Liquid Biopsy: A Multi-Parametric Analysis of Mutation Status, Circulating Tumor Cells and Inflammatory Markers in EGFR-Mutated NSCLC
Abstract
1. Introduction
2. Materials and Methods
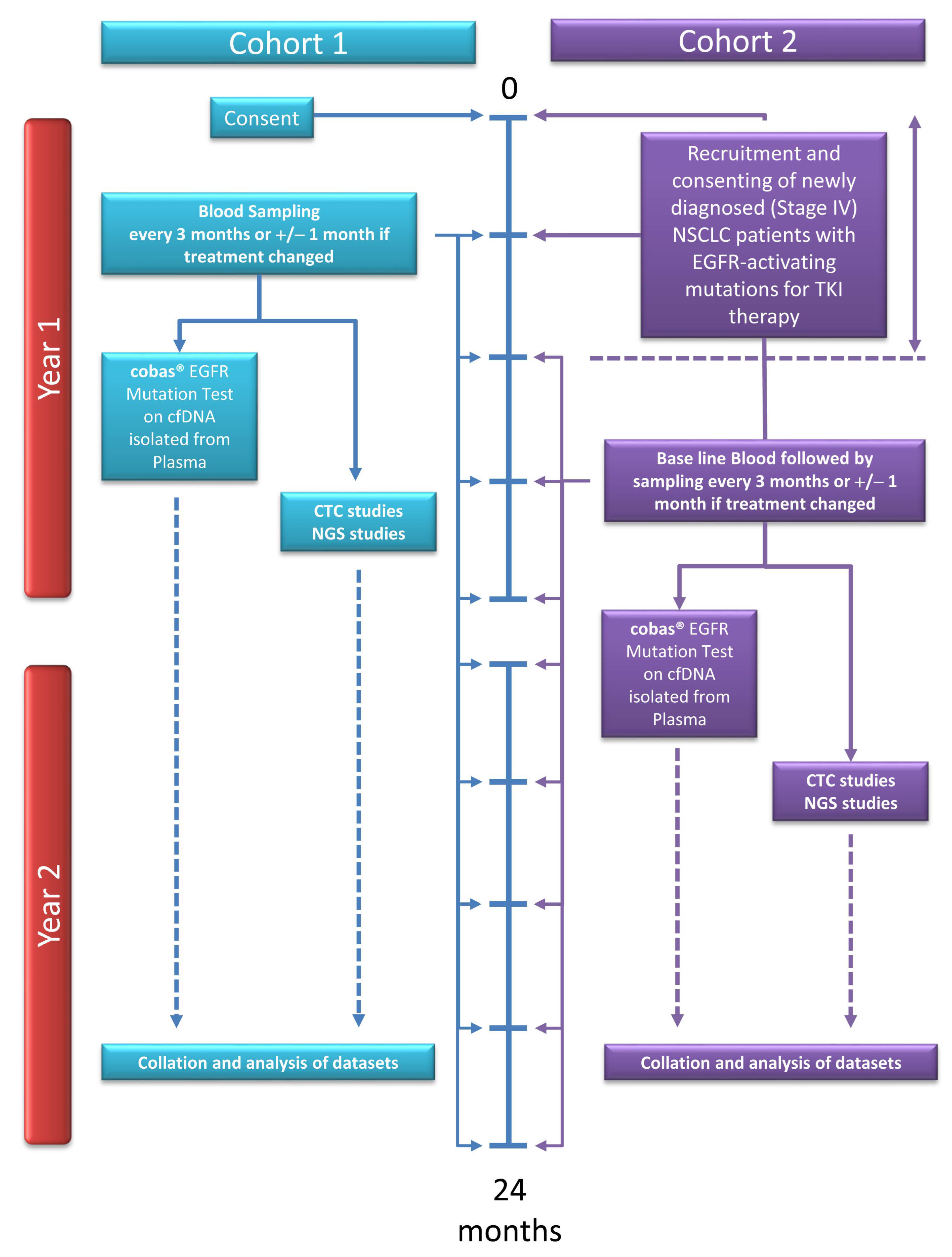
2.1. Patient Recruitment and Sample Collection
2.2. DNA Extraction
2.3. Real-Time RT-PCR
2.4. Circulating Tumor Cell Isolation
2.5. Morphological Staining of ScreenCell® Filters
2.6. AVENIO ctDNA Targeted Panel Sequencing
2.7. Oncomine™ Lung cfDNA Assay
2.8. Inflammatory Marker Analysis
2.9. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Detection of EGFR Mutations in Cell-Free DNA Using Plasma Genotyping
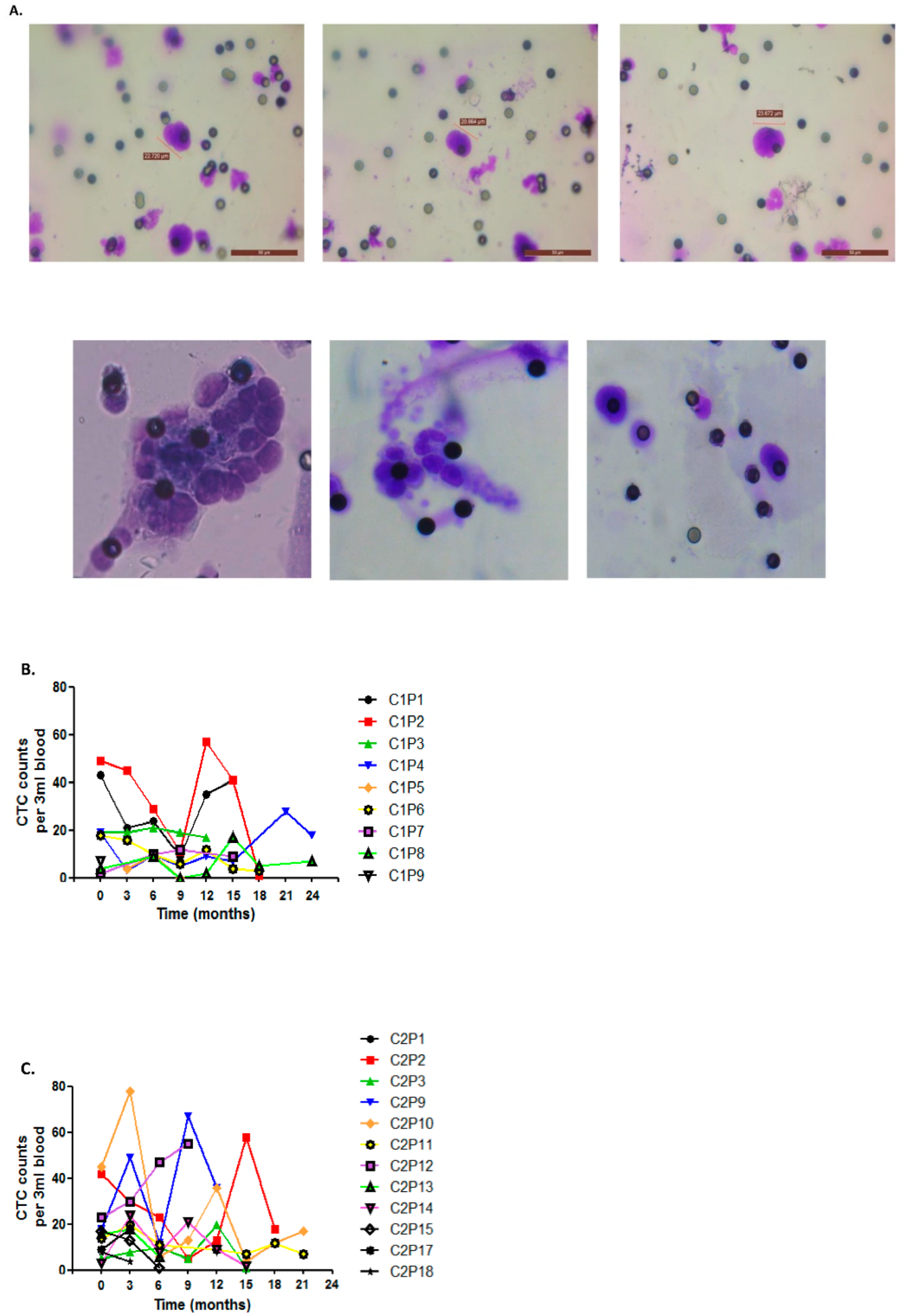
3.3. Circulating Tumor Cells as Markers of Disease Outcome
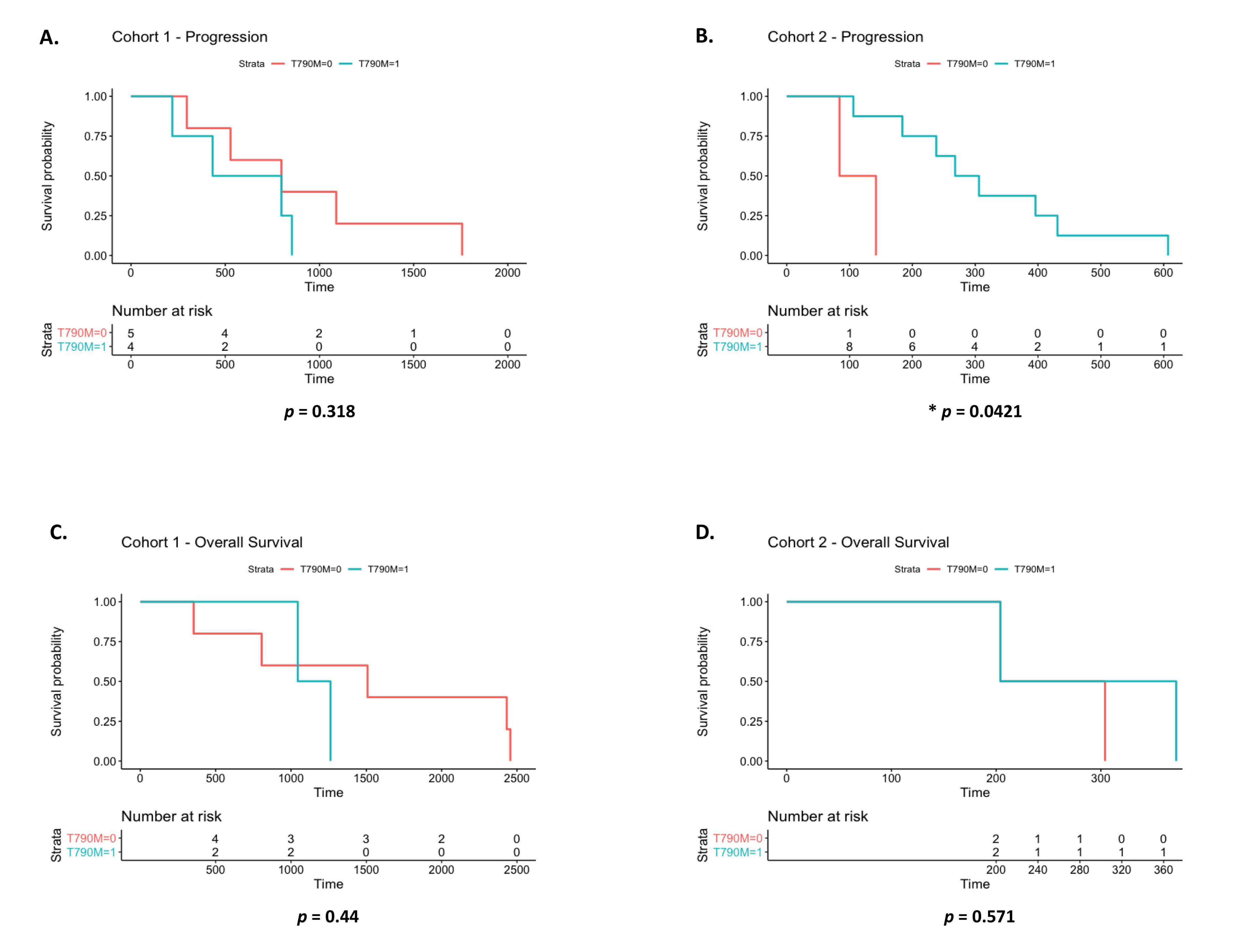
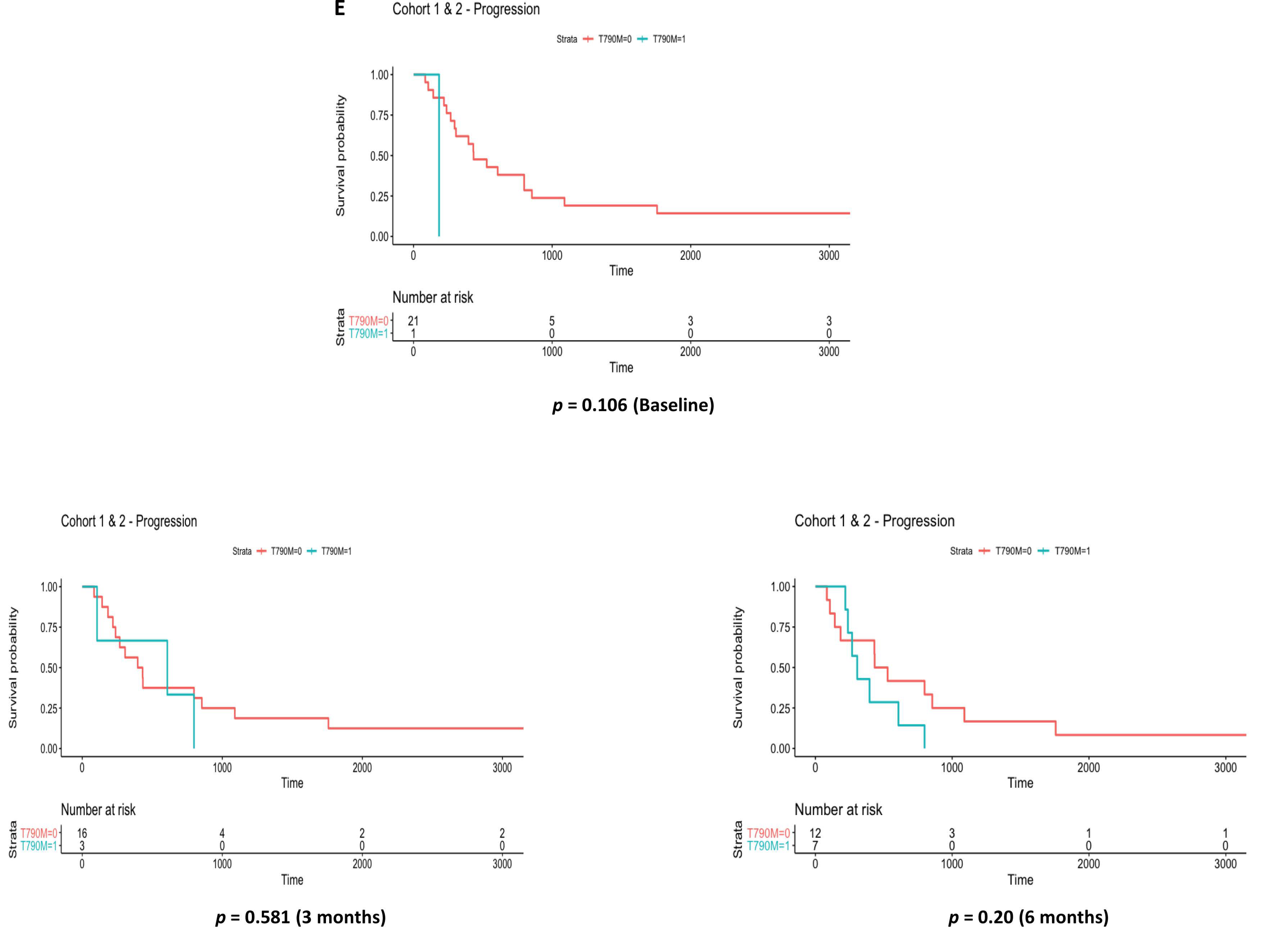
3.4. EGFR Exon 20 p.T790M Detection by Liquid Biopsy vs. Clinical Progression by CT Imaging
3.5. Comparative Assessment of NGS ctDNA Analysis Platforms
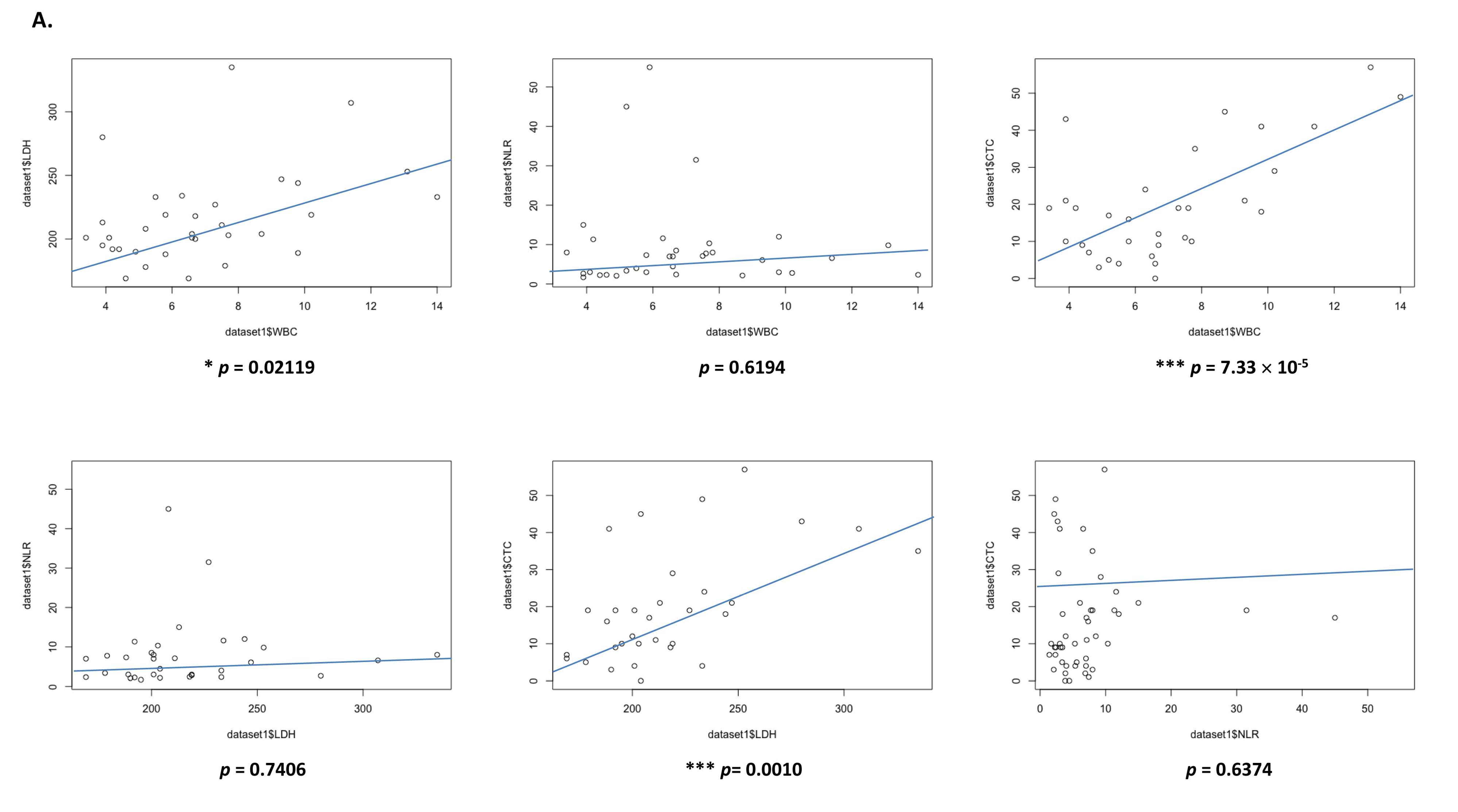
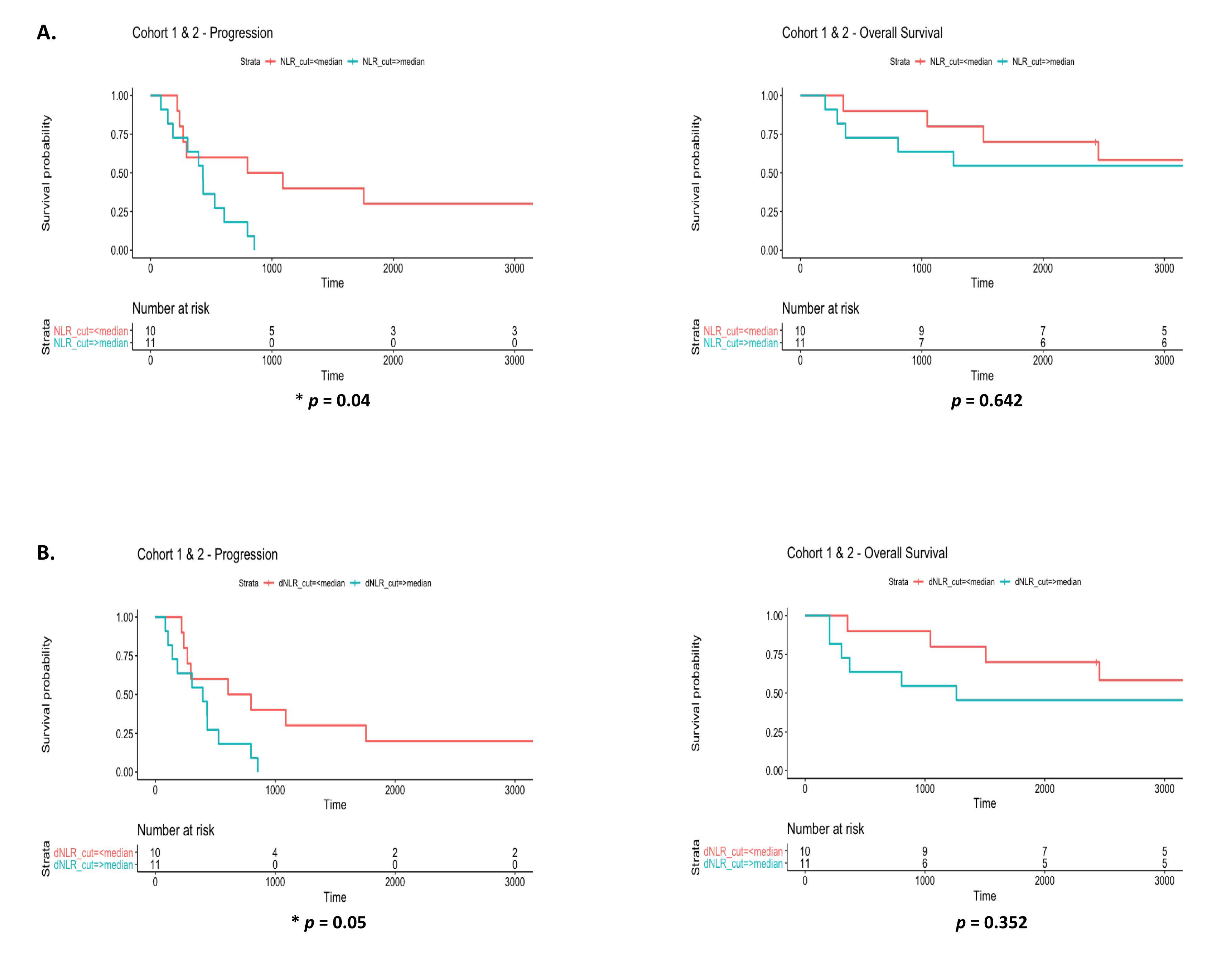
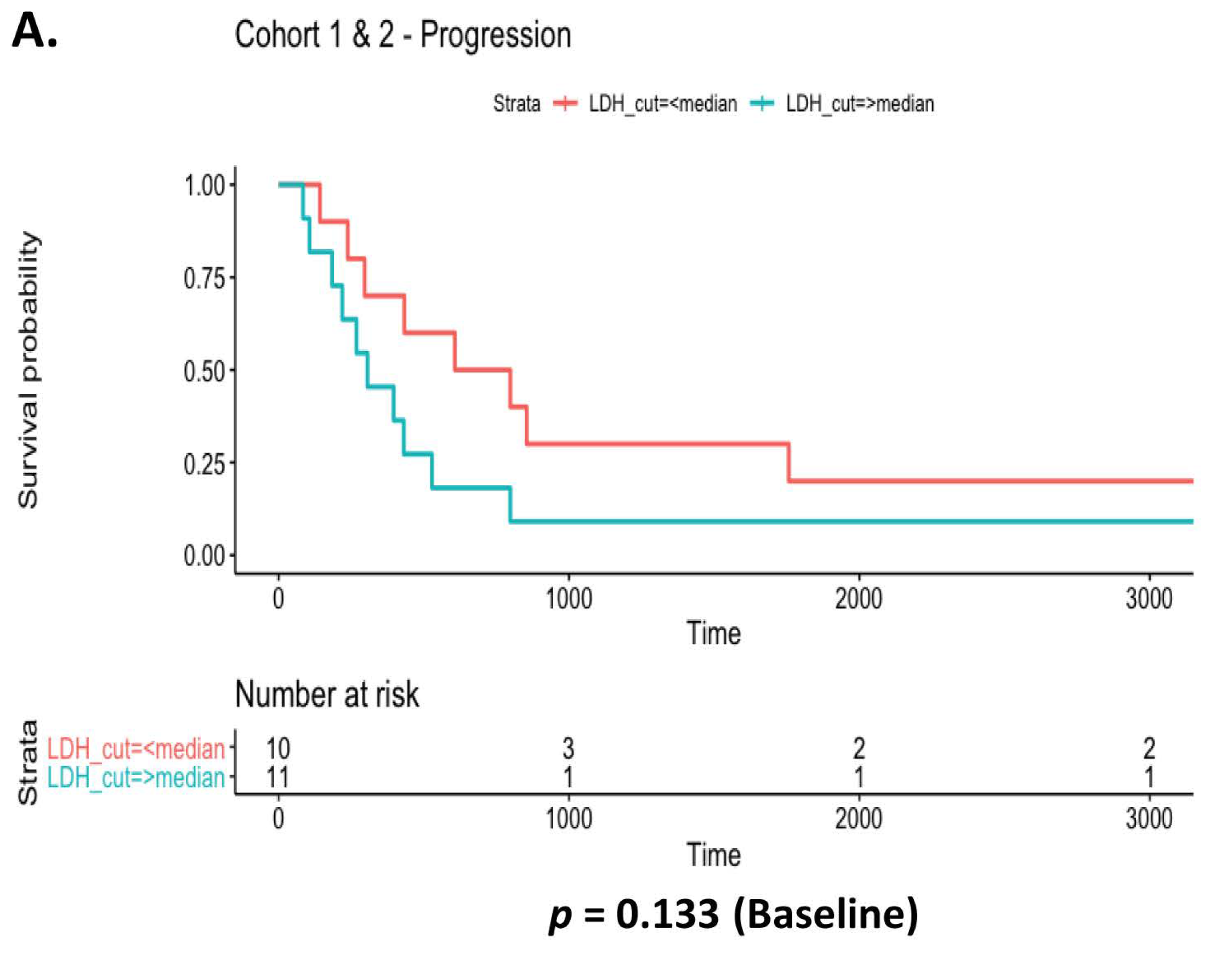
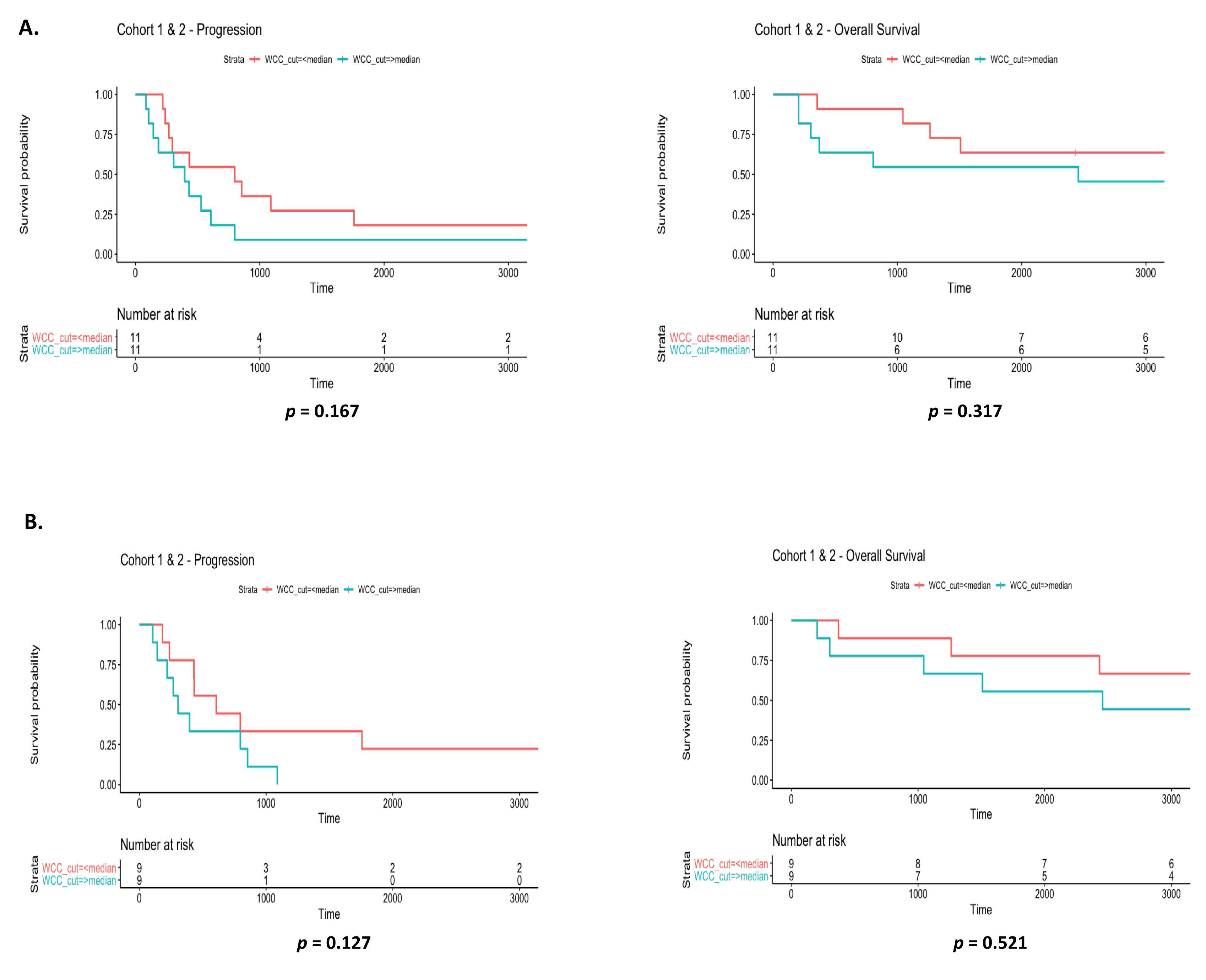
3.6. Prognostic Value of Systemic Inflammatory Markers in EGFR Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kawaguchi, T.; Koh, Y.; Ando, M.; Ito, N.; Takeo, S.; Adachi, H.; Tagawa, T.; Kakegawa, S.; Yamashita, M.; Kataoka, K.; et al. Prospective Analysis of Oncogenic Driver Mutations and Environmental Factors: Japan Molecular Epidemiology for Lung Cancer Study. J. Clin. Oncol. 2016, 34, 2247–2257. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Au, J.S.-K.; Thongprasert, S.; Srinivasan, S.; Tsai, C.-M.; Khoa, M.T.; Heeroma, K.; Itoh, Y.; Cornelio, G.; Yang, P.-C. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small cell lung cancer of adenocarcinoma histology (PIONEER). J. Thorac. Oncol. 2014, 9, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef] [PubMed]
- Paez, J.G.; Jänne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef]
- Pao, W.; Miller, V.; Zakowski, M.; Doherty, J.; Politi, K.; Sarkaria, I.; Singh, B.; Heelan, R.; Rusch, V.; Fulton, L.; et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. USA 2004, 101, 13306–13311. [Google Scholar] [CrossRef]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Mol. Diagn. 2018, 20, 129–159. [Google Scholar] [CrossRef]
- Ciardiello, F.; Tortora, G. EGFR antagonists in cancer treatment. N. Engl. J. Med. 2008, 358, 1160–1174. [Google Scholar] [CrossRef]
- Shigematsu, H.; Gazdar, A.F. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int. J. Cancer. 2006, 118, 257–262. [Google Scholar] [CrossRef]
- Yang, J.C.-H.; Schuler, M.; Popat, S.; Miura, S.; Heeke, S.; Park, K.; Märten, A.; Kim, E.S. Afatinib for the Treatment of NSCLC Harboring Uncommon EGFR Mutations: A Database of 693 Cases. J. Thorac. Oncol. 2020, 15, 803–815. [Google Scholar] [CrossRef]
- Zhang, Q.; Cui, Y.; Zhang, J.; Kenjiabieke, J.; Aerxiding, P. Comparison of the characteristics of uncommon epidermal growth factor receptor (EGFR) mutations and EGFR-tyrosine kinase inhibitor treatment in patients with non-small cell lung cancer from different ethnic groups. Exp. Ther. Med. 2020, 19, 3513–3520. [Google Scholar] [CrossRef]
- Watanabe, S.; Minegishi, Y.; Yoshizawa, H.; Maemondo, M.; Inoue, A.; Sugawara, S.; Isobe, H.; Harada, M.; Ishii, Y.; Gemma, A.; et al. Effectiveness of gefitinib against non-small-cell lung cancer with the uncommon EGFR mutations G719X and L861Q. J. Thorac. Oncol. 2014, 9, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Planchard, D.; Popat, S.; Kerr, K.; Novello, S.; Smit, E.F.; Faivre-Finn, C.; Mok, T.S.; Reck, M.; Van Schil, P.E.; Hellmann, M.D.; et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29 (Suppl. 4), iv192–iv237, reprint in Ann. Oncol. 2019, 30, 863–870. [Google Scholar] [CrossRef]
- Kobayashi, S.; Boggon, T.J.; Dayaram, T.; Jänne, P.A.; Kocher, O.; Meyerson, M.; Johnson, B.E.; Eck, M.J.; Tenen, D.G.; Halmos, B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005, 352, 786–792. [Google Scholar] [CrossRef] [PubMed]
- Arcila, M.E.; Oxnard, G.R.; Nafa, K.; Riely, G.J.; Solomon, S.B.; Zakowski, M.F.; Kris, M.G.; Pao, W.; Miller, V.A.; Ladanyi, M. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin. Cancer Res. 2011, 17, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Kim, L.; Tsao, M.S. Tumour tissue sampling for lung cancer management in the era of personalised therapy: What is good enough for molecular testing? Eur. Respir. J. 2014, 44, 1011–1022. [Google Scholar] [CrossRef]
- Chougule, A.; Basak, S. Epidermal growth factor receptor T790M testing in progressed lung cancer: A review of sensitive methods for analysis of tissue and liquid biopsy samples. Indian J. Cancer 2017, 54, S45–S54. [Google Scholar] [CrossRef] [PubMed]
- Dearden, S.; Brown, H.; Jenkins, S.; Thress, K.S.; Cantarini, M.; Cole, R.; Ranson, M.; Jänne, P.A. EGFR T790M mutation testing within the osimertinib AURA Phase I study. Lung Cancer 2017, 109, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-L.; Chen, Y.-L.; Yang, S.-C.; Ho, C.-L.; Wei, F.; Wong, D.T.; Su, W.-C.; Lin, C.-C. Liquid biopsy genotyping in lung cancer: Ready for clinical utility? Oncotarget 2017, 8, 18590–18608. [Google Scholar] [CrossRef]
- Jenkins, S.; Yang, J.C.-H.; Ramalingam, S.S.; Yu, K.; Patel, S.; Weston, S.; Hodge, R.; Cantarini, M.; Jänne, P.A.; Mitsudomi, T.; et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 1061–1070. [Google Scholar] [CrossRef]
- Wang, W.; Song, Z.; Zhang, Y. A Comparison of ddPCR and ARMS for detecting EGFR T790M status in ctDNA from advanced NSCLC patients with acquired EGFR-TKI resistance. Cancer Med. 2017, 6, 154–162. [Google Scholar] [CrossRef]
- Wu, Y.L.; Zhou, C.; Liam, C.K.; Wu, G.; Liu, X.; Zhong, Z.; Zuo, Y. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: Analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol. 2015, 26, 1883–1889. [Google Scholar] [CrossRef] [PubMed]
- Goss, G.; Tsai, C.-M.; A Shepherd, F.; Bazhenova, L.; Lee, J.S.; Chang, G.-C.; Crino, L.; Satouchi, M.; Chu, Q.; Hida, T.; et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): A multicentre, open-label, single-arm, phase 2 study. Lancet Oncol. 2016, 17, 1643–1652. [Google Scholar] [CrossRef]
- Yousefi, M.; Ghaffari, P.; Nosrati, R.; Dehghani, S.; Salmaninejad, A.; Abarghan, Y.J.; Ghaffari, S.H. Prognostic and therapeutic significance of circulating tumor cells in patients with lung cancer. Cell Oncol. 2020, 43, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Simonaggio, A.; ElAidi, R.; Fournier, L.; Fabre, E.; Ferrari, V.; Borchiellini, D.; Thouvenin, J.; Barthelemy, P.; Thibault, C.; Tartour, E.; et al. Variation in neutrophil to lymphocyte ratio (NLR) as predictor of outcomes in metastatic renal cell carcinoma (mRCC) and non-small cell lung cancer (mNSCLC) patients treated with nivolumab. Cancer Immunol. Immunother. 2020, 69, 2513–2522. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, X.; Shen, Y.; Cao, Y.; Fang, X.; Chen, J.; Yuan, Y. A new prognostic score based on the systemic inflammatory response in patients with inoperable non-small-cell lung cancer. Onco. Targets Ther. 2016, 9, 4879–4886. [Google Scholar] [CrossRef]
- McMillan, D.C. The systemic inflammation-based Glasgow Prognostic Score: A decade of experience in patients with cancer. Cancer Treat Rev. 2013, 39, 534–540. [Google Scholar] [CrossRef]
- Hayes, B.; Brady, L.; Sheill, G.; Baird, A.-M.; Guinan, E.; Stanfill, B.; Dunne, J.; Holden, D.; Vlajnic, T.; Casey, O.; et al. Circulating Tumour Cell Numbers Correlate with Platelet Count and Circulating Lymphocyte Subsets in Men with Advanced Prostate Cancer: Data from the ExPeCT Clinical Trial (CTRIAL-IE 15-21). Cancers. 2021, 13, 4690. [Google Scholar] [CrossRef]
- Templeton, A.J.; Mcnamara, M.G.; Šeruga, B.; Vera-Badillo, F.E.; Aneja, P.; Ocaña, A.; Leibowitz-Amit, R.; Sonpavde, G.; Knox, J.J.; Tran, B.; et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: A systematic review and meta-analysis. J. Natl. Cancer Inst. 2014, 106, dju124. [Google Scholar] [CrossRef]
- Mezquita, L.; Auclin, E.; Ferrara, R.; Charrier, M.; Remon, J.; Planchard, D.; Ponce, S.; Ares, L.P.; Leroy, L.; Audigier-Valette, C.; et al. Association of the Lung Immune Prognostic Index with Immune Checkpoint Inhibitor Outcomes in Patients with Advanced Non-Small Cell Lung Cancer. JAMA Oncol. 2018, 4, 351–357. [Google Scholar] [CrossRef]
- Ferrucci, P.F.; Ascierto, P.A.; Pigozzo, J.; Del Vecchio, M.; Maio, M.; Cappellini, G.C.A.; Guidoboni, M.; Queirolo, P.; Savoia, P.; Mandalà, M.; et al. Baseline neutrophils and derived neutrophil-to-lymphocyte ratio: Prognostic relevance in metastatic melanoma patients receiving ipilimumab. Ann. Oncol. 2016, 27, 732–738. [Google Scholar] [CrossRef]
- Alama, A.; Truini, A.; Coco, S.; Genova, C.; Grossi, F. Prognostic and predictive relevance of circulating tumor cells in patients with non-small-cell lung cancer. Drug Discov. Today 2014, 19, 1671–1676. [Google Scholar] [CrossRef] [PubMed]
- Kowalik, A.; Kowalewska, M.; Góźdź, S. Current approaches for avoiding the limitations of circulating tumor cells detection methods-implications for diagnosis and treatment of patients with solid tumors. Transl. Res. 2017, 185, 58–84.e15. [Google Scholar] [CrossRef] [PubMed]
- Yucel, S.; Bilgin, B. The prognostic values of systemic immune-inflammation index and derived neutrophil-lymphocyte ratio in EGFR-mutant advanced non-small cell lung cancer. J. Oncol. Pharm. Pract. 2021, 27, 71–77. [Google Scholar] [CrossRef]
- Douillard, J.-Y.; Ostoros, G.; Cobo, M.; Ciuleanu, T.; Cole, R.; McWalter, G.; Walker, J.; Dearden, S.; Webster, A.; Milenkova, T.; et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: Circulating-free tumor DNA as a surrogate for determination of EGFR status. J. Thorac. Oncol. 2014, 9, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Lee, V.; Liam, C.-K.; Lu, S.; Park, K.; Srimuninnimit, V.; Wang, J.; Zhou, C.; Appius, A.; Button, P.; et al. ENSURE FASTACT-2 and ASPIRATION Investigators: Clinical utility of a blood-based EGFR mutation test in patients receiving first-line erlotinib therapy in the ENSURE, FASTACT-2, and ASPIRATION studies. Lung Cancer 2018, 126, 1–8. [Google Scholar] [CrossRef]
- Jenkins, S.; Yang, J.C.-H.; Jänne, P.A.; Thress, K.S.; Yu, K.; Hodge, R.; Weston, S.; Dearden, S.; Patel, S.; Cantarini, M.; et al. EGFR Mutation Analysis for Prospective Patient Selection in Two Phase II Registration Studies of Osimertinib. J. Thorac. Oncol. 2017, 12, 1247–1256. [Google Scholar] [CrossRef]
- Wu, S.-G.; Liu, Y.-N.; Tsai, M.-F.; Chang, Y.-L.; Yu, C.-J.; Yang, P.-C.; Yang, J.C.-H.; Wen, Y.-F.; Shih, J.-Y. The mechanism of acquired resistance to irreversible EGFR tyrosine kinase inhibitor-afatinib in lung adenocarcinoma patients. Oncotarget 2016, 7, 12404–12413. [Google Scholar] [CrossRef]
- Liang, H.; Pan, Z.; Wang, W.; Guo, C.; Chen, D.; Zhang, J.; Zhang, Y.; Tang, S.; He, J.; Liang, W.; et al. The alteration of T790M between 19 del and L858R in NSCLC in the course of EGFR-TKIs therapy: A literature-based pooled analysis. J. Thorac. Dis. 2018, 10, 2311–2320. [Google Scholar] [CrossRef]
- Del Re, M.; Petrini, I.; Mazzoni, F.; Valleggi, S.; Gianfilippo, G.; Pozzessere, D.; Chella, A.; Crucitta, S.; Rofi, E.; Restante, G.; et al. Incidence of T790M in Patients With NSCLC Progressed to Gefitinib, Erlotinib, and Afatinib: A Study on Circulating Cell-free DNA. Clin. Lung Cancer 2020, 21, 232–237. [Google Scholar] [CrossRef]
- Otsuka, T.; Mori, M.; Yano, Y.; Uchida, J.; Nishino, K.; Kaji, R.; Hata, A.; Hattori, Y.; Urata, Y.; Kaneda, T.; et al. Effectiveness of Tyrosine Kinase Inhibitors in Japanese Patients with Non-small Cell Lung Cancer Harboring Minor Epidermal Growth Factor Receptor Mutations: Results from a Multicenter Retrospective Study (HANSHIN Oncology Group 0212). Anticancer Res. 2015, 35, 3885–3891. [Google Scholar]
- Kancha, R.K.; Peschel, C.; Duyster, J. The epidermal growth factor receptor-L861Q mutation increases kinase activity without leading to enhanced sensitivity toward epidermal growth factor receptor kinase inhibitors. J. Thorac. Oncol. 2011, 6, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.; Sequist, L.V.; Geater, S.L.; Tsai, C.-M.; Mok, T.S.K.; Schuler, M.; Yamamoto, N.; Yu, C.-J.; I Ou, S.-H.; Zhou, C.; et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: A combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015, 16, 830–838. [Google Scholar] [CrossRef]
- Costa, C.; Muinelo-Romay, L.; Cebey-López, V.; Pereira-Veiga, T.; Martínez-Pena, I.; Abreu, M.; Abalo, A.; Lago-Lestón, R.M.; Abuín, C.; Palacios, P.; et al. Analysis of a Real-World Cohort of Metastatic Breast Cancer Patients Shows Circulating Tumor Cell Clusters (CTC-clusters) as Predictors of Patient Outcomes. Cancers 2020, 29, 1111. [Google Scholar] [CrossRef] [PubMed]
- Pernot, S.; Badoual, C.; Terme, M.; Castan, F.; Cazes, A.; Bouche, O.; Bennouna, J.; Francois, E.; Ghiringhelli, F.; De La Fouchardiere, C.; et al. Dynamic evaluation of circulating tumour cells in patients with advanced gastric and oesogastric junction adenocarcinoma: Prognostic value and early assessment of therapeutic effects. Eur. J. Cancer. 2017, 79, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Khattak, M.A.; Reid, A.; Freeman, J.; Pereira, M.; McEvoy, A.; Lo, J.; Frank, M.H.; Meniawy, T.; Didan, A.; Spencer, I.; et al. PD-L1 Expression on Circulating Tumor Cells May Be Predictive of Response to Pembrolizumab in Advanced Melanoma: Results from a Pilot Study. Oncologist 2019, 25, e520–e527. [Google Scholar] [CrossRef]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.-M.; Greystoke, A.; Ward, T.H.; Ferraldeschi, R.; Hughes, A.; Clack, G.; et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 2011, 29, 1556–1563. [Google Scholar] [CrossRef]
- Jiang, T.; Zhao, J.; Zhao, C.; Li, X.; Shen, J.; Zhou, J.; Ren, S.; Su, C.; Zhou, C.; O’Brien, M. Dynamic Monitoring and Predictive Value of Circulating Tumor Cells in EGFR-Mutated Advanced Non-Small-Cell Lung Cancer Patients Treated with First-Line EGFR Tyrosine Kinase Inhibitors. Clin. Lung Cancer. 2019, 20, 124–133.e2. [Google Scholar] [CrossRef]
- Wu, Z.-X.; Liu, Z.; Jiang, H.-L.; Pan, H.-M.; Han, W.-D. Circulating tumor cells predict survival benefit from chemotherapy in patients with lung cancer. Oncotarget 2016, 7, 67586–67596. [Google Scholar] [CrossRef]
- He, W.; Li, W.; Jiang, B.; Chang, L.; Jin, C.; Tu, C.; Li, Y. Correlation between epidermal growth factor receptor tyrosine kinase inhibitor efficacy and circulating tumor cell levels in patients with advanced non-small cell lung cancer. Onco. Targets Ther. 2016, 9, 7515–7520. [Google Scholar] [CrossRef]
- Sabour, L.; Sabour, M.; Ghorbian, S. Clinical Applications of Next-Generation Sequencing in Cancer Diagnosis. Pathol. Oncol. Res. 2017, 23, 225–234. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Esagian, S.M.; Grigoriadou, G.I.; Nikas, I.P.; Boikou, V.; Sadow, P.M.; Won, J.-K.; Economopoulos, K.P. Comparison of liquid-based to tissue-based biopsy analysis by targeted next generation sequencing in advanced non-small cell lung cancer: A comprehensive systematic review. J. Cancer Res. Clin. Oncol. 2020, 146, 2051–2066. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.; Meldgaard, P.; Hager, H.; Wu, L.; Wei, W.; Tsai, J.; Khalil, A.; Nexo, E.; Sorensen, B.S. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 2014, 14, 294. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Ye, X.; Dong, Z.; Lu, Y.C.; Sun, Y.; Liu, Y.; McCormack, R.; Gu, Y.; Liu, X. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell-Free DNA from Patients with Advanced Non-Small Cell Lung Cancer. J. Mol. Diagn. 2015, 17, 265–272. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Paramanathan, A.; Saxena, A.; Morris, D.L. A systematic review and meta-analysis on the impact of pre-operative neutrophil lymphocyte ratio on long term outcomes after curative intent resection of solid tumours. Surg. Oncol. 2014, 23, 31–39. [Google Scholar] [CrossRef]
- Song, Y.-J.; Wang, L.-X.; Hong, Y.-Q.; Lu, Z.-H.; Tong, Q.; Fang, X.-Z.; Tan, J. Lymphocyte to monocyte ratio is associated with response to first-line platinum-based chemotherapy and prognosis of early-stage non-small cell lung cancer patients. Tumour Biol. 2016, 37, 5285–5293. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Lai, C.-H.; Chang, H.-C.; Chao, T.-Y.; Tseng, C.-C.; Fang, W.-F.; Wang, C.-C.; Chung, Y.-H.; Wang, Y.-H.; Su, M.-C.; et al. Baseline and Trend of Lymphocyte-to-Monocyte Ratio as Prognostic Factors in Epidermal Growth Factor Receptor Mutant Non-Small Cell Lung Cancer Patients Treated with First-Line Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors. PLoS ONE 2015, 10, e0136252. [Google Scholar] [CrossRef]
- Minami, S.; Ihara, S.; Komuta, K. Pretreatment Lung Immune Prognostic Index Is a Prognostic Marker of Chemotherapy and Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor. World J. Oncol. 2019, 10, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Inomata, M.; Hayashi, R.; Tanaka, H.; Shimokawa, K.; Tokui, K.; Taka, C.; Okazawa, S.; Kambara, K.; Ichikawa, T.; Yamada, T.; et al. Elevated levels of plasma lactate dehydrogenase is an unfavorable prognostic factor in patients with epidermal growth factor receptor mutation-positive non-small cell lung cancer, receiving treatment with gefitinib or erlotinib. Mol. Clin. Oncol. 2016, 4, 774–778. [Google Scholar] [CrossRef] [PubMed]
- Fiala, O.; Pesek, M.; Finek, J.; Topolcan, O.; Racek, J.; Svatoň, M.; Kucera, R.; Minarik, M.; Benesova, L.; Bortlicek, Z.; et al. Change in Serum Lactate Dehydrogenase Is Associated with Outcome of Patients with Advanced-stage NSCLC Treated with Erlotinib. Anticancer Res. 2016, 36, 2459–2465. [Google Scholar] [PubMed]
- Diem, S.; Kasenda, B.; Spain, L.; Martin-Liberal, J.; Marconcini, R.; Gore, M.; Larkin, J. Serum lactate dehydrogenase as an early marker for outcome in patients treated with anti-PD-1 therapy in metastatic melanoma. Br. J. Cancer 2016, 114, 256–261. [Google Scholar] [CrossRef] [PubMed]








| Patient Baseline Characteristics | ||
|---|---|---|
| Characteristics | Number of Patients | Percentage (%) |
| Age, years | ||
| Median (Range) | 66 (40−85) | |
| Gender | ||
| Male | 5 | 23.81 |
| Female | 16 | 76.19 |
| Smoking status | ||
| Current | 0 | 0 |
| Former smokers | 10 | 47.62 |
| Never smokers | 11 | 52.38 |
| Histology | ||
| Adenocarcinoma | 20 | 95.24 |
| Squamous cell carcinoma | 1 | 4.76 |
| Alive or Dead | ||
| Alive | 10 | 47.62 |
| Dead | 11 | 52.38 |
| ECOG status | ||
| 0 | 3 | 14.29 |
| 1 | 12 | 57.14 |
| 2 | 5 | 23.81 |
| 3 | 1 | 4.76 |
| Previous lines of treatment | ||
| 0 | 5 | 23.81 |
| 1 | 8 | 38.10 |
| 2 | 5 | 23.81 |
| 3 | 1 | 4.76 |
| 4 | 2 | 9.52 |
| Study ID | Time Difference (Days) between T790M Detection & Disease Progression |
|---|---|
| C1P1-02 | 545 |
| C1P3-01 | −16 |
| C1P4-04 | 97 |
| C1P8-05 | −177 |
| C2P2-02 | −66 |
| C2P3-02 | −201 |
| C2P9-00 | −203 |
| C2P10-05 | 40 |
| C2P11-02 | −33 |
| C2P13-01 | 4 |
| C2P14-02 | −153 |
| C2P15-01 | −481 |
 |  | |||
|---|---|---|---|---|
| Study ID | Tissue Biopsyy (NGS) | Liquid Biopsyy (Oncomine™ NGS) | Liquid Biopsyy (cobas®) | |
| C1P6-00-E | Ex19del | Ex19del | Ex19del | |
| C1P6-00-A48 | Ex19del | NMD | Ex19del | |
| C1P6-00-A72 | Ex19del | Ex19del | Ex19del | |
| C1P2-00-E | Ex19del | NMD | NMD | |
| C1P2-00-A48 | Ex19del | NMD | Ex19del | |
| C1P2-00-A72 | Ex19del | NMD | NMD | |
| C2P2-02-A48 | Ex19del | Ex19del, T790M | Ex19del, T790M | |
| C2P13-01-A72 | Ex19del | Ex19del | Ex19del, T790M | |
| C2P2090662-E | Ex19del | Ex19del | Ex19del, T790M | |
| C2P12-00-A48 | L858R | L858R | L858R | |
| C2P9-00-A48 | L858R | NMD* | L858R, T790M | |
| C1P7-00-A48 | L858R | NMD* | NMD | |
 | |||
|---|---|---|---|
| Study ID | Tissue Biopsy (NGS) | Liquid Biopsyy (Avenio NGS) | Liquid Biopsy (cobas®) |
| C1P9-00-E | Ex19del | Ex19del | L861Q |
| C1P3-04-A48 | Ex19del | Ex19del | Ex19del |
| C1P4-04-E | Ex19del | Ex19del, T790M | Ex19del, T790M |
| C2P9-03-E | L858R | L858R | L858R |
| C1P1-04-E | Ex19del | Ex19del, T790M | Ex19del, T790M |
| C1P6-04-A48 | Ex19del | Ex19del | NMD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barr, M.P.; Baird, A.-M.; Halliday, S.; Martin, P.; Allott, E.H.; Phelan, J.; Korpanty, G.; Coate, L.; O’Brien, C.; Gray, S.G.; et al. Liquid Biopsy: A Multi-Parametric Analysis of Mutation Status, Circulating Tumor Cells and Inflammatory Markers in EGFR-Mutated NSCLC. Diagnostics 2022, 12, 2360. https://doi.org/10.3390/diagnostics12102360
Barr MP, Baird A-M, Halliday S, Martin P, Allott EH, Phelan J, Korpanty G, Coate L, O’Brien C, Gray SG, et al. Liquid Biopsy: A Multi-Parametric Analysis of Mutation Status, Circulating Tumor Cells and Inflammatory Markers in EGFR-Mutated NSCLC. Diagnostics. 2022; 12(10):2360. https://doi.org/10.3390/diagnostics12102360
Chicago/Turabian StyleBarr, Martin P., Anne-Marie Baird, Sophia Halliday, Petra Martin, Emma H. Allott, James Phelan, Greg Korpanty, Linda Coate, Cathal O’Brien, Steven G. Gray, and et al. 2022. "Liquid Biopsy: A Multi-Parametric Analysis of Mutation Status, Circulating Tumor Cells and Inflammatory Markers in EGFR-Mutated NSCLC" Diagnostics 12, no. 10: 2360. https://doi.org/10.3390/diagnostics12102360
APA StyleBarr, M. P., Baird, A.-M., Halliday, S., Martin, P., Allott, E. H., Phelan, J., Korpanty, G., Coate, L., O’Brien, C., Gray, S. G., Sui, J. S. Y., Hayes, B., Cuffe, S., & Finn, S. P. (2022). Liquid Biopsy: A Multi-Parametric Analysis of Mutation Status, Circulating Tumor Cells and Inflammatory Markers in EGFR-Mutated NSCLC. Diagnostics, 12(10), 2360. https://doi.org/10.3390/diagnostics12102360







