Identification of Laminar Composition in Cerebral Cortex Using Low-Resolution Magnetic Resonance Images and Trust Region Optimization Algorithm
Abstract
1. Introduction
2. Materials and Methods
2.1. Fitting Problem
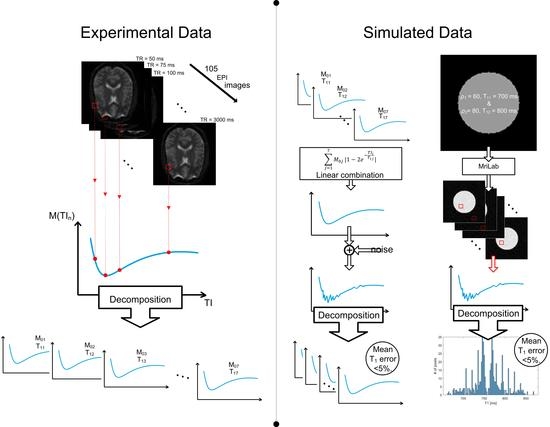
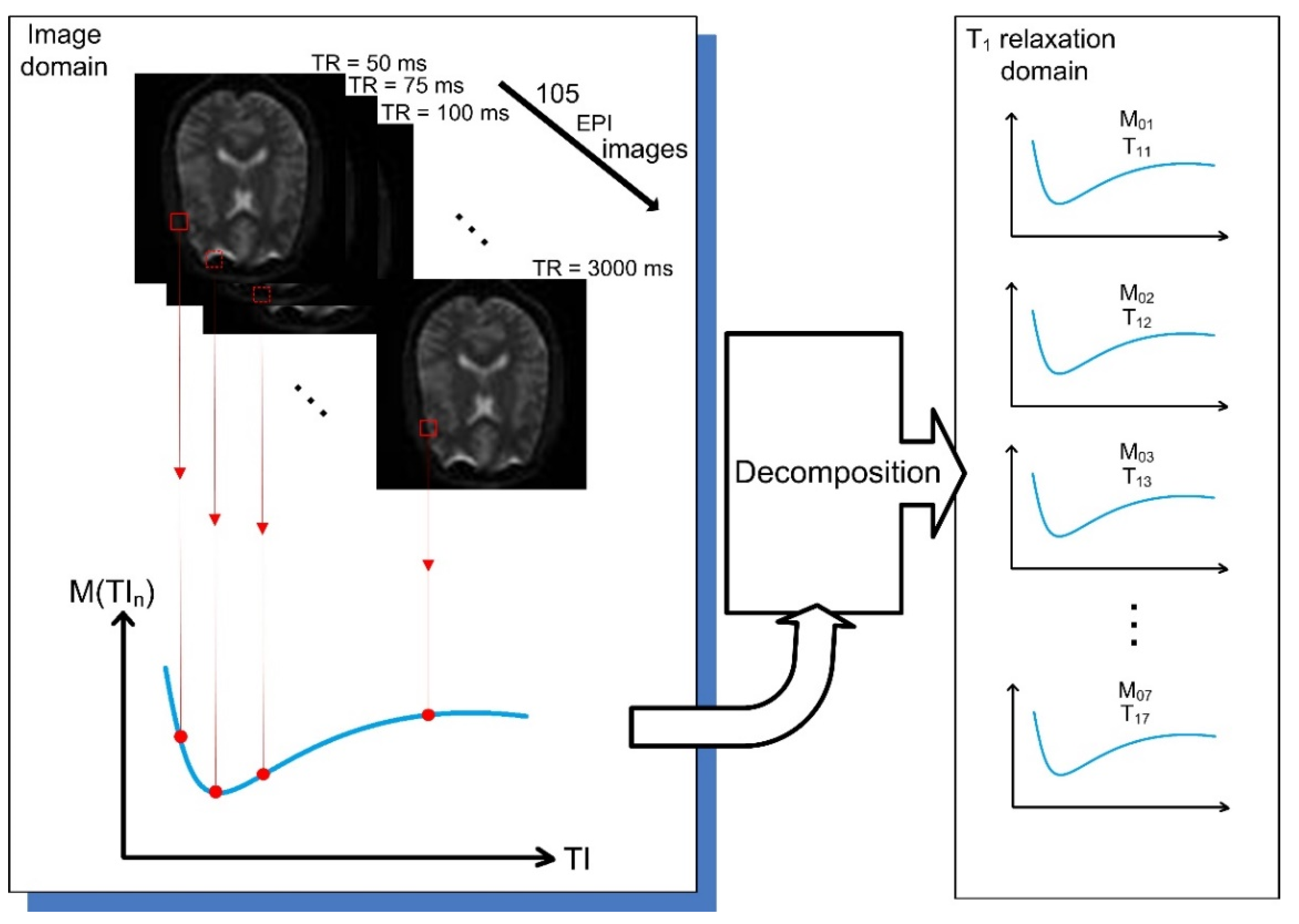
2.2. Experimental MRI Data
- Low-resolution modified echo-planar sequence with the following parameters: TR/TE = 1200/39 ms, 105 inversion times from the interval of 50–3000 ms, with the resolution of 3 × 3 × 3 mm3. The size of the image obtained from this sequence was 64 × 64 × 42 voxels.
- High-resolution MPRAGE sequence with the following parameters: TR/TE = 2150/2.5 ms, TI = 1100 ms, with the resolution of 1 × 1 × 1 mm3. The size of the image obtained from this sequence was 160 × 256 × 256 voxels.
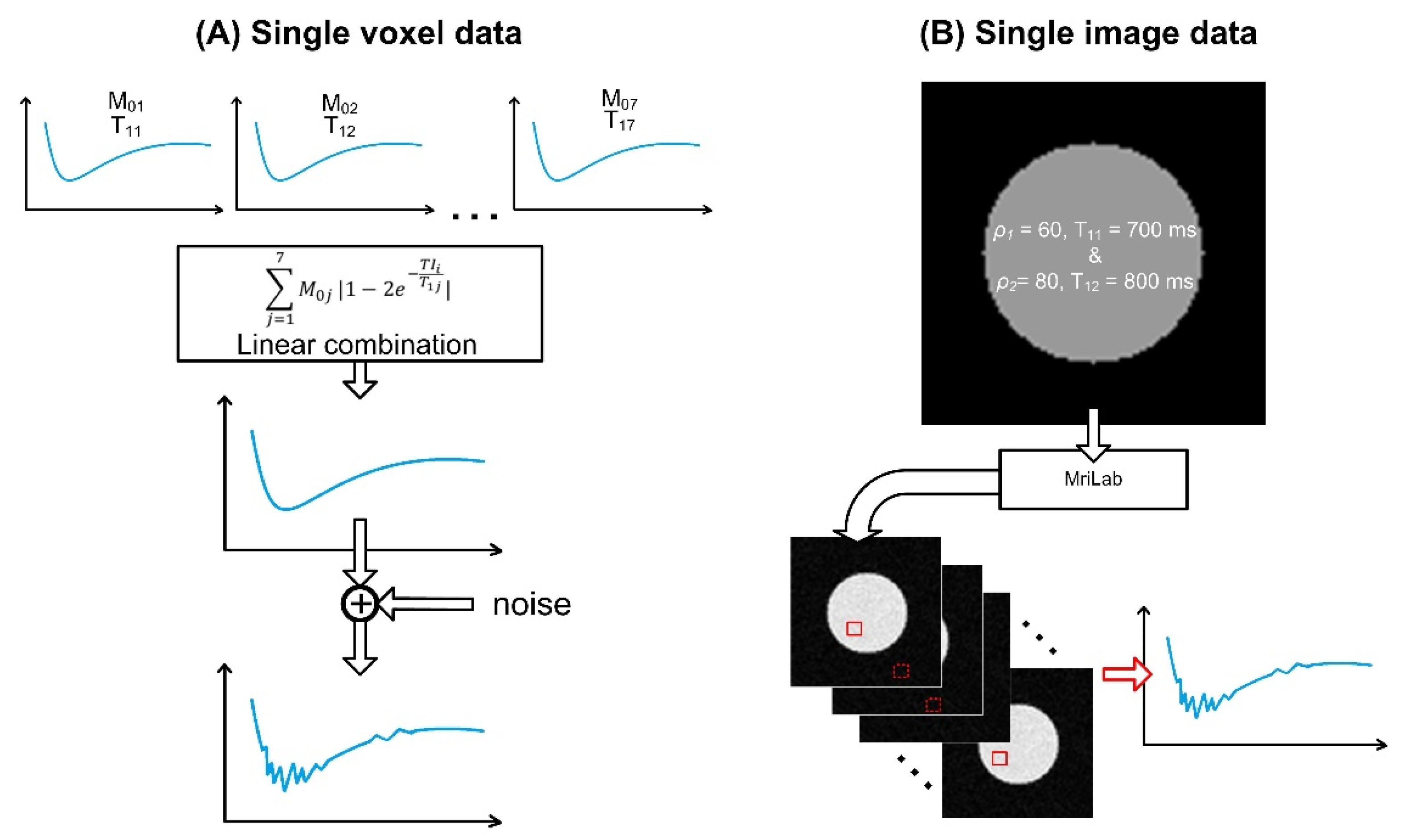
2.3. Simulated Data
2.4. Modified Trust Region Algorithm
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Brodmann, K. Vergleichende Lokalisationslehre der Großhirnrinde in Ihren Prinzipeien Dargestellt auf Grund des Zellenbaues; Barth: Leipzig, Germany, 1909. [Google Scholar]
- Von Economo, C.; Triarhou, L.C. Cellular Structure of the Human Cerebral Cortex; Karger Medical and Scientific Publishers: Basel, Switzerland, 2009. [Google Scholar] [CrossRef]
- Clark, V.P.; Courchesne, E.; Grafe, M. In Vivo Myeloarchitectonic Analysis of Human Striate and Extrastriate Cortex Using Magnetic Resonance Imaging. Cereb. Cortex 1992, 2, 417–424. [Google Scholar] [CrossRef]
- Clare, S.; Jezzard, P.; Matthews, P. Identification of the Myelinated Layers in Striate Cortex on High Resolution MRI at 3 Tesla. In Proceedings of the 10th Annual Meeting of ISMRM, Honolulu, HI, USA, 18–24 May 2002; p. 1465. [Google Scholar]
- Bridge, H.; Clare, S.; Jenkinson, M.; Jezzard, P.; Parker, A.J.; Matthews, P.M. Independent Anatomical and Functional Measures of the V1/V2 Boundary in Human Visual Cortex. J. Vis. 2005, 5, 93–102. [Google Scholar] [CrossRef][Green Version]
- Clare, S.; Bridge, H. Methodological Issues Relating to in Vivo Cortical Myelography Using MRI. Hum. Brain Mapp. 2005, 26, 240–250. [Google Scholar] [CrossRef]
- Turner, R.; Oros-Peusquens, A.-M.; Romanzetti, S.; Zilles, K.; Shah, N.J. Optimised in Vivo Visualisation of Cortical Structures in the Human Brain at 3 T Using IR-TSE. Magn. Reson. Imaging 2008, 26, 935–942. [Google Scholar] [CrossRef]
- Duyn, J.H.; van Gelderen, P.; Li, T.-Q.; de Zwart, J.A.; Koretsky, A.P.; Fukunaga, M. High-Field MRI of Brain Cortical Substructure Based on Signal Phase. Proc. Natl. Acad. Sci. USA 2007, 104, 11796–11801. [Google Scholar] [CrossRef] [PubMed]
- Trampel, R.; Ott, D.V.M.; Turner, R. Do the Congenitally Blind Have a Stria of Gennari? First Intracortical Insights In Vivo. Cereb. Cortex 2011, 21, 2075–2081. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Panchuelo, R.M.; Francis, S.T.; Schluppeck, D.; Bowtell, R.W. Correspondence of Human Visual Areas Identified Using Functional and Anatomical MRI In Vivo at 7 T. J. Magn. Reson. Imaging 2012, 35, 287–299. [Google Scholar] [CrossRef]
- Walters, N.B.; Egan, G.F.; Kril, J.J.; Kean, M.; Waley, P.; Jenkinson, M.; Watson, J.D.G. In Vivo Identification of Human Cortical Areas Using High-Resolution MRI: An Approach to Cerebral Structure-Function Correlation. Proc. Natl. Acad. Sci. USA 2003, 100, 2981–2986. [Google Scholar] [CrossRef]
- Dick, F.; Tierney, A.T.; Lutti, A.; Josephs, O.; Sereno, M.I.; Weiskopf, N. In Vivo Functional and Myeloarchitectonic Mapping of Human Primary Auditory Areas. J. Neurosci. 2012, 32, 16095–16105. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, J.J.M.; Hendrikse, J.; Luijten, P.R. Generalized Multiple-Layer Appearance of the Cerebral Cortex with 3D FLAIR 7.0-T MR Imaging. Radiology 2012, 262, 995–1001. [Google Scholar] [CrossRef][Green Version]
- De Martino, F.; Moerel, M.; Xu, J.; van de Moortele, P.-F.; Ugurbil, K.; Goebel, R.; Yacoub, E.; Formisano, E. High-Resolution Mapping of Myeloarchitecture In Vivo: Localization of Auditory Areas in the Human Brain. Cereb. Cortex 2015, 25, 3394–3405. [Google Scholar] [CrossRef]
- Fracasso, A.; van Veluw, S.J.; Visser, F.; Luijten, P.R.; Spliet, W.; Zwanenburg, J.J.M.; Dumoulin, S.O.; Petridou, N. Lines of Baillarger in Vivo and Ex Vivo: Myelin Contrast across Lamina at 7T MRI and Histology. NeuroImage 2016, 133, 163–175. [Google Scholar] [CrossRef]
- Lema Dopico, A.; Choi, S.; Hua, J.; Li, X.; Harrison, D.M. Multi-Layer Analysis of Quantitative 7 T Magnetic Resonance Imaging in the Cortex of Multiple Sclerosis Patients Reveals Pathology Associated with Disability. Mult. Scler. J. 2021, 27, 2040–2051. [Google Scholar] [CrossRef] [PubMed]
- Barazany, D.; Assaf, Y. Visualization of Cortical Lamination Patterns with Magnetic Resonance Imaging. Cereb. Cortex 2012, 22, 2016–2023. [Google Scholar] [CrossRef] [PubMed]
- Lifshits, S.; Tomer, O.; Shamir, I.; Barazany, D.; Tsarfaty, G.; Rosset, S.; Assaf, Y. Resolution Considerations in Imaging of the Cortical Layers. NeuroImage 2018, 164, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Shamir, I.; Tomer, O.; Baratz, Z.; Tsarfaty, G.; Faraggi, M.; Horowitz, A.; Assaf, Y. A Framework for Cortical Laminar Composition Analysis Using Low-Resolution T1 MRI Images. Brain Struct. Funct. 2019, 224, 1457–1467. [Google Scholar] [CrossRef]
- González Ballester, M.Á.; Zisserman, A.P.; Brady, M. Estimation of the Partial Volume Effect in MRI. Med. Image Anal. 2002, 6, 389–405. [Google Scholar] [CrossRef]
- Barral, J.K.; Gudmundson, E.; Stikov, N.; Etezadi-Amoli, M.; Stoica, P.; Nishimura, D.G. A Robust Methodology for In Vivo T1 Mapping. Magn. Reson. Med. 2010, 64, 1057–1067. [Google Scholar] [CrossRef]
- Istratov, A.; Vyvenko, O. Exponential Analysis in Physical Phenomena. Rev. Sci. Instrum. 1999, 70, 1233–1257. [Google Scholar] [CrossRef]
- Mitchell, J.; Gladden, L.F.; Chandrasekera, T.C.; Fordham, E.J. Low-Field Permanent Magnets for Industrial Process and Quality Control. Prog. Nucl. Magn. Reson. Spectrosc. 2014, 76, 1–60. [Google Scholar] [CrossRef]
- Washburn, K.E.; McCarney, E.R. Improved Quantification of Nuclear Magnetic Resonance Relaxometry Data via Partial Least Squares Analysis. Appl. Magn. Reson. 2018, 49, 429–464. [Google Scholar] [CrossRef]
- Berman, P.; Levi, O.; Parmet, Y.; Saunders, M.; Wiesman, Z. Laplace Inversion of Low-Resolution NMR Relaxometry Data Using Sparse Representation Methods. Concepts Magn. Reson. Part A 2013, 42, 72–88. [Google Scholar] [CrossRef] [PubMed]
- Fordham, E.J.; Venkataramanan, L.; Mitchell, J.; Valori, A. What Are, and What Are Not, Inverse Laplace Transforms. Diffus. Fundam. 2018, 29, 1–8. [Google Scholar]
- Wright, P.J.; Mougin, O.E.; Totman, J.J.; Peters, A.M.; Brookes, M.J.; Coxon, R.; Morris, P.E.; Clemence, M.; Francis, S.T.; Bowtell, R.W.; et al. Water Proton T1 Measurements in Brain Tissue at 7, 3, and 1.5 T Using IR-EPI, IR-TSE, and MPRAGE: Results and Optimization. Magn. Reson. Mater. Phys. Biol. Med. 2008, 21, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Bojorquez, J.Z.; Bricq, S.; Acquitter, C.; Brunotte, F.; Walker, P.M.; Lalande, A. What Are Normal Relaxation Times of Tissues at 3 T? Magn. Reson. Imaging 2017, 35, 69–80. [Google Scholar] [CrossRef]
- Liu, F.; Velikina, J.V.; Block, W.F.; Kijowski, R.; Samsonov, A.A. Fast Realistic MRI Simulations Based on Generalized Multi-Pool Exchange Tissue Model. IEEE Trans. Med. Imaging 2017, 36, 527–537. [Google Scholar] [CrossRef]
- Conn, A.R.; Gould, N.I.M.; Toint, P.L. Trust-Region Methods; MPS-SIAM series on optimization; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2000; ISBN 978-0-89871-460-9. [Google Scholar]
- Byrd, R.H.; Schnabel, R.B.; Shultz, G.A. Approximate Solution of the Trust Region Problem by Minimization over Two-Dimensional Subspaces. Math. Program. 1988, 40, 247–263. [Google Scholar] [CrossRef]
- Coleman, T.F.; Li, Y. An Interior Trust Region Approach for Nonlinear Minimization Subject to Bounds. SIAM J. Optim. 1996, 6, 418–445. [Google Scholar] [CrossRef]
- Gudbjartsson, H.; Patz, S. The Rician Distribution of Noisy MRI Data. Magn. Reson. Med. 1995, 34, 910–914. [Google Scholar] [CrossRef]
- Sonderer, C.M.; Chen, N. Improving the Accuracy, Quality, and Signal-To-Noise Ratio of MRI Parametric Mapping Using Rician Bias Correction and Parametric-Contrast-Matched Principal Component Analysis (PCM-PCA). Yale J. Biol. Med. 2018, 91, 207–214. [Google Scholar] [PubMed]

| Nstarting points = 1 | Nstarting points = 100 | |||||
|---|---|---|---|---|---|---|
| Min. Error [%] | Mean Error [%] | Max. Error [%] | Min. Error [%] | Mean Error [%] | Max. Error [%] | |
| M0 | 0.00 | 44.60 | 604.00 | 0.00 | 0.00 | 0.00 |
| T1 | 0.00 | 6.11 | 36.00 | 0.00 | 0.00 | 0.00 |
| Noise Variance | SNR [dB] | Min. M0 Error [%] | Mean M0 Error [%] | Max. M0 Error [%] | Min. T1 Error [%] | Mean T1 Error [%] | Max. T1 Error [%] |
|---|---|---|---|---|---|---|---|
| 0 | Inf | 0 | 0 | 0 | 0 | 0 | 0 |
| 0.1 | 61 | 0 | 2 | 4 | 0 | 0 | 0 |
| 1 | 51 | 2 | 5 | 14 | 0 | 0 | 1 |
| 5 | 45 | 2 | 28 | 109 | 0 | 2 | 4 |
| 10 | 41 | 3 | 18 | 48 | 0 | 1 | 3 |
| 25 | 38 | 3 | 28 | 86 | 0 | 2 | 5 |
| 50 | 34 | 7 | 80 | 268 | 1 | 10 | 25 |
| 100 | 31 | 18 | 60 | 131 | 2 | 11 | 26 |
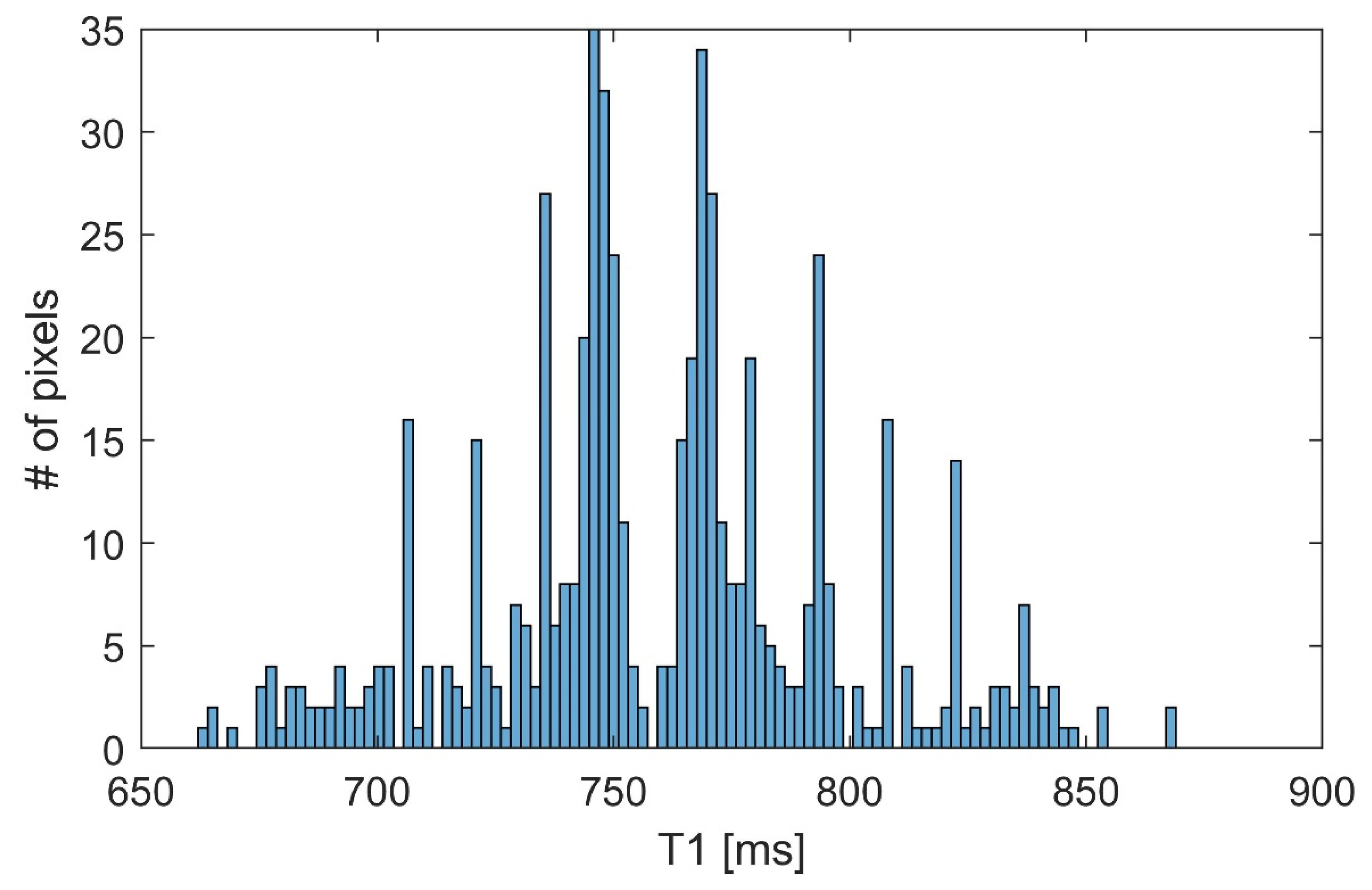
| T1 | Min. Error [%] | Mean Error [%] | Max. Error [%] |
|---|---|---|---|
| 700 | 0.05 | 4.86 | 8.65 |
| 800 | 0.12 | 2.98 | 8.59 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamárik, J.; Vojtíšek, L.; Churová, V.; Kašpárek, T.; Schwarz, D. Identification of Laminar Composition in Cerebral Cortex Using Low-Resolution Magnetic Resonance Images and Trust Region Optimization Algorithm. Diagnostics 2022, 12, 24. https://doi.org/10.3390/diagnostics12010024
Jamárik J, Vojtíšek L, Churová V, Kašpárek T, Schwarz D. Identification of Laminar Composition in Cerebral Cortex Using Low-Resolution Magnetic Resonance Images and Trust Region Optimization Algorithm. Diagnostics. 2022; 12(1):24. https://doi.org/10.3390/diagnostics12010024
Chicago/Turabian StyleJamárik, Jakub, Lubomír Vojtíšek, Vendula Churová, Tomáš Kašpárek, and Daniel Schwarz. 2022. "Identification of Laminar Composition in Cerebral Cortex Using Low-Resolution Magnetic Resonance Images and Trust Region Optimization Algorithm" Diagnostics 12, no. 1: 24. https://doi.org/10.3390/diagnostics12010024
APA StyleJamárik, J., Vojtíšek, L., Churová, V., Kašpárek, T., & Schwarz, D. (2022). Identification of Laminar Composition in Cerebral Cortex Using Low-Resolution Magnetic Resonance Images and Trust Region Optimization Algorithm. Diagnostics, 12(1), 24. https://doi.org/10.3390/diagnostics12010024