Molecular Detection of Human Papillomaviruses in Formalin Fixed Paraffin Embedded Sections from Different Anogenital Lesions in Duhok-Iraq
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Ethical Approval
2.3. Sample Collection
2.4. DNA Extraction
2.5. HPV Detection and Genotyping
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Geng, Y.; Liu, L. Human papillomavirus genotypes and infection among women in Changzhou, China. Hum. Vaccines Immunother. 2019, 15, 1884–1888. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gu, Y.; Wang, H.; Chen, J.; Zheng, Y.; Cui, B.; Yang, X. Distribution of cervical lesions in high-risk HPV (hr-HPV) positive women with ASC-US: A retrospective single-center study in China. Virol. J. 2020, 17, 185. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Rosas, F.; Orozco-Hernández, E.; Maza-Sánchez, L.; Salgado-García, P.C.; Navarro-Vidal, E.; de León-Bautista, M.P. Prevalence and correlation of human papillomavirus genotypes with clinical factors in cervical samples from Mexican women. Exp. Biol. Med. 2020, 246, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Al-Shabanah, O.; Hafez, M.M.; Hassan, Z.K.; Sayed-Ahmed, M.M.; Abozeed, W.N.; Al-Rejaie, S.S.; A Alsheikh, A. Human papillomavirus genotyping and integration in ovarian cancer Saudi patients. Virol. J. 2013, 10, 343. [Google Scholar] [CrossRef] [PubMed]
- Shafaghi, B.; Jarollahi, A.; Yousefzadeh, B.; Ameri, A.; Moghadam, S.; Mostafavi, M. Human papilloma virus prevalence and types among Iranian women attending regular gynecological visits. Rep. Radiother. Oncol. 2013, 1, e2389. [Google Scholar]
- Jalilian, S.; Izadi, B.; Madani, S.H.; Mohajeri, P. The Prevalence and Genotype Distribution of Human Papillomavirus Types in the General Female Population in West of Iran. Jundishapur J. Microbiol. 2017, 10, e40855. [Google Scholar] [CrossRef]
- Kamal, M.; Lameiras, S.; Deloger, M.; Morel, A.; Vacher, S.; Lecerf, C.; Dupain, C.; Jeannot, E.; Girard, E.; Baulande, S.; et al. Human papilloma virus (HPV) integration signature in Cervical Cancer: Identification of MACROD2 gene as HPV hot spot integration site. Br. J. Cancer 2021, 124, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Hasan, U.A.; Bates, E.; Takeshita, F.; Biliato, A.; Accardi, R.; Bouvard, V.; Mansour, M.; Vincent, I.; Gissmann, L.; Iftner, T.; et al. TLR9 Expression and Function Is Abolished by the Cervical Cancer-Associated Human Papillomavirus Type 16. J. Immunol. 2007, 178, 3186–3197. [Google Scholar] [CrossRef] [PubMed]
- Eslami, G.; Golshani, M.; Rakhshan, M.; Fallah, F.; Goudarzi, H.; Taghavi, A. PCR detection and high risk typing of human papillomavirus DNA in cervical cancer in Iranian women. Cancer Ther. 2008, 6, 361–366. [Google Scholar]
- Mirghani, H.; Amen, F.; Moreau, F.; Guigay, J.; Ferchiou, M.; Melkane, A.E.; Hartl, D.M.; Guily, J.L.S. Human papilloma virus testing in oropharyngeal squamous cell carcinoma: What the clinician should know. Oral Oncol. 2014, 50, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Westra, W.H. Detection of human papillomavirus (HPV) in clinical samples: Evolving methods and strategies for the accurate determination of HPV status of head and neck carcinomas. Oral Oncol. 2014, 50, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Greer, C.; Lund, J.K.; Manos, M.M. PCR amplification from paraffin-embedded tissues: Recommendations on fixatives for long-term storage and prospective studies. Genome Res. 1991, 1, 46–50. [Google Scholar] [CrossRef]
- Greer, C.; Wheeler, C.M.; Manos, M.M. Sample preparation and PCR amplification from paraffin-embedded tissues. Genome Res. 1994, 3, S113–S122. [Google Scholar] [CrossRef] [PubMed]
- Donà, M.G.; Ronchetti, L.; Giuliani, M.; Carosi, M.; Rollo, F.; Congiu, M.; Mazza, D.; Pescarmona, E.; Vocaturo, A.; Benevolo, M. Performance of the Linear Array HPV Genotyping Test on Paired Cytological and Formalin-Fixed, Paraffin-Embedded Cervical Samples. J. Mol. Diagn. 2013, 15, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Bruni, L.; Albero, G.; Serrano, B.; Mena, M.; Gómez, D.; Muñoz, J.; Bosch, F.X.; de Sanjosé, S. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). In Human Papillomavirus and Related Diseases in the World; Summary Report 22 October 2021; Institut Català d’Oncologia: L’Hospitalet de Llobregat, Spain, 2021. [Google Scholar]
- Al–Taiee, A.; Alizi, S.; Kadhim, H.; Al-Khalidy, W. Human papilloma virus genotyping in a group of iraqi women by using reproducible DNA extraction from formalin fixed paraffin embedded sections. World J. Pharm. Res. 2016, 5805, 232–240. [Google Scholar] [CrossRef]
- Freij, M.A.; Saleh, H.H.; Farsakh, H.A.A.; Khadra, M.M.; Ijmail, A.A.; Rahal, B.O.; Waldali, M.H.; Najeeb, N.S.; Tahtamouni, L.H. Type-specific prevalence of human papillomavirus among women with abnormal cytology in Jordan. Eur. J. Gynaecol. Oncol. 2017, 38, 901–904. [Google Scholar] [CrossRef]
- Krashias, G.; Koptides, D.; Christodoulou, C. HPV prevalence and type distribution in Cypriot women with cervical cytological abnormalities. BMC Infect. Dis. 2017, 17, 346. [Google Scholar] [CrossRef]
- Alobaid, A.; Al-Badawi, I.A.; Alkadri, H.M.; Gopala, K.; Kandeil, W.; Quint, W.; Al-Aker, M.; DeAntonio, R. Human papillomavirus prevalence and type distribution among women attending routine gynecological examinations in Saudi Arabia. BMC Infect. Dis. 2014, 14, 643. [Google Scholar] [CrossRef]
- Bansal, D.; Elmi, A.A.; Skariah, S.; Haddad, P.; Abu-Raddad, L.J.; Al Hamadi, A.H.; Mohamed-Nady, N.; Affifi, N.M.; Ghedira, R.; Hassen, E.; et al. Molecular epidemiology and genotype distribution of Human Papillomavirus (HPV) among Arab women in the state of Qatar. J. Transl. Med. 2014, 12, 9. [Google Scholar] [CrossRef]
- Guettiti, H.; Ennaifer, E.; Attia, L.; Chelly, D.; Ben Alaya, N.; Ben Aissa, R.; Laassili, T.; Boubaker, S. Pre-vaccination Prevalence and Genotype Distribution of Human Papillomavirus Infection among Women from Urban Tunis: A Cross-sectional Study. Asian Pac. J. Cancer Prev. 2014, 15, 9361–9365. [Google Scholar] [CrossRef][Green Version]
- Giuliani, L.; Coletti, A.; Syrjänen, K.; Favalli, C.; Ciotti, M. Comparison of DNA sequencing and Roche Linear array in human papillomavirus (HPV) genotyping. Anticancer Res. 2006, 26, 3939–3941. [Google Scholar] [PubMed]
- Burchell, A.N.; Winer, R.L.; de Sanjosé, S.; Franco, E. Chapter 6: Epidemiology and transmission dynamics of genital HPV infection. Vaccine 2006, 24, S52–S61. [Google Scholar] [CrossRef] [PubMed]
- Sellors, J.W.; Mahony, J.B.; Kaczorowski, J.; Lytwyn, A.; Bangura, H.; Chong, S.; Lorincz, A.; Dalby, D.M.; Janjusevic, V.; Keller, J.L.; et al. Prevalence and predictors of human papillomavirus infection in women in Ontario, Canada. CMAJ 2000, 163, 503–508. [Google Scholar]
- Oztürk, S.; Kaleli, I.; Kaleli, B.; Bir, F. Servikal orneklerde insan papillomavirus DNA varliğinin hibrid yakalama yöntemiyle araştirilmasi [Investigation of human papillomavirus DNA in cervical specimens by hybrid capture assay]. Mikrobiyol. Bul. 2004, 38, 223–232. [Google Scholar]
- Pity, I.S.; Abdo, H.M.; Goreal, A.A. Human Papillomavirus Genotyping among Different Cervical Smears in Duhok/Iraq. Asian Pac. J. Cancer Prev. 2019, 20, 2059. [Google Scholar] [CrossRef]
- Ismail, A.T.; Ahmed, N.Y.; Hameed, A.A. Prevalence of HPV immunostaining in benign, preneoplastic and neoplastic cervical lesions of kurdish women in Erbil City/Kurdistan of Iraq. Am. J. Res. Commun. 2014, 2, 67–74. [Google Scholar]
- Al-Ahdal, M.N.; Al-Arnous, W.K.; Bohol, M.F.; Abuzaid, S.M.; Shoukri, M.; Elrady, K.S.; Firdous, N.; Aliyan, R.; Taseer, R.; A Al-Hazzani, A.; et al. Human papillomaviruses in cervical specimens of women residing in Riyadh, Saudi Arabia: A hospital-based study. J. Infect. Dev. Ctries. 2014, 8, 320–325. [Google Scholar] [CrossRef]
- Al-Awadhi, R.; Chehadeh, W.; Al-Jassar, W.; Al-Harmi, J.; Al-Saleh, E.; Kapila, K. Viral load of human papillomavirus in women with normal and abnormal cervical cytology in Kuwait. J. Infect. Dev. Ctries. 2013, 7, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Pity, I.S.; Shamdeen, M.Y.; Wais, S.A. Follow up of atypical squamous cell Pap smears in Iraqi women. Asian Pac. J. Cancer Prev. 2012, 13, 3455–3460. [Google Scholar] [CrossRef] [PubMed]
- Bitarafan, F.; Hekmat, M.R.; Khodaeian, M.; Razmara, E.; Ashrafganjoei, T.; Gilani, M.M.; Mohit, M.; Aminimoghaddam, S.; Cheraghi, F.; Khalesi, R.; et al. Prevalence and Genotype Distribution of Human Papillomavirus Infection among 12 076 Iranian Women. Int. J. Infect. Dis. 2021, 111, 295–302. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, C.M.; Fregnani, J.H.T.G.; Carvalho, J.P.; Longatto-Filho, A.; Levi, J.E. Human papillomavirus genotypes distribution in 175 invasive cervical cancer cases from Brazil. BMC Cancer 2013, 13, 357. [Google Scholar] [CrossRef]
- Alacam, S.; Bakir, A. Human Papillomavirus Prevalence and Genotype Distribution in Cervical Swab Samples in Istanbul, Turkey. J. Infect. Dev. Ctries. 2021, 15, 1190–1196. [Google Scholar] [CrossRef] [PubMed]
- Faik, A.J.F.; Saber, M.Q.S.; Mohammed, W.J.M.; Ibraheem, B.Z.I.; Lateef, K.R.L.; Hassen, A.S.H.A.S. Genotyping of High-risk Human Papilloma virus (HPV) among Iraqi women in Baghdad by Multiplex PCR. J. Biotechnol. Res. Cent. 2015, 9, 38–45. [Google Scholar] [CrossRef]
- Islami, F.; Ferlay, J.; Lortet-Tieulent, J.; Bray, F.; Jemal, A. International trends in anal cancer incidence rates. Int. J. Epidemiol. 2017, 46, 924–938. [Google Scholar] [CrossRef] [PubMed]
- Fife, K.H.; Cramer, H.M.; Schroeder, J.M.; Brown, D.R. Detection of multiple human papillomavirus types in the lower genital tract correlates with cervical dysplasia. J. Med. Virol. 2001, 64, 550–559. [Google Scholar] [CrossRef] [PubMed]
- An, H.J.; Cho, N.H.; Lee, S.Y.; Kim, I.H.; Lee, C.; Kim, S.J.; Mun, M.S.; Kim, S.H.; Jeong, J.K. Correlation of cervical carcinoma and precancerous lesions with human papillomavirus (HPV) genotypes detected with the HPV DNA chip microarray method. Cancer 2003, 97, 1672–1680. [Google Scholar] [CrossRef]
- Del Río-Ospina, L.; León, S.C.S.-D.; Camargo, M.; Moreno-Pérez, D.A.; Sánchez, R.; Pérez-Prados, A.; Patarroyo, M.E. The DNA load of six high-risk human papillomavirus types and its association with cervical lesions. BMC Cancer 2015, 15, 100. [Google Scholar] [CrossRef] [PubMed]
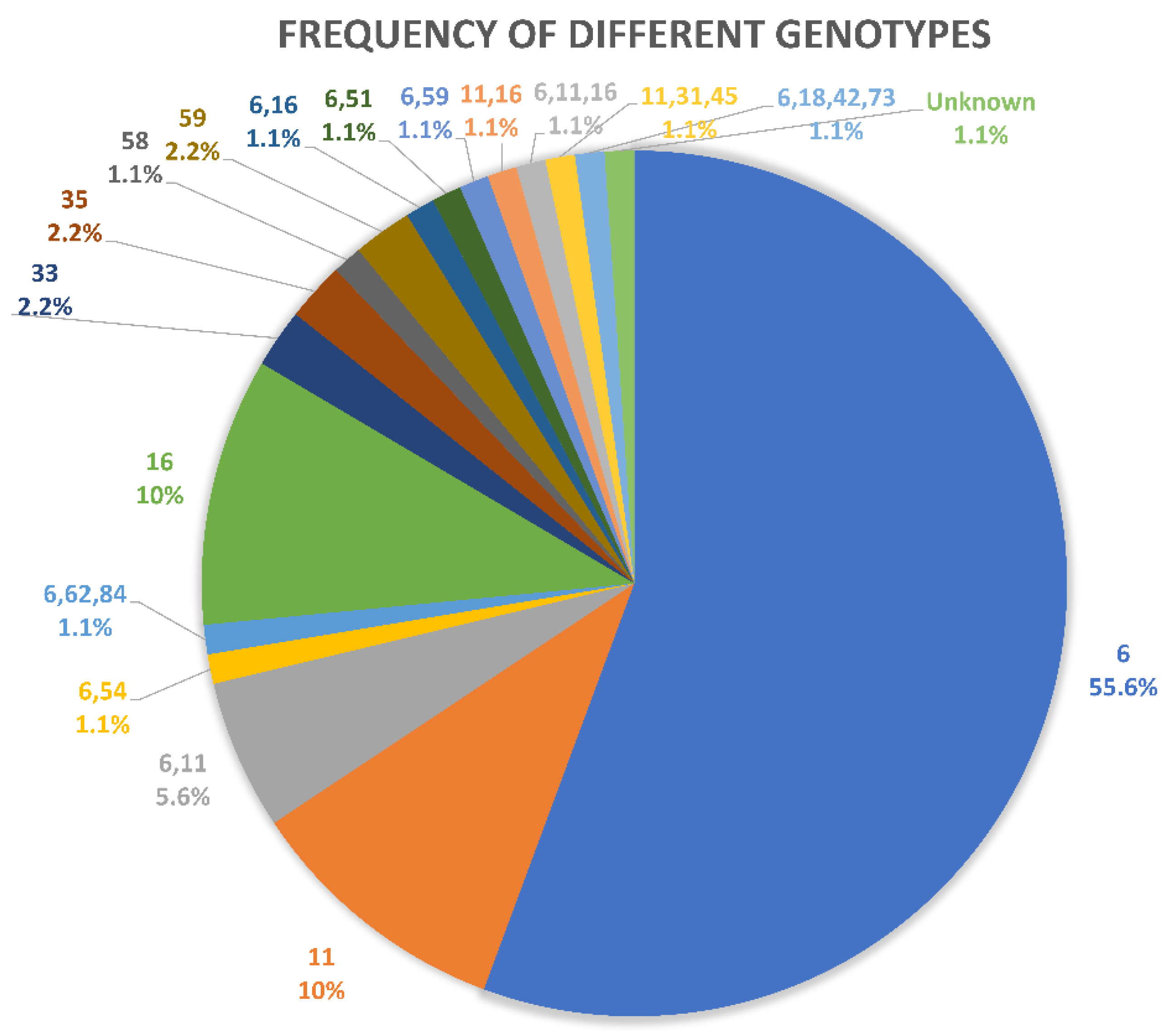
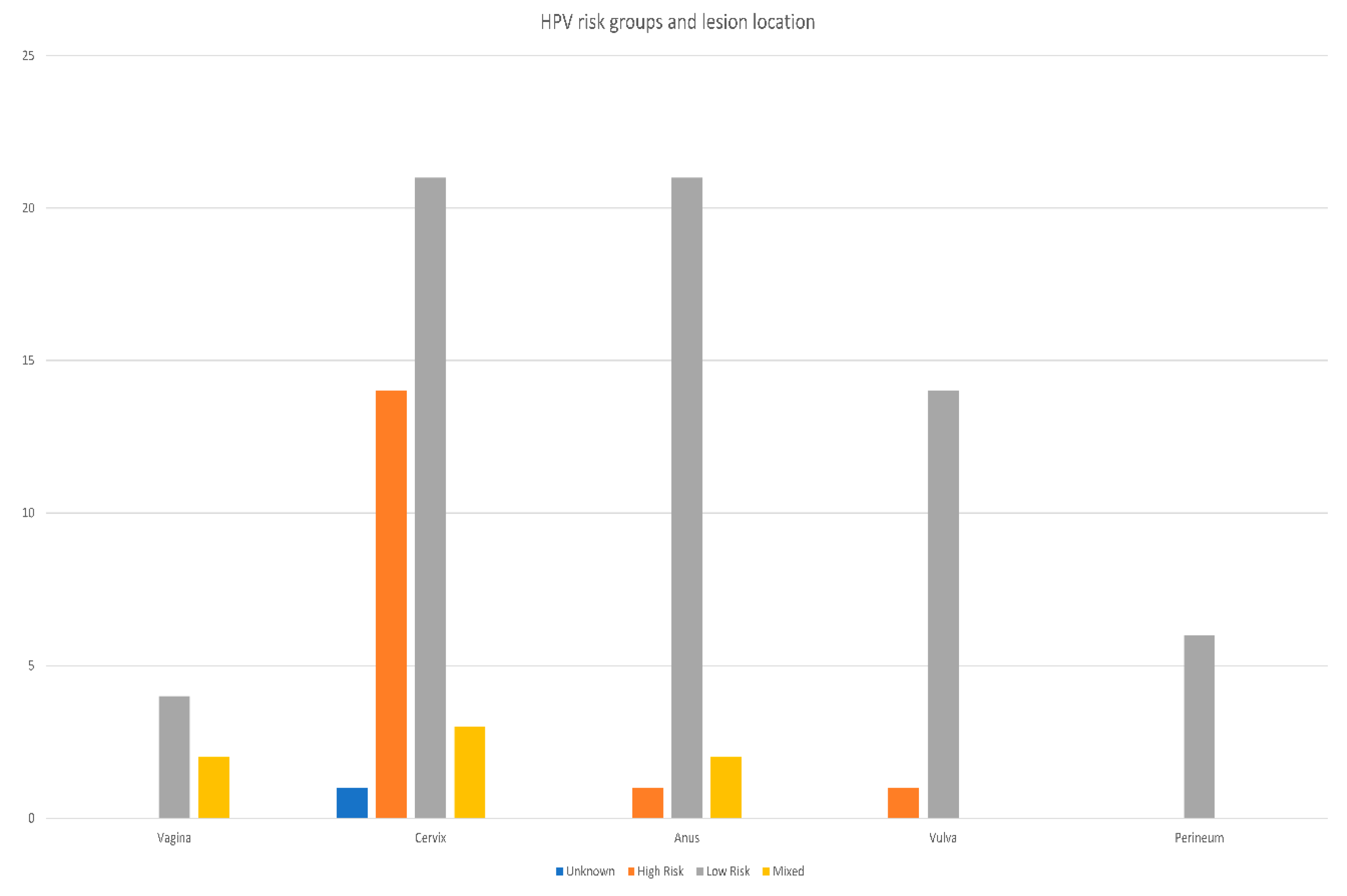
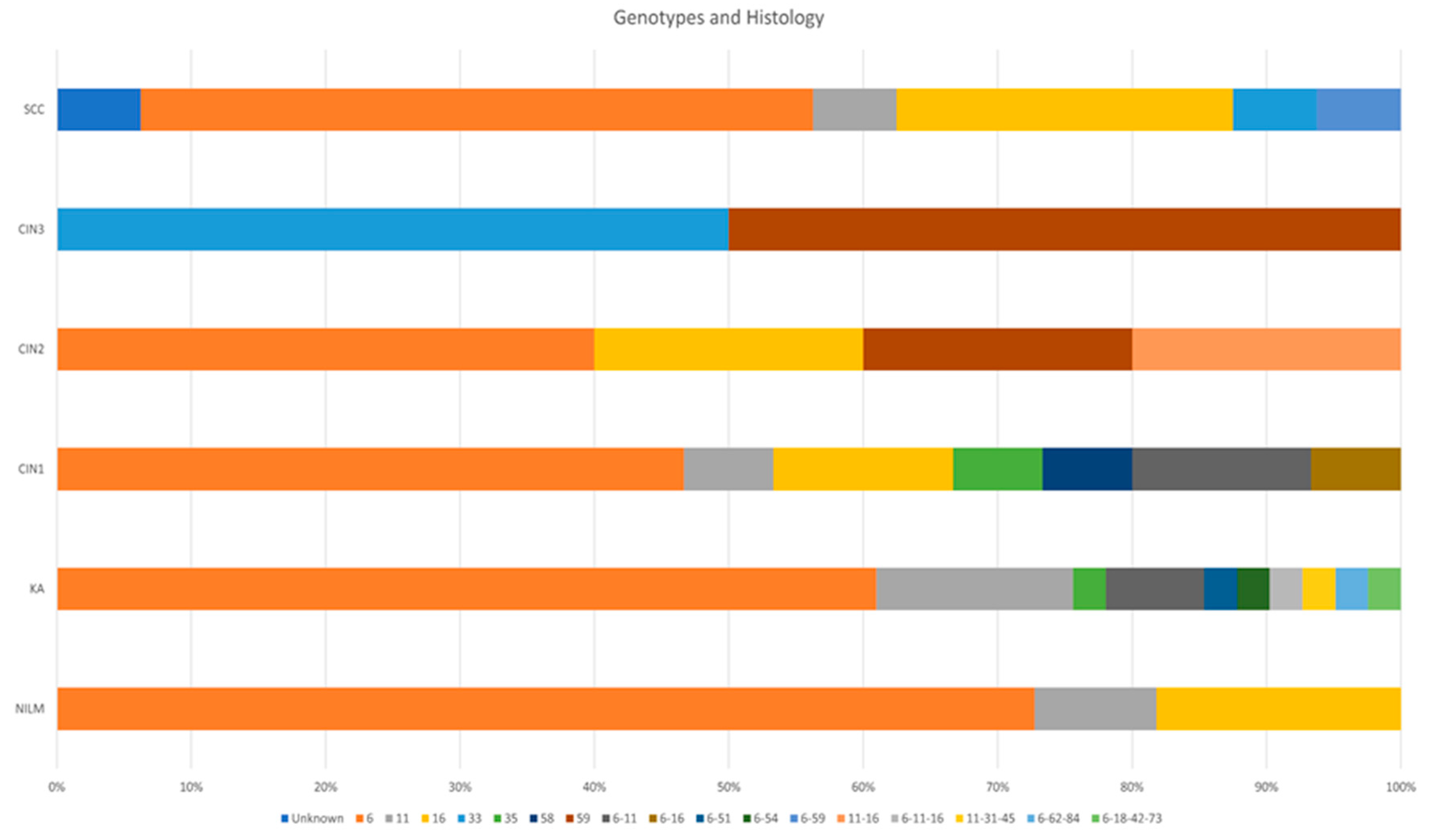
| Result | NILM | KA | CIN1 | CIN2 | CIN3 | SCC | Total | |
|---|---|---|---|---|---|---|---|---|
| Positive | Count | 11 | 41 | 15 | 5 | 2 | 16 | 90 |
| % within Result | 12.2% | 45.6% | 16.7% | 5.6% | 2.2% | 17.8% | 100.0% | |
| % within Histology Diagnosis | 44.0% | 83.7% | 83.3% | 100.0% | 66.7% | 64.0% | 72.0% | |
| % of Total | 8.8% | 32.8% | 12.0% | 4.0% | 1.6% | 12.8% | 72.0% | |
| Negative | Count | 14 | 8 | 3 | 0 | 1 | 9 | 35 |
| % within Result | 40.0% | 22.9% | 8.6% | 0.0% | 2.9% | 25.7% | 100.0% | |
| % within Histology Diagnosis | 56.0% | 16.3% | 16.7% | 0.0% | 33.3% | 36.0% | 28.0% | |
| % of Total | 11.2% | 6.4% | 2.4% | 0.0% | 0.8% | 7.2% | 28.0% | |
| Total | Count | 25 | 49 | 18 | 5 | 3 | 25 | 125 |
| % within Result | 20.0% | 39.2% | 14.4% | 4.0% | 2.4% | 20.0% | 100.0% | |
| % within Histology Diagnosis | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| % of Total | 20.0% | 39.2% | 14.4% | 4.0% | 2.4% | 20.0% | 100.0% | |
| Age Groups | Risk Group | ||||
|---|---|---|---|---|---|
| Unknown | High-Risk | Low-Risk | Mixed | Total | |
| 11–20 | 0 (0.0%) | 0 (0.0%) | 3 (3.3%) | 1 (1.1%) | 4 (4.4%) |
| 21–30 | 0 (0.0%) | 2 (2.2%) | 18 (20.0%) | 2 (2.2%) | 22 (24.4%) |
| 31–40 | 0 (0.0%) | 6 (6.7%) | 18 (20.0%) | 1 (1.1%) | 25 (27.8%) |
| 41–50 | 0 (0.0%) | 6 (6.7%) | 13 (14.4%) | 2 (2.2%) | 21 (23.3%) |
| 51–60 | 1 (1.1%) | 0 (0.0%) | 9 (10.0%) | 0 (0.0%) | 10 (11.1%) |
| 61–70 | 0 (0.0%) | 2 (2.2%) | 3 (3.3%) | 1 (1.1%) | 6 (6.7%) |
| 71–80 | 0 (0.0%) | 0 (0.0%) | 2 (2.2%) | 0 (0.0%) | 2 (2.2%) |
| Total | 1 (1.1%) | 16 (17.8%) | 66 (73.3%) | 7 (7.8%) | 90 (100.0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Othman, A.; Goreal, A.; Pity, I. Molecular Detection of Human Papillomaviruses in Formalin Fixed Paraffin Embedded Sections from Different Anogenital Lesions in Duhok-Iraq. Diagnostics 2022, 12, 2496. https://doi.org/10.3390/diagnostics12102496
Othman A, Goreal A, Pity I. Molecular Detection of Human Papillomaviruses in Formalin Fixed Paraffin Embedded Sections from Different Anogenital Lesions in Duhok-Iraq. Diagnostics. 2022; 12(10):2496. https://doi.org/10.3390/diagnostics12102496
Chicago/Turabian StyleOthman, Adil, Amer Goreal, and Intisar Pity. 2022. "Molecular Detection of Human Papillomaviruses in Formalin Fixed Paraffin Embedded Sections from Different Anogenital Lesions in Duhok-Iraq" Diagnostics 12, no. 10: 2496. https://doi.org/10.3390/diagnostics12102496
APA StyleOthman, A., Goreal, A., & Pity, I. (2022). Molecular Detection of Human Papillomaviruses in Formalin Fixed Paraffin Embedded Sections from Different Anogenital Lesions in Duhok-Iraq. Diagnostics, 12(10), 2496. https://doi.org/10.3390/diagnostics12102496




