Performance Evaluation of Developed Bangasure™ Multiplex rRT-PCR Assay for SARS-CoV-2 Detection in Bangladesh: A Blinded Observational Study at Two Different Sites
Abstract
1. Introduction
2. Methods
2.1. Ethical Approval
2.2. Primer and Probes
2.3. Sample Collection and Preparation
2.4. Optimization of Bangasure™ rRT-PCR Assay
2.5. Limit of Detection (LoD) Determination
2.6. Performance Evaluation of the In-House Assay
2.7. Accelerated Stability Testing
2.8. Data Analysis
3. Results
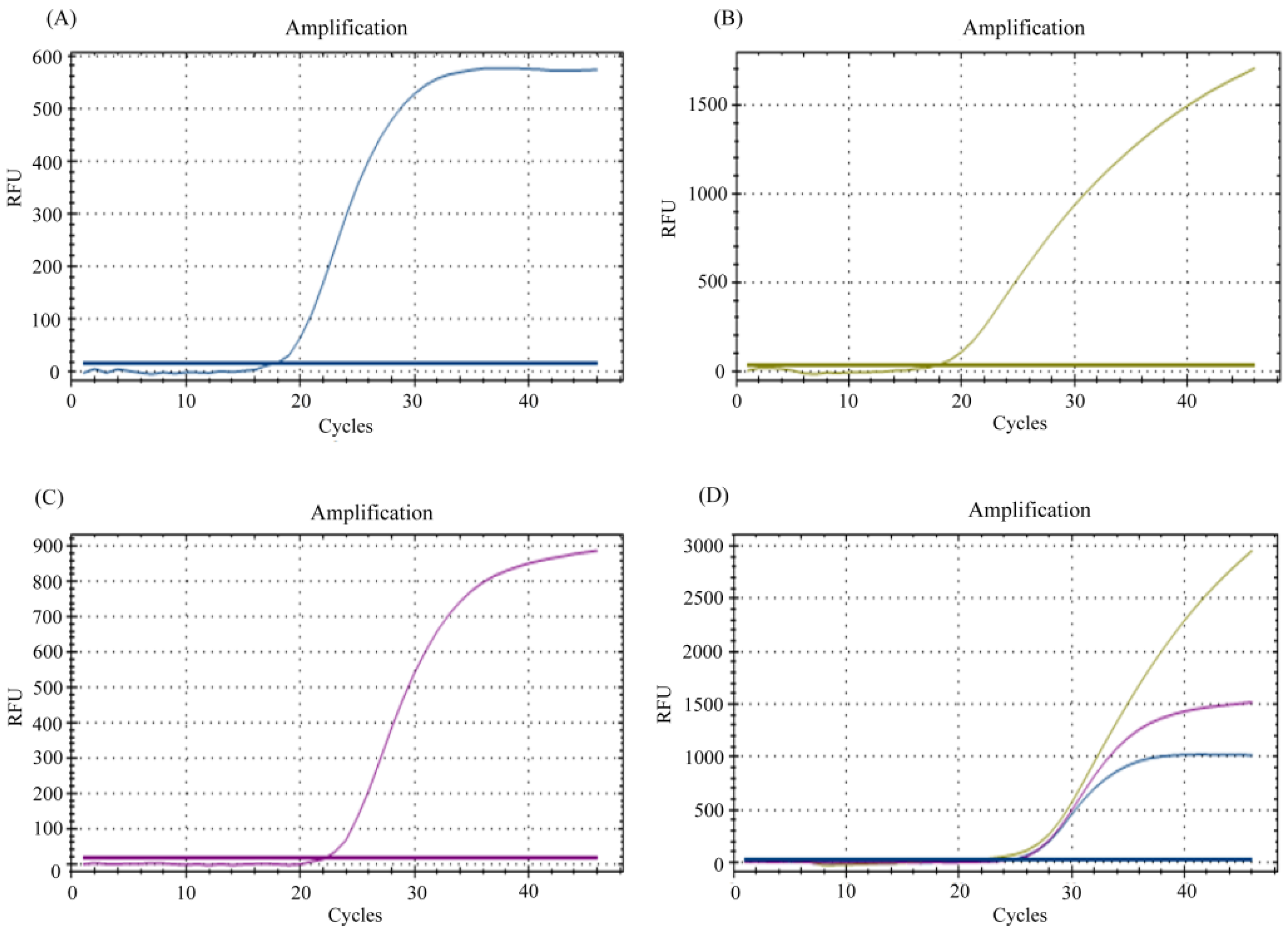
3.1. Multiplexing of E, N, and RP Genes for Detection of SARS-CoV-2 RNA
3.2. Limit of Detection (LoD) Determination of the In-House Multiplex Assay
3.3. Efficiency of In-House Multiplex rRT-PCR Assay
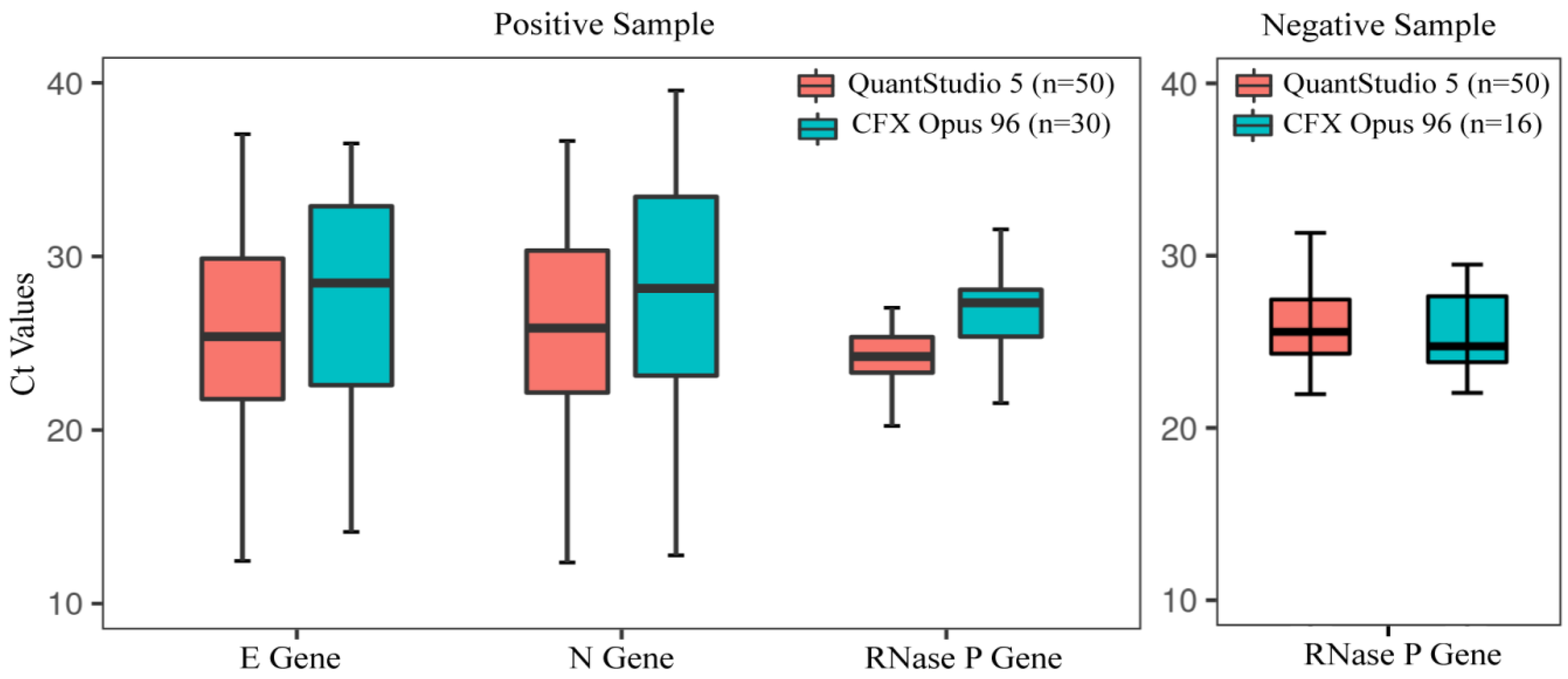
3.4. Performance Evaluation of In-House Assay Using Clinical Specimens
3.5. Determination of Assay Reproducibility and Stability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Du, W.; Han, S.; Li, Q.; Zhang, Z. Epidemic update of COVID-19 in Hubei Province compared with other regions in China. Int. J. Infect. Dis. 2020, 95, 321–325. [Google Scholar] [CrossRef]
- COVID Live Coronavirus Statistics—Worldometer n.d. Available online: https://www.worldometers.info/coronavirus/ (accessed on 19 January 2022).
- COVID-19 Dynamic Dashboard for Bangladesh. Available online: https://dghs-dashboard.com/pages/covid19.php (accessed on 19 January 2022).
- Ma, Q.; Liu, J.; Liu, Q.; Kang, L.; Liu, R.; Jing, W.; Wu, Y.; Liu, M. Global percentage of asymptomatic SARS-CoV-2 infections among the tested population and individuals with confirmed COVID-19 diagnosis: A systematic review and meta-analysis. JAMA Netw. Open 2021, 4, e2137257. [Google Scholar] [CrossRef] [PubMed]
- Eis-Hübinger, A.M.; Hönemann, M.; Wenzel, J.J.; Berger, A.; Widera, M.; Schmidt, B.; Aldabbagh, S.; Marx, B.; Streeck, H.; Ciesek, S.; et al. Ad hoc laboratory-based surveillance of SARS-CoV-2 by real-time RT-PCR using minipools of RNA prepared from routine respiratory samples. J. Clin. Virol. 2020, 127, 104381. [Google Scholar] [CrossRef] [PubMed]
- Hatcher, E.L.; Zhdanov, S.A.; Bao, Y.; Blinkova, O.; Nawrocki, E.P.; Ostapchuck, Y.; Schäffer, A.A.; Brister, J.R. Virus Variation Resource—Improved response to emergent viral outbreaks. Nucleic Acids Res. 2017, 45, D482–D490. [Google Scholar] [CrossRef] [PubMed]
- Kucirka, L.M.; Lauer, S.A.; Laeyendecker, O.; Boon, D.; Lessler, J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann. Intern. Med. 2020, 173, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Wikramaratna, P.S.; Paton, R.S.; Ghafari, M.; Lourenço, J. Estimating the false-negative test probability of SARS-CoV-2 by RT-PCR. Eurosurveillance 2020, 25, 2000568. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Yan, L.; Wang, N.; Yang, S.; Wang, L.; Tang, Y.; Gao, G.; Wang, S.; Ma, C.; Xie, R.; et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 2020, 71, 793–798. [Google Scholar] [CrossRef]
- Hasan, M.M.; Rocha, I.C.N.; Ramos, K.G.; Cedeño, T.D.D.; dos Santos Costa, A.C.; Tsagkaris, C.; Billah, M.M.; Ahmad, S.; Essar, M.Y. Emergence of highly infectious SARS-CoV-2 variants in Bangladesh: The need for systematic genetic surveillance as a public health strategy. Trop. Med. Health 2021, 49, 69. [Google Scholar] [CrossRef]
- Rahman, M.; Shirin, T.; Rahman, S.; Rahman, M.M.; Hossain, M.E.; Khan, M.H.; Rahman, M.Z.; el Arifeen, S.; Ahmed, T. The emergence of SARS-CoV-2 variants in Dhaka city, Bangladesh. Transbound. Emerg. Dis. 2021, 68, 3000–3001. [Google Scholar] [CrossRef]
- van Dorp, L.; Acman, M.; Richard, D.; Shaw, L.P.; Ford, C.E.; Ormond, L.; Owen, C.J.; Pang, J.; Tan, C.C.S.; Boshier, F.A.T.; et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020, 83, 104351. [Google Scholar] [CrossRef]
- Kaushal, N.; Gupta, Y.; Goyal, M.; Khaiboullina, S.F.; Baranwal, M.; Verma, S.C. Mutational frequencies of SARS-CoV-2 genome during the beginning months of the outbreak in USA. Pathogens 2020, 9, 565. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Z.; Chen, Z.; Huang, X.; Xu, M.; He, T.; Zhang, Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020, 92, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.; Nasrullah, M.; Hosen, M.J. COVID-19 and Bangladesh: Challenges and how to address them. Front. Public Health 2020, 8, 154. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- CDC. Real-Time RT- PCR Primers and Probes for COVID-19. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (accessed on 21 January 2022).
- Zhen, W.; Berry, G.J. Design of a novel multiplex real time RT-PCR assay for SARS-CoV-2 detection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cook, R.J.; Dickens, B.M.; Fathalla, M.F. World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. In Reproductive Health and Human Rights; Oxford University Press: Oxford, UK, 2003. [Google Scholar] [CrossRef]
- Bajaj, S.; Singla, D.; Sakhuja, N. Stability testing of pharmaceutical products. J. Appl. Pharm. Sci. 2012, 30, 129–138. [Google Scholar]
- MedCalc’s Diagnostic Test Evaluation Calculator. Available online: https://www.medcalc.org/calc/diagnostic_test.php (accessed on 11 February 2022).
- Eberle, U.; Wimmer, C.; Huber, I.; Neubauer-Juric, A.; Valenza, G.; Ackermann, N.; Sing, A. Comparison of nine different commercially available molecular assays for detection of SARS-CoV-2 RNA. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1303–1308. [Google Scholar] [CrossRef]
- Total COVID-19 Tests Per 1000 People. Available online: https://ourworldindata.org/grapher/full-list-cumulative-total-tests-per-thousand?tab=table (accessed on 11 February 2022).
- Tombuloglu, H.; Sabit, H.; Al-Suhaimi, E.; Al Jindan, R.; Alkharsah, K.R. Development of multiplex real-time RT-PCR assay for the detection of SARS-CoV-2. PLoS ONE 2021, 16, e0250942. [Google Scholar] [CrossRef]
- Anantharajah, A.; Helaers, R.; Defour, J.-P.; Olive, N.; Kabera, F.; Croonen, L.; Deldime, F.; Vaerman, J.; Barbée, C.; Bodéus, M.; et al. How to choose the right real-time RT-PCR primer sets for the SARS-CoV-2 genome detection? J. Virol. Methods 2021, 295, 114197. [Google Scholar] [CrossRef]
- Cao, L.; Xu, T.; Liu, X.; Ji, Y.; Huang, S.; Peng, H.; Li, C.; Guo, D. The Impact of Accumulated Mutations in SARS-CoV-2 Variants on the qPCR Detection Efficiency. Front. Cell. Infect. Microbiol. 2022, 12. [Google Scholar] [CrossRef]
- Freire-Paspuel, B.; Garcia-Bereguiain, M.A. Analytical sensitivity and clinical performance of a triplex RT-qPCR assay using CDC N1, N2, and RP targets for SARS-CoV-2 diagnosis. Int. J. Infect. Dis. 2021, 102, 14–16. [Google Scholar] [CrossRef] [PubMed]
- Nalla, A.K.; Casto, A.M.; Huang, M.-L.W.; Perchetti, G.A.; Sampoleo, R.; Shrestha, L.; Wei, Y.; Zhu, H.; Jerome, K.R.; Greninger, A.L.; et al. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020, 58, e00557-20. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Won, J.; Choi, B.Y.; Lee, C.J. Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using PCR and real-time PCR. Exp. Mol. Med. 2020, 52, 963–977. [Google Scholar] [CrossRef]
- Vogels, C.B.; Brito, A.F.; Wyllie, A.L.; Fauver, J.R.; Ott, I.M.; Kalinich, C.C.; Petrone, M.E.; Casanovas-Massana, A.M.; Muenker, C.; Moore, A.J.; et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020, 5, 1299–1305. [Google Scholar] [CrossRef] [PubMed]
- Noor, F.A.; Safain, K.S.; Hossain, M.W.; Arafath, K.; Mannoor, K.; Kabir, M. Development and performance evaluation of the first in-house multiplex rRT-PCR assay in Bangladesh for highly sensitive detection of SARS-CoV-2. J. Virol. Methods 2021, 293, 114147. [Google Scholar] [CrossRef] [PubMed]
- Ishige, T.; Murata, S.; Taniguchi, T.; Miyabe, A.; Kitamura, K.; Kawasaki, K.; Nishimura, M.; Igari, H.; Matsushita, K. Highly sensitive detection of SARS-CoV-2 RNA by multiplex rRT-PCR for molecular diagnosis of COVID-19 by clinical laboratories. Clin. Chim. Acta 2020, 507, 139–142. [Google Scholar] [CrossRef]
- Banko, A.; Petrovic, G.; Miljanovic, D.; Loncar, A.; Vukcevic, M.; Despot, D.; Cirkovic, A. Comparison and sensitivity evaluation of three different commercial real-time quantitative PCR kits for SARS-CoV-2 detection. Viruses 2021, 13, 1321. [Google Scholar] [CrossRef]
- Wu, S.; Shi, X.; Chen, Q.; Jiang, Y.; Zuo, L.; Wang, L.; Jiang, M.; Lin, Y.; Fang, S.; Peng, B.; et al. Comparative evaluation of six nucleic acid amplification kits for SARS-CoV-2 RNA detection. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 38. [Google Scholar] [CrossRef]
- Onwuamah, C.K.; Okwuraiwe, A.P.; Salu, O.B.; Shaibu, J.O.; Ndodo, N.; Amoo, S.O.; Okoli, L.C.; Ige, F.A.; Ahmed, R.A.; Bankole, M.A.; et al. Comparative performance of SARS-CoV-2 real-time PCR diagnostic assays on samples from Lagos, Nigeria. PLoS ONE 2021, 16, e0246637. [Google Scholar] [CrossRef]
- Tetzner, R.; Dietrich, D.; Distler, J. Control of carry-over contamination for PCR-based DNA methylation quantification using bisulfite treated DNA. Nucleic Acids Res. 2007, 35, e4. [Google Scholar] [CrossRef]





| Target Gene | Primer/Probe | Oligonucleotide Sequence (5′–3′) |
|---|---|---|
| E gene | E_SarbecoF_primer | ACAGGTACGTTAATAGTTAATAGCGT |
| E_SarbecoR_Primer | ATATTGCAGCAGTACGCACACA | |
| Probe_E | FAM-ACACTAGCCATCCTTACTGCGCTTCG-BHQ1 | |
| N gene | N_cdcF_Primer | TTACAAACATTGGCCGCAAA |
| N_cdcFR_Primer | GCGCGACATTCCGAAGAA | |
| Probe_N | VIC-ACAATTTGCCCCCAGCGCTTCAG-BHQ1 | |
| RNase P | RP_F_Primer | AGATTTGGACCTGCGAGCG |
| RP_R_Primer | GAGCGGCTGTCTCCACAAGT | |
| Probe_RNase P | CY5-TTCTGACCTGAAGGCTCTGCGCG-BHQ-1 |
| Virus Copy/mL | BangasureTM | Sansure Biotec. | 1copy | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detection Rate, % | Target Gene | Detection Rate, % | Target Gene | Detection Rate, % | Target Gene | ||||||||||||
| E | N | ORF1ab | N | E | N | RdRp | |||||||||||
| Ct Value | Ct Value | Ct Value | Ct Value | Ct Value | Ct Value | Ct Value | |||||||||||
| Mean | %CV | Mean | %CV | Mean | %CV | Mean | %CV | Mean | %CV | Mean | %CV | Mean | %CV | ||||
| 100,000 | 100 | 26.35 | 2.87 | 26.83 | 1.22 | 100 | 27.64 | 0.48 | 26.46 | 0.24 | 100 | 28.31 | 0.83 | 27.77 | 0.27 | 28.99 | 0.34 |
| 10,000 | 100 | 30.01 | 0.65 | 30.21 | 0.62 | 100 | 30.81 | 0.21 | 29.56 | 0.45 | 100 | 31.54 | 0.87 | 31.34 | 1.07 | 32.35 | 0.81 |
| 1000 | 100 | 33.52 | 1.29 | 34.48 | 2.40 | 100 | 34.77 | 0.81 | 33.65 | 1.97 | 100 | 34.99 | 0.82 | 34.90 | 1.63 | 35.43 | 0.96 |
| 100 | 100 | 37.49 | 1.34 | 37.71 | 1.59 | 100 | 38.38 | 1.57 | 37.11 | 1.34 | 0 | UND | UND | UND | UND | UND | UND |
| 10 | 0 | UND | UND | UND | UND | 0 | UND | UND | UND | UND | 0 | UND | UND | UND | UND | UND | UND |
| 1 | 0 | UND | UND | UND | UND | 0 | UND | UND | UND | UND | 0 | UND | UND | UND | UND | UND | UND |
| (A) | |||||
|---|---|---|---|---|---|
| Information of Clinical Samples | Number of Samples | Tested by BangasureTM RT-PCR Kit | Tested by Sansure COVID-19 rRT-PCR Kit | Tested by 1copy 4plex Kit | |
| Samples Tested positive, n = 50 | High (Ct < 26) | 24 | 24 | 24 | 24 |
| Moderate (26 < Ct ≤ 32) | 18 | 18 | 18 | 18 | |
| Low (32 < Ct ≤ 38) | 8 | 8 | 8 | 8 | |
| Total | 50 | 50 | 50 | 50 | |
| Samples tested negative, n = 50 | 50 | 50 | 50 | 50 | |
| Sensitivity,%(95% CI) | 100 (92.89–100) | 100 (92.89–100) | 100 (92.89–100) | ||
| Specificity,%(95% CI) | 100 (92.89–100) | 100 (92.89–100) | 100 (92.89–100) | ||
| PPV,% | 100 | 100 | 100 | ||
| NPV,% | 100 | 100 | 100 | ||
| Accuracy,% (95% CI) | 100 (96.38–100) | 100 (96.38–100) | 100 (96.38–100) | ||
| (B) | |||||
| Information of Clinical Samples | Number of Samples | Tested by BangasureTM RT-PCR Kit | Tested by Sansure COVID-19 rRT-PCR Kit | ||
| Sample Tested positive, n = 30 | High (Ct < 26) | 10 | 10 | 10 | |
| Moderate (26 < Ct ≤ 32) | 10 | 10 | 10 | ||
| Low (32 < Ct ≤ 38) | 10 | 10 | 10 | ||
| Total | 30 | 30 | 30 | ||
| Sample tested negative, n = 16 | 16 | 16 | 16 | ||
| Sensitivity, % (95% CI) | 100 (88.4–100) | 100 (88.4–100) | |||
| Specificity, % (95% CI) | 100 (79.4–100) | 100 (79.4–100) | |||
| PPV, % | 100 | 100 | |||
| NPV, % | 100 | 100 | |||
| Accuracy, % (95%CL) | 100 (92.29–100) | 100 (92.29–100) | |||
| Site 1 | Site 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ct Values for Multiplex PCR at 4 °C | Ct Values for Multiplex PCR at 4 °C | Ct Values for Multiplex PCR at −20 °C | |||||||
| E | N | RNase P | E | N | RNase P | E | N | RNase P | |
| Week 1 | 21.91 | 22.75 | 23.99 | 23.96 | 21.52 | 27.24 | 25.61 | 21.52 | 23.73 |
| Week 2 | 21.44 | 23.73 | 24.42 | 25.068 | 25.35 | 25.54 | 25.35 | 25.35 | 25.63 |
| Week 3 | 23.50 | 23.66 | 24.76 | 25.938 | 24.77 | 25.94 | 25.96 | 24.78 | 25.84 |
| Week 4 | 22.96 | 24.35 | 23.74 | 26.17 | 25.13 | 25.73 | 26.25 | 24.89 | 25.95 |
| Week 5 | 23.44 | 24.31 | 23.71 | 27.256 | 25.37 | 26.23 | 26.89 | 25.12 | 25.75 |
| Mean (SD) | 22.65 (0.93) | 23.76 (0.65) | 24.12 (0.45) | 25.68 (1.24) | 24.43 (1.64) | 26.14 (0.67) | 26.01(0.59) | 24.33 (1.58) | 25.38 (0.93) |
| CV (%) | 4.11 | 2.73 | 1.88 | 4.82 | 6.73 | 2.56 | 2.29 | 6.52 | 3.66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razu, M.H.; Ahmed, Z.B.; Hossain, M.I.; Rabbi, M.F.A.; Nayem, M.R.; Hassan, M.A.; Paul, G.K.; Khan, M.R.; Moniruzzaman, M.; Karmaker, P.; et al. Performance Evaluation of Developed Bangasure™ Multiplex rRT-PCR Assay for SARS-CoV-2 Detection in Bangladesh: A Blinded Observational Study at Two Different Sites. Diagnostics 2022, 12, 2617. https://doi.org/10.3390/diagnostics12112617
Razu MH, Ahmed ZB, Hossain MI, Rabbi MFA, Nayem MR, Hassan MA, Paul GK, Khan MR, Moniruzzaman M, Karmaker P, et al. Performance Evaluation of Developed Bangasure™ Multiplex rRT-PCR Assay for SARS-CoV-2 Detection in Bangladesh: A Blinded Observational Study at Two Different Sites. Diagnostics. 2022; 12(11):2617. https://doi.org/10.3390/diagnostics12112617
Chicago/Turabian StyleRazu, Mamudul Hasan, Zabed Bin Ahmed, Md. Iqbal Hossain, Mohammad Fazle Alam Rabbi, Maksudur Rahman Nayem, Md. Akibul Hassan, Gobindo Kumar Paul, Md. Robin Khan, Md. Moniruzzaman, Pranab Karmaker, and et al. 2022. "Performance Evaluation of Developed Bangasure™ Multiplex rRT-PCR Assay for SARS-CoV-2 Detection in Bangladesh: A Blinded Observational Study at Two Different Sites" Diagnostics 12, no. 11: 2617. https://doi.org/10.3390/diagnostics12112617
APA StyleRazu, M. H., Ahmed, Z. B., Hossain, M. I., Rabbi, M. F. A., Nayem, M. R., Hassan, M. A., Paul, G. K., Khan, M. R., Moniruzzaman, M., Karmaker, P., & Khan, M. (2022). Performance Evaluation of Developed Bangasure™ Multiplex rRT-PCR Assay for SARS-CoV-2 Detection in Bangladesh: A Blinded Observational Study at Two Different Sites. Diagnostics, 12(11), 2617. https://doi.org/10.3390/diagnostics12112617






