.; Mendis, P. Artificial intelligence and smart vision for building and construction 4.0: Machine and deep learning methods and applications. Autom. Constr. 2022, 141, 104440. [Google Scholar] [CrossRef]
- Cunningham, P.; Cord, M.; Delany, S.J. Supervised learning. In Machine Learning Techniques for Multimedia; Springer: Berlin/Heidelberg, Germany, 2008; pp. 21–49. [Google Scholar]
- Li, N.; Shepperd, M.; Guo, Y. A systematic review of unsupervised learning techniques for software defect prediction. Inf. Softw. Technol. 2020, 122, 106287. [Google Scholar] [CrossRef]
- Czech, J. Distributed methods for reinforcement learning survey. In Reinforcement Learning Algorithms: Analysis and Applications; Springer: Cham, Switzerland, 2021; pp. 151–161. [Google Scholar]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Janiesch, C.; Zschech, P.; Heinrich, K. Machine learning and deep learning. Electron. Mark. 2021, 31, 685–695. [Google Scholar] [CrossRef]
- Pereira, C.; Pereira, D.; Papa, J.; Rosa, G.; Yang, X. Convolutional Neural Networks Applied for Parkinson’s Disease Identification. InMachine Learning for Health Informatics; Springer: Cham, Switzerland, 2016; pp. 377–390. [Google Scholar]
- Shaban, M. Deep Convolutional Neural Network for Parkinson’s Disease Based Handwriting Screening. In Proceedings of the IEEE International Symposium on Biomedical Imaging, Iowa City, IA, USA, 4 April 2020. [Google Scholar]
- Frid, A.; Kantor, A.; Svechin, D.; Manevitz, L. Diagnosis of Parkinson’s Disease from Continuous Speech Using Deep Convolutional Networks without Manual Selection of Features. In Proceedings of the IEEE International Conference on the Science of Electrical Engineering, Eilat, Israel, 16–18 November 2016. [Google Scholar]
- Charalambous, C. Conjugate gradient algorithm for efficient training of artificial neural networks. IEE Proc. G (Circuits Devices Syst.) 1992, 139, 301–310. [Google Scholar] [CrossRef]
- O’Shea, K.; Nash, R. An introduction to convolutional neural networks. arXiv 2015, arXiv:1511.08458. [Google Scholar]
- Saba, L.; Biswas, M.; Kuppili, V.; Godia, E.C.; Suri, H.S.; Edla, D.R.; Omerzu, T.; Laird, J.R.; Khanna, N.N.; Mavrogeni, S.; et al. The present and future of deep learning in radiology. Eur. J. Radiol. 2019, 114, 14–24. [Google Scholar] [CrossRef]
- Rao, K.M.M.; Reddy, M.S.N.; Teja, V.R.; Krishnan, P.; Aravindhar, D.J.; Sambath, M. Parkinson’s Disease Detection Using Voice and Spiral Drawing Dataset. In Proceedings of the 2020 Third International Conference on Smart Systems and Inventive Technology (ICSSIT), Online, 20–22 August 2020. [Google Scholar]
- Jena, B.; Saxena, S.; Nayak, G.K.; Saba, L.; Sharma, N.; Suri, J.S. Artificial intelligence-based hybrid deep learning models for image classification: The first narrative review. Comput. Biol. Med. 2021, 137, 104803. [Google Scholar] [CrossRef] [PubMed]
- Raees, P.C.M.; Thomas, V. Automated detection of Alzheimer’s Disease using Deep Learning in MRI. J. Phys. Conf. Ser. 2021, 1921, 012024. [Google Scholar] [CrossRef]
- Oriol, J.D.V.; Vallejo, E.E.; Estrada, K.; Taméz Peña, J.G.; Disease Neuroimaging Initiative. Benchmarking machine learning models for late-onset alzheimer’s disease prediction from genomic data. BMC Bioinform. 2019, 20, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Suri, J.S.; Agarwal, S.; Gupta, S.K.; Puvvula, A.; Viskovic, K.; Suri, N.; Alizad, A.; El-Baz, A.; Saba, L.; Fatemi, M.; et al. Systematic Review of Artificial Intelligence in Acute Respiratory Distress Syndrome for COVID-19 Lung Patients: A Biomedical Imaging Perspective. IEEE J. Biomed. Health Inform. 2021, 25, 4128–4139. [Google Scholar] [CrossRef] [PubMed]
- Perlmutter, J.S. Assessment of Parkinson disease manifestations. Curr. Protoc. Neurosci. 2009, 10, Unit10.1. [Google Scholar] [CrossRef] [PubMed]
- Rissardo, J.P.; Caprara, A.F. Parkinson’s disease rating scales: A literature review. Ann. Mov. Disord. 2020, 3, 3. [Google Scholar] [CrossRef]
- Samantaray, T.; Saini, J.; Gupta, C.N. Meta-Analysis of Clinical Symptoms and Data Driven Subtyping Approaches in Parkinson’s Disease. In Proceedings of the Brain Conference 2021, Online, 4 March 2021; Available online: https://thebrainconference.co.uk/wp-content/uploads/2021/02/R2_51_Samantaray_Tanmayee_MovementDisorders_51.png (accessed on 12 September 2022).
- Mohammed, F.; He, X.; Lin, Y. An easy-to-use deep-learning model for highly accurate diagnosis of Parkison’s disease using SPECT images. Comput. Med. Imaging Graph. 2020, 87, 101810. [Google Scholar] [CrossRef]
- Sveinbjornsdottir, S. The clinical symptoms of Parkinson’s disease. J. Neurochem. 2016, 139, 318–324. [Google Scholar] [CrossRef]
- Bikias, T.; Iakovakis, D.; Hadjidimitriou, S.; Charisis, V.; Hadjileontiadis, L.J. Deepfog: An IMU-based detection of freezing of gait episodes in Parkison’s disease patients via deep learning. Front. Robot. AI 2021, 8, 537384. [Google Scholar] [CrossRef]
- Samantaray, T.; Saini, J.; Gupta, C.N. Subgrouping and Structural Brain Connectivity of Parkinson’s Disease—Past Studies and Future Directions. Neurosci. Inform. 2022, 2, 100100. [Google Scholar] [CrossRef]
- Morris, M.; Iansek, R.; Matyas, T.; Summers, J. Abnormalities in the stride length-cadence relation in parkinsonian gait. Mov. Disord. 1998, 13, 61–69. [Google Scholar] [CrossRef]
- Aita, J.F. Why patients with Parkinson’s disease fall. JAMA 1982, 247, 515–516. [Google Scholar] [CrossRef]
- Koller, W.C.; Glatt, S.; Vetere-Overfield, B.; Hassanein, R. Falls and Parkinson’s disease. Clin. Neuropharmacol. 1989, 12, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.E.; Iansek, R.; Matyas, T.A.; Summers, J.J. Stride length regulation in Parkinson’s disease. Normalization strategies and underlying mechanisms. Brain 1996, 119 Pt 2, 551–568. [Google Scholar] [CrossRef] [PubMed]
- Hausdorff, J.M.; Cudkowicz, M.E.; Firtion, R.; Wei, J.Y.; Goldberger, A.L. Gait variability and basal ganglia disorders: Stride-to-stride variations of gait cycle timing in Parkinson’s disease and Huntington’s disease. Mov. Disord. 1998, 13, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Vieregge, P.; Stolze, H.; Klein, C.; Heberlein, I. Gait quantitation in Parkinson’s disease—Locomotor disability and correlation to clinical rating scales. Jo. Neural Transm. 1997, 104, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Zijlstra, W.; Rutgers, A.W.; Van Weerden, T.W. Voluntary and involuntary adaptation of gait in Parkinson’s disease. Gait Posture 1998, 7, 53–63. [Google Scholar] [CrossRef]
- Dashtipour, K.; Tafreshi, A.; Lee, J.; Crawley, B. Speech disorders in Parkinson’s disease: Pathophysiology, medical management and surgical approaches. Neurodegener. Dis. Manag. 2018, 8, 337–348. [Google Scholar] [CrossRef]
- Available online: https://www.parkinson.org/understanding-parkinsons/symptoms/non-movement-symptoms/small-handwriting (accessed on 25 October 2022).
- Available online: https://www.healthline.com/health/parkinsons/parkinsons-mri (accessed on 25 October 2022).
- Available online: https://www.aans.org/en/Patients/Neurosurgical-Conditions-and-Treatments/Parkinsons-Disease (accessed on 12 September 2022).
- Iarkov, A.; Barreto, G.E.; Grizzell, J.A.; Echeverria, V. Strategies for the Treatment of Parkinson’s Disease: Beyond Dopamine. Front. Aging Neurosci. 2020, 12, 4. [Google Scholar] [CrossRef]
- Salamon, A.; Zádori, D.; Szpisjak, L.; Klivényi, P.; Vécsei, L. What is the impact of catechol-O-methyltransferase (COMT) on Parkinson’s disease treatment? InExpert Opinion on Pharmacotherapy; Taylor & Francis: Abingdon, UK, 2022. [Google Scholar] [CrossRef]
- Gallazzi, M.; Mauri, M.; Bianchi, M.L.; Riboldazzi, G.; Cariddi, L.P.; Carimati, F.; Rebecchi, V.; Versino, M. Selegiline reduces daytime sleepiness in patients with Parkinson’s disease. Brain Behav. 2021, 11, e01880. [Google Scholar] [CrossRef]
- Marzoughi, S.; Banerjee, A.; Jutzeler, C.R.; Prado, M.A.; Rosner, J.; Cragg, J.J.; Cashman, N. Tardive neurotoxicity of anticholinergic drugs: A review. J. Neurochem. 2021, 158, 1334–1344. [Google Scholar] [CrossRef]
- Marmol, S.; Feldman, M.; Singer, C.; Margolesky, J. Amantadine Revisited: A Contender for Initial Treatment in Parkinson’s disease? CNS Drugs 2021, 35, 1141–1152. [Google Scholar] [CrossRef]
- Avuçlu, E.; Elen, A. Evaluation of train and test performance of machine learning algorithms and Parkinson diagnosis with statistical measurements. Med. Biol. Eng. Comput. 2020, 58, 2775–2788. [Google Scholar] [CrossRef] [PubMed]
- KarimiRouzbahani, H.; Daliri, M.R. Diagnosis of Parkinson’s Disease in Human Using Voice Signals. BCN 2011, 2, 12–20. [Google Scholar]
- Khamparia, A.; Gupta, D.; Nguyen, N.G.; Khanna, A.; Pandey, B.; Tiwari, P. Sound Classification Using Convolutional Neural Network and Tensor Deep Stacking Network. IEEE Access 2019, 7, 7717–7727. [Google Scholar] [CrossRef]
- Bourouhou, A.; Jilbab, A.; Nacir, C.; Hammouch, A. Comparison of classification methods to detect the parkinson disease. In Proceedings of the 2016 International Conference on Electrical and Information Technologies (ICEIT), Tangiers, Morocco, 4–7 May 2016; pp. 421–424. [Google Scholar]
- Sharma, A.; Giri, R.N. Automatic Recognition of Parkinson’s Disease via Artificial Neural Network and Support Vector Machine. Int. J. Innov. Technol. Explor. Eng. (IJITEE) 2014, 4, 7. [Google Scholar]
- Purwins, H.; Li, B.; Virtanen, T.; Schluter, J.; Chang, S.; Sainath, T. Deep Learning for Audio Signal Processing. IEEE J. Select. Top.Signal Process. 2019, 13, 206–219. [Google Scholar] [CrossRef]
- Zhang, L.; Qu, Y.; Jin, B.; Jing, L.; Gao, Z.; Liang, Z. An intelligent mobile-enabled system for diagnosing Parkinson disease: Development and validation of a speech impairment detection system. JMIR Med. Inform. 2020, 8, e18689. [Google Scholar] [CrossRef]
- Kadiri, S.R.; Kethireddy, R.; Alku, P. Parkinson’s disease detection from speech using single frequency filtering cepstral Coefficients. In Proceedings of the Interspeech, Shanghai, China, 25–29 October 2020; pp. 4971–4975. [Google Scholar]
- Pramanik, M.; Pradhan, R.; Nandy, P.; Bhoi, A.K.; Barsocchi, P. Machine learning methods with decision forests for Parkinson’s detection. Appl. Sci. 2021, 11, 581. [Google Scholar] [CrossRef]
- Gunduz, H. Deep Learning-Based Parkinson’s Disease Classification Using Vocal Feature Sets. IEEE Access 2019, 7, 115540–115551. [Google Scholar] [CrossRef]
- Available online: https://www.anjusoftware.com/about/all-news/ai-clinical-trials (accessed on 5 September 2022).
- Available online: https://www.dataversity.net/improving-clinical-insights-machine-learning/# (accessed on 5 September 2022).
- Sakar, C.O.; Serbes, G.; Gunduz, A.; Tunc, H.C.; Nizam, H.; Sakar, B.E.; Tutuncu, M.; Aydin, T.; Isenkul, M.E.; Apaydin, H. A comparative analysis of speech signal processing algorithms for Parkinson’s disease classification and the use of the tunable-factor wavelet transform. Appl. Soft Comput. 2019, 74, 255–263. [Google Scholar] [CrossRef]
- Yasar, A.; Saritas, I.; Sahman, M.A.; Cinar, A.C. Classification of Parkinson disease data with artificial neural networks. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2019; Volume 675, p. 012031. [Google Scholar]
- Ouhmida, A.; Raihani, A.; Cherradi, B.; Terrada, O. A Novel Approach for Parkinson’s Disease Detection Based on Voice Classification and Features Selection Techniques. International Journal of Online & Biomedical Engineering. Int. J. Online Biomed. Eng. 2021, 17. [Google Scholar]
- Marar, S.; Swain, D.; Hiwarkar, V.; Motwani, N.; Awari, A. Predicting the occurrence of Parkinson’s Disease using various Classification Models. In Proceedings of the 2018 International Conference on Advanced Computation and Telecommunication (ICACAT), Bhopal, India, 28–29 December 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Sheibani, R.; Nikookar, E.; Alavi, S.E. An Ensemble Method for Diagnosis of Parkinson’s Disease Based on Voice Measurements. J. Med. Signals Sens. 2019, 9, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Tracy, J.M.; Özkanca, Y.; Atkins, D.C.; Ghomi, R.H. Investigating voice as a biomarker: Deep phenotyping methods for early detection of Parkinson’s disease. J. Biomed. Inform. 2020, 104, 103362. [Google Scholar] [CrossRef]
- Cibulka, M.; Brodnanova, M.; Grendar, M.; Grofik, M.; Kurca, E.; Pilchova, I.; Osina, O.; Tatarkova, Z.; Dobrota, D.; Kolisek, M. SNPs rs11240569, rs708727, and rs823156 in SLC41A1 Do Not Discriminate between Slovak Patients with Idiopathic Parkinson’s Disease and Healthy Controls: Statistics and Machine-Learning Evidence. Int. J. Mol. Sci. 2019, 20, 4688. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-Y.; Lin, H.-C.; Chen, T.-B.; Du, W.-C.; Hsu, Y.-H.; Wu, Y.-C.; Tu, P.-W.; Huang, Y.-H.; Chen, H.-Y. Feasible Classified Models for Parkinson Disease from 99mTc-TRODAT-1 SPECT Imaging. Sensors 2019, 19, 1740. [Google Scholar] [CrossRef] [PubMed]
- Drotár, P.; Mekyska, J.; Rektorová, I.; Masarová, L.; Smékal, Z.; Faundez-Zanuy, M. Evaluation of handwriting kinematics and pressure for differential diagnosis of Parkinson’s disease. Artif. Intell. Med. 2016, 67, 39–46. [Google Scholar] [CrossRef]
- Maass, F.; Michalke, B.; Willkommen, D.; Leha, A.; Schulte, C.; Tönges, L.; Mollenhauer, B.; Trenkwalder, C.; Rückamp, D.; Börger, M.; et al. Elemental fingerprint: Reassessment of a cerebrospinal fluid biomarker for Parkinson’s disease. Neurobiol. Dis. 2020, 134, 104677. [Google Scholar] [CrossRef] [PubMed]
- Mucha, J.; Mekyska, J.; Faundez-Zanuy, M.; Lopez-De-Ipina, K.; Zvoncak, V.; Galaz, Z.; Kiska, T.; Smekal, Z.; Brabenec, L.; Rektorova, I. Advanced Parkinson’s Disease Dysgraphia Analysis Based on Fractional Derivatives of Online Handwriting. In Proceedings of the 2018 10th International Congress on Ultra Modern Telecommunications and Control Systems and Workshops (ICUMT), Moscow, Russia, 5–9 November 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Wenzel, M.; Milletari, F.; Krüger, J.; Lange, C.; Schenk, M.; Apostolova, I.; Klutmann, S.; Ehrenburg, M.; Buchert, R. Automatic classification of dopamine transporter SPECT: Deep convolutional neural networks can be trained to be robust with respect to variable image characteristics. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2800–2811. [Google Scholar] [CrossRef]
- Segovia, F.; Górriz, J.M.; Ramírez, J.; Martínez-Murcia, F.J.; Castillo-Barnes, D. Assisted diagnosis of Parkinsonism based on the striatal morphology. Int. J. Neural Syst. 2019, 29, 1950011. [Google Scholar] [CrossRef]
- Ye, Q.; Xia, Y.; Yao, Z. Classification of gait patterns in patients with neurodegenerative disease using adaptive neuro-fuzzy inference system. Comput. Math. Methods Med. 2018, 2018, 9831252. [Google Scholar] [CrossRef]
- Klomsae, A.; Auephanwiriyakul, S.; Theera-Umpon, N. String grammar unsupervised possibilistic fuzzy c-medians for gait pattern classification in patients with neurodegenerative diseases. Comput. Intell. Neurosci. 2018, 2018, 1869565. [Google Scholar] [CrossRef]
- Felix, J.P.; Vieira, F.H.T.; Cardoso, A.A.; Ferreira, M.V.G.; Franco, R.A.P.; Ribeiro, M.A.; Araujo, S.G.; Correa, H.P.; Carneiro, M.L. A Parkinson’s Disease Classification Method: An Approach Using Gait Dynamics and Detrended Fluctuation Analysis. In Proceedings of the 2019 IEEE Canadian Conference of Electrical and Computer Engineering (CCECE), Edmonton, AB, Canada, 5–8 May 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Andrei, A.-G.; Tăuțan, A.-M.; Ionescu, B. Parkinson’s Disease Detection from Gait Patterns. In Proceedings of the 2019 E-Health and Bioengineering Conference (EHB), Iasi, Romania, 21–23 November 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Priya, S.J.; Rani, A.J.; Subathra, M.S.P.; Mohammed, M.A.; Damaševičius, R.; Ubendran, N. Local Pattern Transformation Based Feature Extraction for Recognition of Parkinson’s Disease Based on Gait Signals. Diagnostics 2021, 11, 1395. [Google Scholar] [CrossRef] [PubMed]
- Yurdakul, O.C.; Subathra, M.; George, S.T. Detection of Parkinson’s Disease from gait using Neighborhood Representation Local Binary Patterns. Biomed. Signal Process. Control 2020, 62, 102070. [Google Scholar] [CrossRef]
- Li, B.; Yao, Z.; Wang, J.; Wang, S.; Yang, X.; Sun, Y. Improved Deep Learning Technique to Detect Freezing of Gait in Parkinson’s Disease Based on Wearable Sensors. Electronics 2020, 9, 1919. [Google Scholar] [CrossRef]
- Gong, B.; Shi, J.; Ying, S.; Dai, Y.; Zhang, Q.; Dong, Y.; Zhang, Y. Neuroimaging-based diagnosis of Parkinson’s disease with deep neural mapping large margin distribution machine. Neurocomputing 2018, 320, 141–149. [Google Scholar] [CrossRef]
- Chakraborty, S.; Aich, S.; Kim, H.-C. 3D textural, morphological and statistical analysis of voxel of interests in 3T MRI scans for the detection of Parkinson’s disease using artificial neural networks. Healthcare 2020, 8, 34. [Google Scholar] [CrossRef]
- Bhan, A.; Kapoor, S.; Gulati, M.; Goyal, A. Early diagnosis of Parkinson’s disease in brain MRI using deep learning algorithm. In Proceedings of the 2021 Third International Conference on Intelligent Communication Technologies and Virtual Mobile Networks, Tirunelveli, India, 4–6 February 2021; pp. 1467–1470. [Google Scholar]
- Kumar, R.; Gupta, A.; Arora, H.S.; Raman, B. IBRDM: An intelligent framework for brain tumor classification using radiomics-and DWT-based fusion of MRI sequences. ACM Trans. Internet Technol. (TOIT) 2021, 22, 1–30. [Google Scholar] [CrossRef]
- Pang, Y.; Christenson, J.; Jiang, F.; Lei, T.; Rhoades, R.; Kern, D.; Thompson, J.A.; Liu, C. Automatic detection and quantification of hand movements toward development of an objective assessment of tremor and bradykinesia in Parkison’s disease. J. Neurosci. Methods 2020, 333, 108576. [Google Scholar] [CrossRef]
- Radziunas, A.; Deltuva, V.P.; Tamasauskas, A.; Gleizniene, R.; Pranckeviciene, A.; Petrikonis, K.; Bunevicius, A. Brain MRI morphometric analysis in Parkinson’s disease patients with sleep disturbances. BMC Neurol. 2018, 18, 88. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, J.; Liu, X.; Chen, J.; Liu, B. Aberrant brain network efficiency in Parkinson’s disease patients with tremor: A multi-modality study. Front. Aging Neurosci. 2015, 7, 169. [Google Scholar] [CrossRef]
- Kiryu, S.; Yasaka, K.; Akai, H.; Nakata, Y.; Sugomori, Y.; Hara, S.; Seo, M.; Abe, O.; Ohtomo, K. Deep learning to differentiate parkinsonian disorders separately using single midsagittal MR imaging: A proof of concept study. Eur. Radiol. 2019, 29, 6891–6899. [Google Scholar] [CrossRef]
- Magesh, P.R.; Myloth, R.D.; Tom, R.J. An explainable machine learning model for early detection of Parkinson’s disease using LIME on DaTSCAN imagery. Comput. Biol. Med. 2020, 126, 104041. [Google Scholar] [CrossRef] [PubMed]
- Mabrouk, R.; Chikhaoui, B.; Bentabet, L. Machine learning based classification using clinical and DaTSCAN SPECT imaging features: A study on Parkinson’s disease and SWEDD. IEEE Trans. Radiat. Plasma Med. Sci. 2018, 3, 170–177. [Google Scholar] [CrossRef]
- Quan, J.; Xu, L.; Xu, R.; Tong, T.; Su, J. DaTscan SPECT Image Classification for Parkinson’s Disease. arXiv 2019, arXiv:1909.04142. [Google Scholar]
- Moon, S.; Song, H.J.; Sharma, V.D.; Lyons, K.E.; Pahwa, R.; Akinwuntan, A.E.; Devos, H. Classification of Parkinson’s disease and essential tremor based on balance and gait characteristics from wearable motion sensors via machine learning techniques: A data-driven approach. J. Neuroeng. Rehabil. 2020, 17, 125. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.P.; Rahmim, A.; Tang, J. Improved motor outcome prediction in Parkinson’s disease applying deep learning to DaTscan SPECT images. Comput. Biol. Med. 2021, 132, 104312. [Google Scholar] [CrossRef]
- Khachnaoui, H.; Khlifa, N.; Mabrouk, R. Machine Learning for Early Parkinson’s Disease Identification within SWEDD Group Using Clinical and DaTSCAN SPECT Imaging Features. J. Imaging 2022, 8, 97. [Google Scholar] [CrossRef]
- Oliveira, F.P.; Faria, D.B.; Costa, D.C.; Castelo-Branco, M.; Tavares, J.M.R. Extraction, selection and comparison of features for an effective automated computer-aided diagnosis of Parkinson’s disease based on [123I] FP-CIT SPECT images. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 1052–1062. [Google Scholar] [CrossRef]
- Saponaro, S.; Giuliano, A.; Bellotti, R.; Lombardi, A.; Tangaro, S.; Oliva, P.; Retico, A. Multi-site harmonization of MRI data uncovers machine-learning discrimination capability in barely separable populations: An example from the ABIDE dataset. NeuroImage Clin. 2022, 35, 103082. [Google Scholar] [CrossRef]
- Tufail, A.B.; Ma, Y.K.; Zhang, Q.N.; Khan, A.; Zhao, L.; Yang, Q.; Adeel, M.; Khan, R.; Ullah, I. 3D convolutional neural networks-based multiclass classification of Alzheimer’s and Parkinson’s diseases using PET and SPECT neuroimaging modalities. Brain Inform. 2021, 8, 1–9. [Google Scholar] [CrossRef]
- Antikainen, E.; Cella, P.; Tolonen, A.; van Gils, M. SPECT Image Features for Early Detection of Parkinson’s Disease using Machine Learning Methods. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Virtual, 1–5 November 2021; pp. 2773–2777. [Google Scholar]
- Salvatore, C.; Cerasa, A.; Castiglioni, I.; Gallivanone, F.; Augimeri, A.; Lopez, M.; Quattrone, A. Machine learning on brain MRI data for differential diagnosis of Parkinson’s disease and Progressive Supranuclear Palsy. J. Neurosci. Methods 2014, 222, 230–237. [Google Scholar] [CrossRef]
- Martínez-Ibañez, M.; Ortiz, A.; Munilla, J.; Salas-Gonzalez, D.; Górriz, J.M.; Ramírez, J. Isosurface modelling of DatSCAN images for parkinson disease diagnosis. In International Work-Conference on the Interplay between Natural and Artificial Computation; Springer: Cham, Switzerland, 2019; pp. 360–368. [Google Scholar]
- Kurmi, A.; Biswas, S.; Sen, S.; Sinitca, A.; Kaplun, D.; Sarkar, R. An Ensemble of CNN Models for Parkinson’s Disease Detection Using DaTscan Images. Diagnostics 2022, 12, 1173. [Google Scholar] [CrossRef] [PubMed]
- Hossen, A. A neural network approach for feature extraction and discrimination between parkinsonian tremor and essential tremor. Technol. Health Care 2013, 21, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Challa KN, R.; Pagolu, V.S.; Panda, G.; Majhi, B. An improved approach for prediction of Parkison’s disease using machine learning techniques. In Proceedings of the 2016 international conference on signal processing, communication, power and embedded system (SCOPES), Paralakhemundi, India, 3–5 October 2016; pp. 1446–1451. [Google Scholar]
- Choi, H.; Ha, S.; Im, H.J.; Paek, S.H.; Lee, D.S. Refining diagnosis of Parkinson’s disease with deep learning-based interpretation of dopamine transporter imaging. NeuroImage Clin. 2017, 16, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Wit, H.; Thurston, M. Artificial intelligence in the diagnosis of Parkison’s disease from ioflupane-123 single-photon emission computed tomography dopamine transporter scans using transfer learning. Nucl. Med. Commun. 2018, 39, 887–893. [Google Scholar] [CrossRef]
- Esmaeilzadeh, S.; Yang, Y.; Adeli, E. End-to-end parkinson disease diagnosis using brain MR-images by 3D-CNN. arXiv 2018, arXiv:1806.05233. [Google Scholar]
- Kim, H.B.; Lee, W.W.; Kim, A.; Lee, H.J.; Park, H.Y.; Jeon, H.S.; Kim, S.K.; Jeon, B.; Park, K.S. Wrist sensor-based tremor severity quantification in Parkison’s disease using convolutional neural network. Comput. Biol. Med. 2018, 95, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Murcia, F.J.; Ortiz, A.; Górriz, J.M.; Ramírez, J.; Segovia, F.; Salas-Gonzalez, D.; Castillo-Barnes, D.; Illán, I.A. A 3D convolutional neural network approach for the diagnosis of Parkison’s disease. In International Work-Conference on the Interplay between Natural and Artificial Computation; Springer: Cham, Switzerland, 2017; pp. 324–333. [Google Scholar]
- Qin, Z.; Jiang, Z.; Chen, J.; Hu, C.; Ma, Y. SEMG-based tremor severity evaluation for Parkison’s disease using a light-weight CNN. IEEE Signal Process. Lett. 2019, 26, 637–641. [Google Scholar] [CrossRef]
- Kollia, I.; Stafylopatis, A.-G.; Kollias, S. Predicting Parkison’s disease using latent information extracted from deep neural networks. In Proceedings of the 2019 International Joint Conference on Neural Networks, Budapest, Hungary, 14–19 July 2019; pp. 1–8. [Google Scholar]
- Szumilas, M.; Lewenstein, K.; Ślubowska, E.; Szlufik, S.; Koziorowski, D. A multimodal approach to the quantification of kinetic tremor in Parkison’s disease. Sensors 2020, 20, 184. [Google Scholar] [CrossRef]
- Oktay, A.B.; Kocer, A. Differential diagnosis of Parkinson and essential tremor with convolutional LSTM networks. Biomed. Signal Process. Control 2020, 56, 101683. [Google Scholar] [CrossRef]
- Shahtalebi, S.; Atashzar, S.F.; Samotus, O.; Patel, R.V.; Jog, M.S.; Mohammadi, A. Phtnet: Characterization and deep mining of involuntary pathological hand tremor using recurrent neural network models. Sci. Rep. 2020, 10, 2195. [Google Scholar] [CrossRef]
- Veeraragavan, S.; Gopalai, A.A.; Gouwanda, D.; Ahmad, S.A. Parkison’s disease diagnosis and severity assessment using ground reaction forces and neural networks. Front. Physiol. 2020, 11, 587057. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.-Y.; Hsu, S.-W.; Lee, T.-L.; Sung, P.-S.; Lin, C.-C. Using artificial neural network to discriminate Parkison’s disease from other parkinsonisms by focusing on putamen of dopamine transporter SPECT images. Biomedicines 2021, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Yasaka, K.; Kamagata, K.; Ogawa, T.; Hatano, T.; Takeshige-Amano, H.; Ogaki, K.; Andica, C.; Akai, H.; Kunimatsu, A.; Uchida, W.; et al. Parkison’s disease: Deep learning with a parameter-weighted structural connectome matrix for diagnosis and neural circuit disorder investigation. Neuroradiology 2021, 63, 1451–1462. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wei, L.; Hu, Y.; Wu, Y.; Hu, L.; Nie, S. Classification of Parkison’s disease based on multi-modal features and stacking ensemble learning. J. Neurosci. Methods 2021, 350, 109019. [Google Scholar] [CrossRef]
- Vyas, T.; Yadav, R.; Solanki, C.; Darji, R.; Desai, S.; Tanwar, S. Deep learning-based scheme to diagnose Parkison’s disease. Expert Syst. 2022, 39, e12739. [Google Scholar] [CrossRef]
- Yadav, S. Bayesian Deep Learning Based Convolutional Neural Network for Classification of Parkison’s Disease Using Functional Magnetic Resonance Images. 2021. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3833760 (accessed on 24 October 2022).
- Mei, J.; Desrosiers, C.; Frasnelli, J. Machine Learning for the Diagnosis of Parkinson’s disease: A Review of Literature. Front. Aging Neurosci. 2021, 13, 633752. [Google Scholar] [CrossRef]
- Khedr, E.M.; El Fetoh, N.A.; Khalifa, H.E.; Ahmed, M.A.; El Beh, K.M. Prevalence of non-motor features in a cohort of Parkinson’s disease patients. Clin. Neurol. Neurosurg. 2013, 115, 673–677. [Google Scholar] [CrossRef]
- Zappia, M.; Annesi, G.; Nicoletti, G.; Arabia, G.; Annesi, F.; Messina, D.; Pugliese, P.; Spadafora, P.; Tarantino, P.; Carrideo, S. Sex differences in clinical and genetic determinants of levodopa peak-dose dyskinesias in Parkinson disease: An exploratory study. Arch. Neurol. 2005, 62, 601–605. [Google Scholar] [CrossRef]
- Ma, X.; Niu, Y.; Gu, L.; Wang, Y.; Zhao, Y.; Bailey, J.; Lu, F. Understanding adversarial attacks on deep learning based medical image analysis systems. Pattern Recogn. 2021, 110, 107332. [Google Scholar] [CrossRef]
- Alluri, R.K.; Vaishnav, A.S.; Sivaganesan, A.; Ricci, L.; Sheha, E.; Qureshi, S.A. Multimodality Intraoperative Neuromonitoring in Lateral Lumbar Interbody Fusion: A Review of Alerts in 628 Patients. Glob. Spine J. 2021. [Google Scholar] [CrossRef]
- Hassan, M.; Chaton, L.; Benquet, P.; Delval, A.; Leroy, C.; Plomhause, L.; Dujardin, K. Functional connectivity disruptions correlate with cognitive phenotypes in Parkinson’s disease. NeuroImage Clin. 2017, 14, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, D.; Shoeibi, A.; Ghassemi, N.; Moridian, P.; Khadem, A.; Alizadehsani, R.; Acharya, U.R. An overview of artificial intelligence techniques for diagnosis of Schizophrenia based on magnetic resonance imaging modalities: Methods, challenges, and future works. Comput. Biol. Med. 2022, 146, 105554. [Google Scholar] [CrossRef] [PubMed]
- Utianski, R.L.; Caviness, J.N.; van Straaten, E.C.; Beach, T.G.; Dugger, B.N.; Shill, H.A.; Hentz, J.G. Graph theory network function in Parkinson’s disease assessed with electroencephalography. Clin. Neurophysiol. 2016, 127, 2228–2236. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.D.; Dong, Z.; Wang, S.H.; Yu, X.; Yao, X.; Zhou, Q.; Gorriz, J.M. Advances in multimodal data fusion in neuroimaging: Overview, challenges, and novel orientation. Inf. Fusion 2020, 64, 149–187. [Google Scholar] [CrossRef] [PubMed]
- Vij, R.; Arora, S. Computer Vision with Deep Learning Techniques for Neurodegenerative Diseases Analysis Using Neuroimaging: A Survey. In International Conference on Innovative Computing and Communications; Springer: Singapore, 2022; pp. 179–189. [Google Scholar]
- Xu, J.; Jiao, F.; Huang, Y.; Luo, X.; Xu, Q.; Li, L.; Zhuang, X. A fully automatic framework for parkinson’s disease diagnosis by multi-modality images. Front. Neurosci. 2019, 13, 874. [Google Scholar] [CrossRef] [PubMed]
- Tăuţan, A.M.; Ionescu, B.; Santarnecchi, E. Artificial intelligence in neurodegenerative diseases: A review of available tools with a focus on machine learning techniques. Artif. Intell. Med. 2021, 117, 102081. [Google Scholar] [CrossRef] [PubMed]
- Yao, A.D.; Cheng, D.L.; Pan, I.; Kitamura, F. Deep learning in neuroradiology: A systematic review of current algorithms and approaches for the new wave of imaging technology. Radiol. Artif. Intell. 2020, 2, e190026. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.L.; Robson, S.E.; Morris, P.G.; Brookes, M.J. The relationship between MEG and fMRI. Neuroimage 2014, 102, 80–91. [Google Scholar] [CrossRef]
- Moridian, P.; Ghassemi, N.; Jafari, M.; Salloum-Asfar, S.; Sadeghi, D.; Khodatars, M.; Acharya, U.R. Automatic Autism Spectrum Disorder Detection Using Artificial Intelligence Methods with MRI Neuroimaging: A Review. arXiv 2022, arXiv:2206.11233. [Google Scholar] [CrossRef]
- Zou, L.; Zheng, J.; Miao, C.; Mckeown, M.J.; Wang, Z.J. 3D CNN based automatic diagnosis of attention deficit hyperactivity disorder using functional and structural MRI. IEEE Access 2017, 5, 23626–23636. [Google Scholar] [CrossRef]
- Amini, M.; Pedram, M.M.; Moradi, A.; Jamshidi, M.; Ouchani, M. Single and combined neuroimaging techniques for Alzheimer’s disease detection. Comput. Intell. Neurosci. 2021, 2021, 9523039. [Google Scholar] [CrossRef] [PubMed]
- Shoeibi, A.; Ghassemi, N.; Khodatars, M.; Moridian, P.; Khosravi, A.; Zare, A.; Acharya, U.R. Automatic Diagnosis of Schizophrenia and Attention Deficit Hyperactivity Disorder in rs-fMRI Modality using Convolutional Autoencoder Model and Interval Type-2 Fuzzy Regression. arXiv 2022, arXiv:2205.15858. [Google Scholar]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019, 17, 195. [Google Scholar] [CrossRef] [PubMed]
- Sivaranjini, S.; Sujatha, C.M. Deep learning based diagnosis of Parkinson’s disease using convolutional neural network. Multimed. Tools Appl. 2020, 79, 15467–15479. [Google Scholar] [CrossRef]
- Yagis, E.; De Herrera, A.G.S.; Citi, L. Generalization performance of deep learning models in neurodegenerative disease classification. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 1692–1698. [Google Scholar]
- Lee, S.; Hussein, R.; McKeown, M.J. A deep convolutional-recurrent neural network architecture for Parkinson’s disease EEG classification. In Proceedings of the 2019 IEEE Global Conference on Signal and Information Processing (GlobalSIP), Ottawa, ON, Canada, 11–14 November 2019; pp. 1–4. [Google Scholar]
- Noor MB, T.; Zenia, N.Z.; Kaiser, M.S.; Mahmud, M.; Mamun, S.A. Detecting neurodegenerative disease from MRI: A brief review on a deep learning perspective. In International Conference on Brain Informatics; Springer: Cham, Switzerland, 2019; pp. 115–125. [Google Scholar]
- Paré, G.; Trudel, M.C.; Jaana, M.; Kitsiou, S. Synthesizing information systems knowledge: A typology of literature reviews. Inf. Manag. 2015, 52, 183–199. [Google Scholar] [CrossRef]
- Li, R.; Zhang, W.; Suk, H.I.; Wang, L.; Li, J.; Shen, D.; Ji, S. Deep learning based imaging data completion for improved brain disease diagnosis. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2014; pp. 305–312. [Google Scholar]
- Nie, D.; Trullo, R.; Lian, J.; Petitjean, C.; Ruan, S.; Wang, Q.; Shen, D. Medical image synthesis with context-aware generative adversarial networks. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2017; pp. 417–425. [Google Scholar]
- Nie, D.; Trullo, R.; Lian, J.; Wang, L.; Petitjean, C.; Ruan, S.; Shen, D. Medical image synthesis with deep convolutional adversarial networks. IEEE Trans. Biomed. Eng. 2018, 65, 2720–2730. [Google Scholar] [CrossRef]
- Cai, L.; Wang, Z.; Gao, H.; Shen, D.; Ji, S. Deep adversarial learning for multi-modality missing data completion. In Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, London, UK, 19–23 August 2018; pp. 1158–1166. [Google Scholar]
- Pan, Y.; Liu, M.; Lian, C.; Zhou, T.; Xia, Y.; Shen, D. Synthesizing missing PET from MRI with cycle-consistent generative adversarial networks for Alzheimer’s disease diagnosis. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2018; pp. 455–463. [Google Scholar]
- Zhi, Y.; Wang, M.; Yuan, Y.S.; Shen, Y.T.; Ma, K.W.; Gan, C.T.; Si, Q.Q.; Wang, L.N.; Cao, S.W.; Zhang, K.Z. The increased gray matter volumes of precentralgyri in Parkinson’s disease patients with diphasic dyskinesia. Aging (Albany NY) 2019, 11, 9661. [Google Scholar] [CrossRef]
- Li, P.; Xu, P.; Liu, J. Biomarkers and Pathogenesis of Alpha-Synuclein in Parkinson’s Disease. Aging Neurosci. 2021, 13, 776873. [Google Scholar] [CrossRef]
- Yuan, Y.S.; Ji, M.; Gan, C.T.; Sun, H.M.; Wang, L.N.; Zhang, K.Z. Impaired Interhemispheric Synchrony in Parkinson’s Disease with Fatigue. J. Personal. Med. 2022, 12, 884. [Google Scholar] [CrossRef]
- Daveau, R.S.; Law, I.; Henriksen, O.M.; Hasselbalch, S.G.; Andersen, U.B.; Anderberg, L.; Højgaard, L.; Andersen, F.L.; Ladefoged, C.N. Deep learning based low-activity PET reconstruction of [11C] PiB and [18F] FE-PE2I in neurodegenerative disorders. Neuroimage 2022, 259, 119412. [Google Scholar] [CrossRef]
- Noor MB, T.; Zenia, N.Z.; Kaiser, M.S.; Mamun, S.A.; Mahmud, M. Application of deep learning in detecting neurological disorders from magnetic resonance images: A survey on the detection of Alzheimer’s disease, Parkinson’s disease and schizophrenia. Brain inform. 2020, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.K.; Seo, S.; Shin, S.A.; Byun, M.S.; Lee, D.Y.; Kim, Y.K.; Lee, J.S. Adaptive template generation for amyloid PET using a deep learning approach. Hum. Brain Mapp. 2018, 39, 3769–3778. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Cheng, D.; Wang, K.; Wang, Y. Multi-modality cascaded convolutional neural networks for Alzheimer’s disease diagnosis. Neuroinformatics 2018, 16, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.; Chung, Y.C.; Kim, K.W.; Kim, W.S.; Oh, I.S. Classification and visualization of Alzheimer’s disease using volumetric convolutional neural network and transfer learning. Sci. Rep. 2019, 9, 18150. [Google Scholar] [CrossRef]
- Wang, S.H.; Phillips, P.; Sui, Y.; Liu, B.; Yang, M.; Cheng, H. Classification of Alzheimer’s disease based on eight-layer convolutional neural network with leaky rectified linear unit and max pooling. J. Med. Syst. 2018, 42, 85. [Google Scholar] [CrossRef]
- Shang, R.; Wang, J.; Jiao, L.; Yang, X.; Li, Y. Spatial feature-based convolutional neural network for PolSAR image classification. Appl. Soft Comput. 2022, 123, 108922. [Google Scholar] [CrossRef]
- Ye, A.; Zhou, X.; Miao, F. Innovative Hyperspectral Image Classification Approach Using Optimized CNN and ELM. Electronics 2022, 11, 775. [Google Scholar] [CrossRef]
- Mohapatra, M.; Parida, A.K.; Mallick, P.K.; Zymbler, M.; Kumar, S. Botanical Leaf Disease Detection and Classification Using Convolutional Neural Network: A Hybrid Metaheuristic Enabled Approach. Computers 2022, 11, 82. [Google Scholar] [CrossRef]
- Hei, Y.; Liu, C.; Li, W.; Ma, L.; Lan, M. CNN Based Hybrid Precoding for MmWave MIMO Systems with Adaptive Switching Module and Phase Modulation Array. IEEE Trans. Wirel. Commun. 2022. [Google Scholar] [CrossRef]
- Aslan, M.F.; Unlersen, M.F.; Sabanci, K.; Durdu, A. CNN-based transfer learning–BiLSTM network: A novel approach for COVID-19 infection detection. Appl. Soft Comput. 2021, 98, 106912. [Google Scholar] [CrossRef]



| Journal Name | No. of Articles | Publisher | Indexing |
|---|---|---|---|
| Sensors | 47 | MDPI | SCIE and Scopus |
| IEEE Access | 32 | IEEE | SCIE |
| Plos One | 23 | Public Library Science | SCIE |
| Frontiers in Neurology | 17 | Frontiers Media Sa | SCIE |
| Frontiers in Neuroscience | 14 | Frontiers Media Sa | SCIE |
| IEEE journal of biomedical and health informatics | 14 | IEEE | SCIE |
| Diagnostics | 13 | MDPI | SCIE and Scopus |
| Applied Sciences Basel | 11 | MDPI | SCIE and Scopus |
| IEEE Transactions on Neural Systems and Rehabilitation Engineering | 11 | IEEE | SCIE |
| IEEE Sensors Journal | 10 | IEEE | SCIE |
| NPJ Parkinson’s Disease | 9 | Nature | SCIE |
| Brain Sciences | 8 | MDPI | SCIE and Scopus |
| IEEE Transactions on Biomedical Engineering | 7 | IEEE | SCIE |
| Multimedia Tools and Applications | 6 | Springer | SCIE |
| Journal of Healthcare Engineering | 5 | Hindawi | SCIE |
| IEEE Journal of Transactional Engineering in Health and Medicine | 4 | IEEE | SCIE |
| Nature Communications | 3 | Nature Portfolio | SCIE |
| Applied Acoustics | 2 | Elsevier | SCIE |
| Electronics | 2 | MDPI | SCIE and Scopus |
| IEEE ACM Transactions on Audio Speech and Language Processing | 2 | IEEE | SCIE |
| IEEE Transactions on Automation Science And Engineering | 2 | IEEE | SCIE |
| IEEE Transactions on Biomedical Circuits and Systems | 2 | IEEE | SCIE |
| IEEE Transactions on Radiations and Plasma Medical Sciences | 2 | IEEE | SCIE |
| Authors | Scope of the Review | Citations | Type of Study |
|---|---|---|---|
| Sibley, KG et al., 2021 [41] | Analysis of Parkinson’s disease severity based on videos | 16 | A brief review |
| Belic, M et al., 2019 [42] | Using artificial intelligence to aid in the diagnosis and evaluation of Parkinson’s disease | 74 | A review |
| Landers, M et al., 2021 [43] | Can artificial intelligence diagnose and treat Parkinson’s disease instead of a movement disorders specialist? | 1 | A review |
| Palumbo, B et al., 2021 [44] | In order to more accurately diagnose Parkinson’s disease and Parkinsonian symptoms, artificial intelligence approaches enhance nuclear medicine modalities. | 3 | A review |
| Saravanan, S et al., 2022 [45] | Artificial intelligence (AI)-based approaches for the diagnosis of Parkinson’s disease. | 1 | A systematic review |
| Perju-Dumbrava, L et al., 2022 [46] | Applications of robotic technology and artificial intelligence in Parkinson’s disease. | 0 | A review |
| Giannakopoulou, KM et al., 2022 [47] | Methods of the Iot technology and machine learning for the detection, monitoring, and management of Parkinson’s disease. | 2 | A systematic review |
| Khachnaoui, H et al., 2020 [48] | PET/SPECT imaging for Parkinson’s disease using machine learning and deep learning. | 5 | A review |
| Narayanan, RR et al., 2022 [49] | The effects of artificial intelligence (AI) on drug discovery and product development. | 0 | A review |
| Termine, A et al., 2021 [50] | A multi-layer view of neurodegenerative diseases: insights from the application of artificial intelligence to big data. | 8 | A review |
| Lim, ACY et al., 2022 [51] | Adult gait analysis and the diagnosis of disorders that modify their stride using artificial intelligence and personalised algorithms with inertial wearable technology. | 1 | A review |
| Xu, JJ et al., 2019 [52] | Parkinson’s disease diagnosis studies using magnetic resonance imaging and artificial intelligence. | 21 | A review |
| Zhang, Z et al., 2021 [53] | Artificial intelligence used to classify human brain neurological and psychiatric disorders using MRI. | 3 | A scoping review |
| Yadav, D et al., 2020 [54] | Intelligent diagnostic tools using mechanobiological and artificial intelligence methods. | 2 | A review |
| Patil, AD et al., 2022 [55] | An understanding of neurodegenerative disease with artificial intelligence in ophthalmology. | 0 | A review |
| Suri, JS et al., 2022 [56] | Using the atherosclerosis pathway and an artificial intelligence paradigm, cardiovascular/stroke risk stratification in Parkinson’s disease patients. | 7 | A systematic review |
| Vitale, A et al., 2021 [57] | Neuroimaging data from Parkinson’s symptoms using artificial intelligence. | 3 | A review |
| Cascianelli, S et al., 2017 [58] | Molecular imaging modalities in neurodegenerative diseases. | 21 | A review |
| Rana, A et al., 2022 [59] | Detection of Parkinson’s disease: the critical role of machine learning algorithms. | 0 | A review |
| Raghavendra, U et al., 2022 [60] | Automated diagnosis of neurological disorders using artificial intelligence techniques. | 81 | A review |
| Singh, AV et al., 2021 [61] | Artificial intelligence and nanorobotics: anew approach to cross the BBB. | 31 | A review |
| Maitin, AM et al., 2022 [62] | Analysis of EEG signals for Parkinson’s disease using machine learning techniques. | 1 | A systematic review |
| Vatansever, S et al., 2021 [63] | Using artificial intelligence and machine learning to help in medication development for illnesses of the central nervous system. | 38 | State–of–the–art |
| Hansen, C et al., 2018 [64] | How electronic health records and mobile health technology will change Parkinson’s disease patient care. | 29 | A review |
| Luis-Martinez, R et al., 2020 [65] | Using digital technology to integrate multidisciplinary care for Parkinson’s disease. | 21 | A review |
| Fiandaca, MS et al., 2020 [66] | Advances in Parkinson’s disease and other neurological diseases gene treatments, approaches, and technology. | 12 | A review |
| Kubota, KJ et al., 2016 [67] | Large-scale wearable sensor data for Parkinson’s disease using machine learning. | 180 | A review |
| a. Speech Parameter | |||||||
| Reference | Modality | Algorithms Used | Objective | Tools | Source of Data | Subjects | Performance |
| Sakar et al., 2019 [121] | Speech | Support Vector Machine | Classification of PD from HC | JupyterLab with python programming language | Collected from participants | 188 PD and 64 HC | Accuracy (ACC.)—86% |
| Yasar A. et al., 2019 [122] | Speech | Artificial Neural Network | Classification of PD from HC | MATLAB | Collected from participants | 40 PD and 40 HC | ACC.—94.93% |
| Ouhmida, A, 2021 [123] | Speech | SVM, K-NN, Decision Tree | Classification of PD from HC | Not mentioned | UCI machine learning repository | Not mentioned | AUC-98.26% |
| Marar et al., 2018 [124] | Speech | Naive Bayes | Classification of PD from HC | R programming | Collected from participants | 23 PD and 8 HC | ACC.—94.87% |
| Sheibani R etal., 2019 [125] | Speech | Ensemble-Based Method | Classification of PD from HC | Python programming | UCI machine learning repository | 23 PD and 8 HC | ACC.—90.6% |
| John M. Tracy etal., 2020 [126] | Speech | Gradient Boosted Trees | Classification of PD from HC | Python | Not mentioned | 246 PD and 2023 HC | ACC.—79.7% |
| b. Handwriting Patterns | |||||||
| Reference | Modality | Algorithms Used | Objective | Tools | Source of Data | Subjects | Performance |
| Cibulka et al., 2019 [127] | Handwriting Patterns | Random Forest | Classification of PD from HC | Not mentioned | Collected from participants | 150 PD and 120 HC | Not mentioned |
| Hsu S-Y et al., 2019 [128] | Handwriting Patterns | Support Vector Machine | Classification of PD from HC | Weka | PACS | 196 PD and 6 HC | ACC.—83.2% |
| Drotár, P et al., 2016 [129] | Handwriting Patterns | K-NN, Ensemble AdaBoostClassifier, Support Vector Machine | Classification of PD from HC | Python [scikit-learn library] | PaHaW database | 37 PD and 38 HC | ACC.—81.3% |
| Fabian Maass etal., 2020 [130] | Handwriting Patterns | Support Vector Machine | Classification of PD from HC | Not mentioned | Collected from participants | 82 PD and 68 HC | Sensitivity—80% |
| J. Mucha et al., 2018 [131] | Handwriting Patterns | Random Forest Classifier | Classification of PD from HC | Not mentioned | Collected from participants | 33 PD and 36 HC | ACC.—90% and Sensitivity—89% |
| Wenzel et al., 2019 [132] | Handwriting Patterns | Convolutional Neural Network | Classification of PD from HC | MATLAB | Not mentioned | 438 PD and 207 HC | ACC.—97.23% |
| Segovia, F. etal., 2019 [133] | Handwriting Patterns | Support Vector Machine | Classification of PD from HC | Python programming | Not mentioned | 95 PD and 94 HC | ACC.—94.2% |
| c. Gait Parameter | |||||||
| Reference | Modality | Algorithms Used | Objective | Tools | Source of Data | Subjects | Performance |
| Ye, Q. et al., 2018 [134] | Gait | Support Vector Machine | Classification of PD from HC | Not mentioned | Collected from participants | 15 PD and 16 HC | ACC.—90.32% |
| Klomsae, A et al., 2018 [135] | Gait | Fuzzy KNN | Classification of PD from HC | Not mentioned | Collected from participants | 15 PD and 16 HC | ACC.—96.43% |
| J. P. Félix et al., 2019 [136] | Gait | Support Vector Machine | Classification of PD from HC | MATLAB | Not mentioned | 15 PD and 16 HC | ACC.—96.8% |
| Andrei et al., 2019 [137] | Gait | SVM | Classification of PD from HC | Not mentioned | Laboratory for Gait & Neurodynamics | 93 PD and 73 HC | ACC.—100% |
| Priya SJ et al., 2021 [138] | Gait | ANN | Classification of PD from HC | MATLAB | Laboratory for Gait & Neurodynamics | 93 PD and 73 HC | ACC.—96.28% |
| Oğul, et al., 2020 [139] | Gait | ANN | Classification of PD from HC | MATLAB | Laboratory for Gait & Neurodynamics | 93 PD and 73 HC | ACC.—98.3% |
| Li B et al., 2020 [140] | Gait | Deep CNN | Classification of PD from HC | Not mentioned | Collected from participants | 10 PD and 10 HC | ACC.—91.9% |
| Reference | Modality | Algorithms Used | Objective | Tools | Source of Data | Subjects | Performance |
|---|---|---|---|---|---|---|---|
| Hosseini and Makki, 2013 [162] | Essential Tremor (ET) | Auto Associative Neural Network | Classification of ET, PD from HC | Not Mentioned | Collected from participants | 20 ET and 20 PD and | ACC.—87.5% |
| Challa et al., 2016 [163] | DaTSCAN SPECT | Boosted Logistic Regression | Classification of PD from HC | Weka | PPMI Database | 402 PD | ACC.—97.159% |
| Choi et al. 2017 [164] | SPECT | Deep Convolutional Neural Network | Classification of PD from HC | MATLAB | PPMI Database | 431 PD, 193 HC, 77 SWEDD | ACC.—98.8% and sensitivity—98.6% |
| Kim, Wit, and Thurston 2018 [165] | SPECT | Inception-V3 (Pre-trained) | Classification of PD from HC | Not mentioned | Not mentioned | 54 PD and 54 HC | Sensitivity—96.3% |
| Esmaeilzadeh et al., 2018 [166] | MRI-T | Convolutional Neural Network | Classification of PD from HC | Not mentioned | PPMI Database | 452 PD and 204 HC | ACC.—100% |
| Kim, Lee, et al., 2018 [167] | Tremor | Convolutional Neural Network | Classification of PD from HC | Not mentioned | Collected from the participants | 92 PD and 95 HC | ACC.—85% |
| Martinez- Murcia et al., 2017 [168] | SPECT | Convolutional Neural Network | Classification of PD from HC | Not mentioned | PPMI Database | 158 PD and 32 SWEDD and 111 HC | ACC.-PD vs HC: 95.5 ± 0.44 and sensitivity-PD vs HC: 96.2 ± 0.051 |
| Qin et al., 2019 [169] | Tremor | Convolutional Neural Network | Classification of PD from HC | Not mentioned | Collected from the participants | 147 PD | ACC.—90.55% |
| Kollia et al., 2019 [170] | MRI and DaTSCAN | Convolutional Neural Network and Recurrent Neural Network | Classification of PD from HC | Not mentioned | Not mentioned | 55 PD and 23HC | ACC.—98% |
| Szumilas et al., 2020 [171] | Tremor | Recurrent Neural Network | Develop a prediction model to evaluate tremor severity in PD patients | Not mentioned | Collected from the participants | 64 PD | Not mentioned |
| Oktay and Kocer 2020 [172] | Tremor | Convolutional Long Short-Term Memory | Classification of PD from HC | C++ with Leap motion API | Medical Faculty Teaching Hospital Neurology Istanbul Medeniyet University | 23 Parkinson’s tremors and 17 ET | ACC.—90% |
| Shahtalebi, S et al., 2020 [173] | Tremor | 3D Convolutional Neural Network | Develop a deep recurrent model to predict and eliminate the PHT component of hand motion | Not mentioned | Collected from the participants | 81 PD | Not mentioned |
| Veeraragavan et al., 2020 [174] | MRI | Artificial Neural Network | Classification of PD from HC | Not mentioned | Collected from the participants | 93 PD and 73 HC | ACC.—97.41% and sensitivity—97.70% |
| Chien et al., 2021 [175] | DAT-SPECT | Artificial Neural Network | Classification of PD from HC | MATLAB 2018B | Collected from the participants | 234 PD | ACC.—99.22% and sensitivity—81.8% |
| Yasaka et al., 2021 [176] | MRI | 2D Convolutional Neural Network | Classification of PD from HC | MATLAB | Collected from the participants from Juntendo University Hospital | 115 PD and 115 HC | Not Mentioned |
| Yang et al., 2021 [177] | MRI + CI | Ensemble (SVM, RF, KNN, ANN, LR) | Classification of PD from HC | Not mentioned | Not mentioned | 65 PD and 36 HC | ACC.—96.88% and sensitivity—95.0% |
| Vyas et al., 2021 [178] | MRI | 2D and 3D Convolutional Neural Network | Classification of PD from HC | Not mentioned | PPMI Database | 236 PD and 82 HC | ACC. from 2D and 3D 88.9% and 72.22%, respectively, and sensitivity—92% and 100%, respectively |
| Yadav 2021 [179] | fMRI | Bayesian 3D-Convolutional Neural Network | Classification of PD from HC | Not mentioned | ADNI Dataset | 15 PD and 15 HC | ACC.—97.92% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rana, A.; Dumka, A.; Singh, R.; Panda, M.K.; Priyadarshi, N. A Computerized Analysis with Machine Learning Techniques for the Diagnosis of Parkinson’s Disease: Past Studies and Future Perspectives. Diagnostics 2022, 12, 2708. https://doi.org/10.3390/diagnostics12112708
Rana A, Dumka A, Singh R, Panda MK, Priyadarshi N. A Computerized Analysis with Machine Learning Techniques for the Diagnosis of Parkinson’s Disease: Past Studies and Future Perspectives. Diagnostics. 2022; 12(11):2708. https://doi.org/10.3390/diagnostics12112708
Chicago/Turabian StyleRana, Arti, Ankur Dumka, Rajesh Singh, Manoj Kumar Panda, and Neeraj Priyadarshi. 2022. "A Computerized Analysis with Machine Learning Techniques for the Diagnosis of Parkinson’s Disease: Past Studies and Future Perspectives" Diagnostics 12, no. 11: 2708. https://doi.org/10.3390/diagnostics12112708
APA StyleRana, A., Dumka, A., Singh, R., Panda, M. K., & Priyadarshi, N. (2022). A Computerized Analysis with Machine Learning Techniques for the Diagnosis of Parkinson’s Disease: Past Studies and Future Perspectives. Diagnostics, 12(11), 2708. https://doi.org/10.3390/diagnostics12112708






