Analysis of Microbial Communities: An Emerging Tool in Forensic Sciences
Abstract
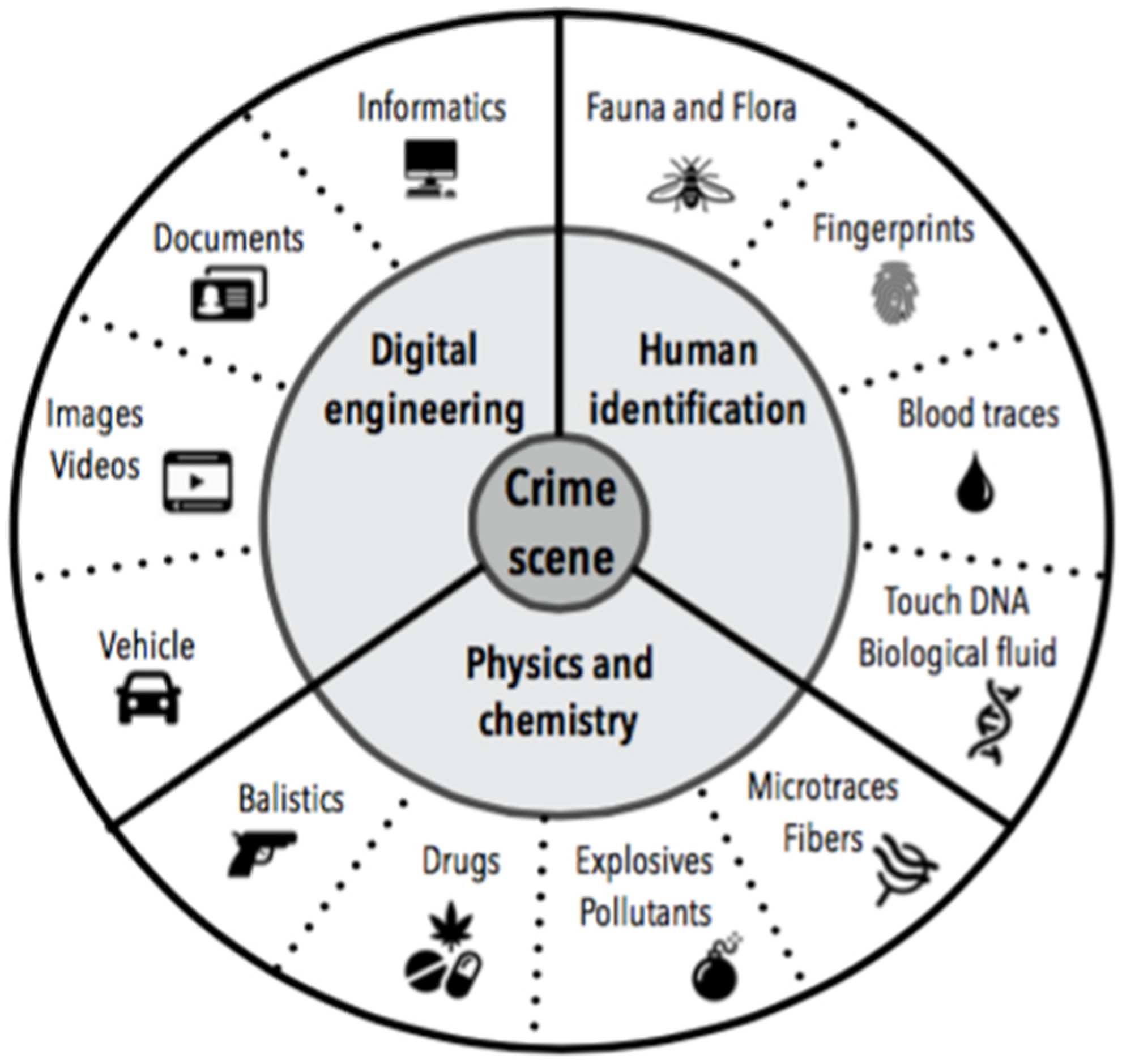
:1. Introduction
2. Use of Microbiome in Forensic Sciences
3. Microbiome Analysis and Individual Determination
3.1. Determination of the Core Microbiome
3.2. Example of Specific Bacterial Communities
3.3. Personal Microbial Cloud
3.4. Cross Transfer between Individuals
4. Interest in the Study of Biological Fluids Microbiomes
4.1. Saliva
4.2. Vaginal Secretions
4.3. Blood Samples
4.4. Fecal Material
4.5. Scalp and Pubic Hairs
4.6. Sperm Sample
4.7. Other Samples
5. Microbiome Determination on Supports and Objects
5.1. Mobile Phones
5.2. Keyboards and Mice
5.3. Shoes
5.4. Identification of Objects Handled by the Decedent before Death
6. Determination of the Post-Mortem Interval (PMI) Using Microbiome Analysis
7. Geolocalisation and Determination of Soil Microbial Communities
8. Factors Influencing Microbiome Analysis
8.1. Storage Conditions
8.2. Sampling and Lab Protocol
8.3. Influence of Hand Washing
8.4. Environmental Conditions
9. Microbiota and Real Cases
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Roux, C.; Talbot-Wright, B.; Robertson, J.; Crispino, F.; Ribaux, O. The end of the (forensic science) world as we know it? The example of trace evidence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Using the Microbiome to Unlock the Secrets of Forensic Evidence: Classification of the Body Source of Origin of Human Traces. Available online: https://www.qiagen.com/us/knowledge-and-support/knowledge-hub/events-and-webinars/webinars/microbiome-to-unlock-the-secrets-of-forensic-evidence (accessed on 20 October 2021).
- Metcalf, J.L.; Parfrey, L.W.; Gonzalez, A.; Lauber, C.L.; Knights, D.; Ackermann, G.; Humphrey, G.C.; Gebert, M.J.; Van Treuren, W.; Berg-Lyons, D.; et al. A microbial clock provides an accurate estimate of the postmortem interval in a mouse model system. eLife 2013, 2, e0110. [Google Scholar] [CrossRef]
- Inman, K.; Rudin, N. The origin of evidence. Forensic Sci. Int. 2002, 126, 11–16. [Google Scholar] [CrossRef]
- Bouslimani, A.; Melnik, A.V.; Xu, Z.; Amir, A.; da Silva, R.R.; Wang, M.; Bandeira, N.; Alexandrov, T.; Knight, R.; Dorrestein, P.C. Lifestyle chemistries from phones for individual profiling. Proc. Natl. Acad. Sci. USA 2016, 113, E7645–E7654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harbison, S.A.; Fleming, R.I. Forensic body fluid identification: State of the art. Res. Rep. Forensic Med. Sci. 2016, 6, 11–23. [Google Scholar] [CrossRef] [Green Version]
- Hanssen, E.N.; Avershina, E.; Rudi, K.; Gill, P.; Snipen, L. Body fluid prediction from microbial patterns for forensic application. Forensic Sci. Int. Genet. 2017, 30, 10–17. [Google Scholar] [CrossRef]
- Dobay, A.; Haas, C.; Fucile, G.; Downey, N.; Morrison, H.G.; Kratzer, A.; Arora, N. Microbiome-based body fluid identification of samples exposed to indoor conditions. Forensic Sci. Int. Genet. 2019, 40, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Ingold, S.; Dørum, G.; Hanson, E.; Berti, A.; Branicki, W.; Brito, P.; Elsmore, P.; Gettings, K.B.; Giangasparo, F.; Gross, T.E.; et al. Body fluid identification using a targeted mRNA massively parallel sequencing approach—Results of a EUROFORGEN/EDNAP collaborative exercise. Forensic Sci. Int. Genet. 2018, 34, 105–115. [Google Scholar] [CrossRef] [Green Version]
- Sauer, E.; Reinke, A.K.; Courts, C. Differentiation of five body fluids from forensic samples by expression analysis of four microRNAs using quantitative PCR. Forensic Sci. Int. Genet. 2016, 22, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Hampton-Marcell, J.; Lopez, J.V.; Gilbert, J.A. The human microbiome: An emerging tool in forensics. Microb. Biotechnol. 2017, 10, 228–230. [Google Scholar] [CrossRef]
- Beans, C. News feature: Can microbes keep time for forensic investigators? Proc. Natl. Acad. Sci. USA 2018, 115, 3–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Dembinski, G.M.; Picard, C.J. Effects of microbial DNA on human DNA profiles generated using the PowerPlex 16 HS system. J. Forensic Leg. Med. 2017, 52, 208–214. [Google Scholar] [CrossRef]
- Arenas, M.; Pereira, F.; Oliveira, M.; Pinto, N.; Lopes, A.M.; Gomes, V.; Carracedo, A.; Amorim, A. Forensic genetics and genomics: Much more than just a human affair. PLoS Genet. 2017, 13, e1006960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuiper, I. Microbial forensics: Next-generation sequencing as catalyst: The use of new sequencing technologies to analyze whole microbial communities could become a powerful tool for forensic and criminal investigations. EMBO Rep. 2016, 17, 1085–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leake, S.L.; Pagni, M.; Falquet, L.; Taroni, F.; Greub, G. The salivary microbiome for differentiating individuals: Proof of principle. Microbes Infect 2016, 18, 399–405. [Google Scholar] [CrossRef] [Green Version]
- Nema, V. Microbial Forensics: Beyond a Fascination. In DNA Fingerprinting: Advancements and Future Endeavors; Dash, H., Shrivastava, P., Mohapatra, B., Das, S., Eds.; Springer: Singapore, 2018; pp. 295–306. [Google Scholar]
- Kodama, W.A.; Xu, Z.; Metcalf, J.L.; Song, S.J.; Harrison, N.; Knight, R.; Carter, D.O.; Happy, C.B. Trace Evidence Potential in Postmortem Skin Microbiomes: From Death Scene to Morgue. J. Forensic Sci. 2019, 64, 791–798. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Zhou, N.; McDonald, N.; Costello, E.K.; Knight, R. Forensic identification using skin bacterial communities. Proc. Natl. Acad. Sci. USA 2010, 107, 6477–6481. [Google Scholar] [CrossRef] [Green Version]
- Schmedes, S.E.; Sajantila, A.; Budowle, B. Expansion of microbial forensics. J. Clin. Microbiol. 2016, 54, 1964–1974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, T.H.; Gomez, A.; Singh, H.; Nelson, K.E.; Brinkac, L.M. Integrating the microbiome as a resource in the forensics toolkit. Forensic Sci. Int. Genet. 2017, 30, 141–147. [Google Scholar] [CrossRef] [Green Version]
- Budowle, B.; Connell, N.D.; Bielecka-Oder, A.; Colwell, R.R.; Corbett, C.R.; Fletcher, J.; Forsman, M.; Kadavy, D.R.; Markotic, A.; Morse, S.A.; et al. Validation of high throughput sequencing and microbial forensics applications. Investig. Genet. 2014, 5, 9. [Google Scholar] [CrossRef] [Green Version]
- Giampaoli, S.; Berti, A.; Valeriani, F.; Gianfranceschi, G.; Piccolella, A.; Buggiotti, L.; Rapone, C.; Valentini, A.; Ripani, L.; Spica, V.R. Molecular identification of vaginal fluid by microbial signature. Forensic Sci. Int. Genet. 2012, 6, 559–564. [Google Scholar] [CrossRef]
- Fierer, N.; Hamady, M.; Lauber, C.L.; Knight, R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 17994–17999. [Google Scholar] [CrossRef] [Green Version]
- Brinkac, L.; Clarke, T.H.; Singh, H.; Greco, C.; Gomez, A.; Torralba, M.G.; Frank, B.; Nelson, K.E. Spatial and Environmental Variation of the Human Hair Microbiota. Sci. Rep. 2018, 8, 9017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, J.; Byrd, A.L.; Park, M.; NISC Comparative Sequencing Program; Kong, H.H.; Segre, J.A. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cullen, C.M.; Aneja, K.K.; Beyhan, S.; Cho, C.E.; Woloszynek, S.; Convertino, M.; McCoy, S.J.; Zhang, Y.; Anderson, M.Z.; Alvarez-Ponce, D.; et al. Emerging Priorities for Microbiome Research. Front. Microbiol. 2020, 11, 136. [Google Scholar] [CrossRef] [Green Version]
- Schmedes, S.E.; Woerner, A.E.; Budowle, B. Forensic human identification using skin microbiomes. Appl. Environ. Microbiol. 2017, 83, e01672-17. [Google Scholar] [CrossRef] [Green Version]
- Schmedes, S.E.; Woerner, A.E.; Novroski, N.M.M.; Wendt, F.R.; King, J.L.; Stephens, K.M.; Budowle, B. Targeted sequencing of clade-specific markers from skin microbiomes for forensic human identification. Forensic Sci. Int. Genet. 2018, 32, 50–61. [Google Scholar] [CrossRef]
- Park, J.; Kim, S.J.; Lee, J.A.; Kim, J.W.; Kim, S.B. Microbial forensic analysis of human-associated bacteria inhabiting hand surface. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, 510–512. [Google Scholar] [CrossRef] [Green Version]
- Flowers, L.; Grice, E.A. The skin microbiota: Balancing risk and reward. Cell Host Microbe 2020, 28, 190–200. [Google Scholar] [CrossRef]
- Edmonds-Wilson, S.L.; Nurinova, N.I.; Zapka, C.A.; Fierer, N.; Wilson, M. Review of human hand microbiome research. J. Dermatol. Sci. 2015, 80, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, D.; Leung, M.H.Y.; Lee, P.K.H. Microbiota fingerprints lose individually identifying features over time. Microbiome 2017, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Meadow, J.F.; Altrichter, A.E.; Bateman, A.C.; Stenson, J.; Brown, G.Z.; Green, J.L.; Bohannan, B.J.M. Humans differ in their personal microbial cloud. PeerJ 2015, 3, e1258. [Google Scholar] [CrossRef] [Green Version]
- Neckovic, A.; van Oorschot, R.A.; Szkuta, B.; Durdle, A. Investigation of direct and indirect transfer of microbiomes between individuals. Forensic Sci. Int. Genet. 2020, 45, 102212. [Google Scholar] [CrossRef]
- Hall, M.W.; Singh, N.; Ng, K.F.; Lam, D.K.; Goldberg, M.B.; Tenenbaum, H.C.; Neufeld, J.D.; Beiko, R.G.; Senadheera, D.B. Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity. NPJ Biofilms Microbiomes 2017, 3, 2. [Google Scholar] [CrossRef]
- Jung, J.Y.; Yoon, H.K.; An, S.; Lee, J.W.; Ahn, E.R.; Kim, Y.J.; Park, H.C.; Lee, K.; Hwang, J.H.; Lim, S.K. Rapid oral bacteria detection based on real-time PCR for the forensic identification of saliva. Sci. Rep. 2018, 8, 10852. [Google Scholar] [CrossRef]
- Miller, E.A.; Beasley, D.E.; Dunn, R.R.; Archie, E.A. Lactobacilli Dominance and Vaginal pH: Why Is the Human Vaginal Microbiome Unique? Front. Microbiol. 2016, 7, 1936. [Google Scholar] [CrossRef]
- Coolen, M.J.L.; Post, E.; Davis, C.C.; Forney, J.L. Characterization of Microbial Communities Found in the Human Vagina by Analysis of Terminal Restriction Fragment Length Polymorphisms of 16S rRNA Genes. Appl. Environ. Microbiol. 2005, 71, 8729–8737. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, J.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Doi, M.; Gamo, S.; Okiura, T.; Nishimukai, H.; Asano, M. A simple identification method for vaginal secretions using relative quantification of Lactobacillus DNA. Forensic Sci. Int. Genet. 2014, 12, 93–99. [Google Scholar] [CrossRef]
- López, C.D.; González, D.M.; Haas, C.; Vidaki, A.; Kayser, M. Microbiome-based body site of origin classification of forensically relevant T blood traces. Forensic Sci. Int. Genet. 2020, 47, 102280. [Google Scholar] [CrossRef]
- Tropini, C.; Earle, K.A.; Huang, K.C.; Sonnenburg, J.L. The gut microbiome: Connecting spatial organization to function. Cell Host Microbe 2017, 21, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Martínez, I.; Muller, C.E.; Walter, J. Long-Term Temporal Analysis of the Human Fecal Microbiota Revealed a Stable Core of Dominant Bacterial Species. PLoS ONE 2013, 8, e69621. [Google Scholar] [CrossRef]
- Conlon, M.A.; Bird, A.R. The Impact of Diet and Lifestyle on Gut Microbiota and Human Health. Nutrients 2015, 7, 17–44. [Google Scholar] [CrossRef]
- Tridico, S.R.; Murray, D.C.; Addison, J.; Kirkbride, K.P.; Bunce, M. Metagenomic analyses of bacteria on human hairs: A qualitative assessment for applications in forensic science. Investig. Genet. 2014, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Williams, D.W.; Gibson, G. Classification of individuals and the potential to detect sexual contact using the microbiome of the pubic region. Forensic Sci. Int. Genet. 2019, 41, 177–187. [Google Scholar] [CrossRef]
- Molina, N.M.; Plaza-Diaz, J.; Vilchez-Vargas, R.; Sola-Leyva, A.; Vargas, E.; Mendoza-Tesarik, R.; Galán-Lázaro, M.; de Guevara, N.M.-L.; Tesarik, J.; Altmäe, S. Assessing the testicular sperm microbiome: A low-biomass site with abundant contamination. Reprod. Biomed. Online 2021, 43, 523–531. [Google Scholar] [CrossRef]
- Farahani, L.; Tharakan, T.; Yap, T.; Ramsay, J.W.; Jayasena, C.N.; Minhas, S. The semen microbiome and its impact on sperm function and male fertility: A systematic review and meta-analysis. Andrology 2021, 9, 115–144. [Google Scholar] [CrossRef]
- Baud, D.; Pattaroni, C.; Vulliemoz, N.; Castella, V.; Marsland, B.J.; Stojanov, M. Sperm Microbiota and Its Impact on Semen Parameters. Front. Microbiol. 2019, 10, 234. [Google Scholar] [CrossRef] [Green Version]
- Quaak, F.C.A.; van Duijn, T.; Hoogenboom, J.; Kloosterman, A.D.; Kuiper, I. Human-associated microbial populations as evidence in forensic casework. Forensic Sci. Int. Genet. 2018, 36, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Meadow, J.F.; Altrichter, A.E.; Green, J.L. Mobile phones carry the personal microbiome of their owners. PeerJ 2014, 2, e447. [Google Scholar] [CrossRef] [Green Version]
- Lax, S.; Hampton-Marcell, J.T.; Gibbons, S.M.; Colares, G.B.; Smith, D.; Eisen, J.A.; Gilbert, J.A. Forensic analysis of the microbiome of phones and shoes. Microbiome 2015, 3, 21. [Google Scholar] [CrossRef] [Green Version]
- Dong, K.; Xin, Y.; Cao, F.; Huang, Z.; Sun, J.; Peng, M.; Liu, W.; Shi, P. Succession of oral microbiota community as a tool to estimate postmortem interval. Sci. Rep. 2019, 9, 13063. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Fu, X.; Liao, H.; Hu, Z.; Long, L.; Yan, W.; Ding, Y.; Zha, L.; Guo, Y.; Yan, J.; et al. Potential use of bacterial community succession for estimating post-mortem interval as revealed by high-throughput sequencing. Sci. Rep. 2016, 6, 24197. [Google Scholar] [CrossRef] [Green Version]
- Cobaugh, K.L.; Schaeffer, S.M.; DeBruyn, J.M. Functional and Structural Succession of Soil Microbial Communities below Decomposing Human Cadavers. PLoS ONE 2015, 10, e0130201. [Google Scholar] [CrossRef]
- Metcalf, J.L. Estimating the postmortem interval using microbes: Knowledge gaps and a path to technology adoption. Forensic Sci. Int. Genet. 2019, 38, 211–218. [Google Scholar] [CrossRef] [Green Version]
- DeBruyn, J.M.; Hauther, K.A. Postmortem succession of gut microbial communities in deceased human subjects. PeerJ 2017, 5, e3437. [Google Scholar] [CrossRef] [Green Version]
- Belk, A.; Xu, Z.Z.; Carter, D.O.; Lynne, A.; Bucheli, S.; Knight, R.; Metcalf, J.L. Microbiome Data Accurately Predicts the Postmortem Interval Using Random Forest Regression Models. Genes 2018, 9, 104. [Google Scholar] [CrossRef] [Green Version]
- Javan, G.T.; Finley, S.J.; Abidin, Z.; Mulle, J.G. The Thanatomicrobiome: A Missing Piece of the Microbial Puzzle of Death. Front. Microbiol. 2016, 7, 225. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Bian, Y. Thanatomicrobiome composition profiling as a tool for forensic investigation. Forensic Sci. Res. 2018, 3, 105–110. [Google Scholar] [CrossRef]
- Hyde, E.R.; Haarmann, D.P.; Lynne, A.M.; Bucheli, S.R.; Petrosino, J.F. The Living Dead: Bacterial Community Structure of a Cadaver at the Onset and End of the Bloat Stage of Decomposition. PLoS ONE 2013, 8, e77733. [Google Scholar]
- Metcalf, J.L.; Carter, D.O.; Knight, R. Microbiology of death. Curr. Biol. 2016, 26, R561–R563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handke, J.; Procopio, N.; Buckley, M.; van der Meer, D.; Williams, G.; Carr, M.; Williams, A. Successive bacterial colonisation of pork and its implications for forensic investigations. Forensic Sci. Int. 2017, 281, 1–8. [Google Scholar] [CrossRef]
- Pechal, J.L.; Schmidt, C.J.; Jordan, H.R.; Benbow, M.E. A large-scale survey of the postmortem human microbiome, and its potential to provide insight into the living health condition. Sci. Rep. 2018, 8, 5724. [Google Scholar] [CrossRef]
- Habtom, H.; Pasternak, Z.; Matan, O.; Azulay, C.; Gafny, R.; Jurkevitch, E. Applying microbial biogeography in soil forensics. Forensic Sci. Int. Genet. 2019, 38, 195–203. [Google Scholar] [CrossRef]
- Demanèche, S.; Schauser, L.; Dawson, L.; Franqueville, L.; Simonet, P. Microbial soil community analyses for forensic application: Application to a blind test. Forensic Sci. Int. 2017, 270, 153–158. [Google Scholar] [CrossRef]
- López, C.D.; Vidaki, A.; Ralf, A.; González, D.M.; Radjabzadeh, D.; Kraaij, R.; Uitterlinden, A.G.; Haas, C.; Lao, O.; Kayser, M. Novel taxonomy-independent deep learning microbiome approach allows for accurate classification of different forensically relevant human epithelial materials. Forensic Sci. Int. Genet. 2019, 41, 72–82. [Google Scholar] [CrossRef]
- Williams, D.W.; Gibson, G. Individualization of pubic hair bacterial communities and the effects of storage time and temperature. Forensic Sci. Int. Genet. 2017, 26, 12–20. [Google Scholar] [CrossRef]
- Song, S.J.; Amir, A.; Metcalf, J.L.; Amato, K.R.; Xu, Z.Z.; Humphrey, G.; Knight, R. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems 2016, 1, e00021-16. [Google Scholar] [CrossRef] [Green Version]
- Hanssen, E.N.; Liland, K.H.; Gill, P.; Snipen, L. Optimizing body fluid recognition from microbial taxonomic profiles. Forensic Sci. Int. Genet. 2018, 37, 13–20. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores, G.E.; Caporaso, J.G.; Henley, J.B.; Rideout, J.R.; Domogala, D.; Chase, J.; Leff, J.W.; Vázquez-Baeza, Y.; Gonzalez, A.; Knight, R.; et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014, 15, 531. [Google Scholar] [CrossRef] [Green Version]
- Chase, J.; Fouquier, J.; Zare, M.; Sonderegger, D.L.; Knight, R.; Kelley, S.T.; Siegel, J.; Caporaso, J.G. Geography and location are the primary drivers of office microbiome composition. mSystems 2016, 1, e00022-16. [Google Scholar] [CrossRef] [Green Version]
- Tims, S.; van Wamel, W.; Endtz, H.P.; van Belkum, A.; Kayser, M. Microbial DNA fingerprinting of human fingerprints: Dynamic colonization of fingertip microflora challenges human host inferences for forensic purposes. Int. J. Leg. Med. 2010, 124, 477–481. [Google Scholar] [CrossRef] [Green Version]
- Flores, G.E.; Bates, S.T.; Caporaso, J.G.; Lauber, C.L.; Leff, J.W.; Knight, R.; Fierer, N. Diversity, distribution and sources of bacteria in residential kitchens. Environ. Microbiol. 2013, 15, 588–596. [Google Scholar] [CrossRef]
- Lax, S.; Smith, D.P.; Hampton-Marcell, J.; Owens, S.M.; Handley, K.M.; Scott, N.M.; Gibbons, S.M.; Larsen, P.; Shogan, B.D.; Weiss, S.; et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 2014, 345, 1048–1052. [Google Scholar] [CrossRef] [Green Version]
- Song, S.J.; Lauber, C.; Costello, E.K.; Lozupone, C.A.; Humphrey, G.; Berg-Lyons, D.; Caporaso, J.G.; Knights, D.; Clemente, J.C.; Nakielny, S.; et al. Cohabiting family members share microbiota with one another and with their dogs. eLife 2013, 2, e00458. [Google Scholar] [CrossRef]
- Quaak, F.C.A.; van de Wal, Y.; Maaskant-van Wijk, P.A.; Kuiper, I. Combining human STR and microbial population profiling: Two case reports. Forensic Sci. Int. Genet. 2018, 37, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Le Pajolec, M.G. French Speaking Working Group of International Society for Forensic Genetics; Personal Communication: Pontoise, France, 2017. [Google Scholar]
- Py, B.; Leonhard, J.; Martinelle, M.; Ménabé, C. ADN et Justice: L’utilisation de L’empreinte Génétique Dans les Procédures Judiciaires; Collection «Santé, qualité de vie et handicap»; Presses Universitaires de Nancy—Editions Universitaires de Lorraine: Nancy, France, 2020; pp. 50–53 and 233–237. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gouello, A.; Dunyach-Remy, C.; Siatka, C.; Lavigne, J.-P. Analysis of Microbial Communities: An Emerging Tool in Forensic Sciences. Diagnostics 2022, 12, 1. https://doi.org/10.3390/diagnostics12010001
Gouello A, Dunyach-Remy C, Siatka C, Lavigne J-P. Analysis of Microbial Communities: An Emerging Tool in Forensic Sciences. Diagnostics. 2022; 12(1):1. https://doi.org/10.3390/diagnostics12010001
Chicago/Turabian StyleGouello, Audrey, Catherine Dunyach-Remy, Christian Siatka, and Jean-Philippe Lavigne. 2022. "Analysis of Microbial Communities: An Emerging Tool in Forensic Sciences" Diagnostics 12, no. 1: 1. https://doi.org/10.3390/diagnostics12010001
APA StyleGouello, A., Dunyach-Remy, C., Siatka, C., & Lavigne, J.-P. (2022). Analysis of Microbial Communities: An Emerging Tool in Forensic Sciences. Diagnostics, 12(1), 1. https://doi.org/10.3390/diagnostics12010001







