Comparison of Next-Generation Sequencing and Fluorescence In Situ Hybridization for Detection of Segmental Chromosomal Aberrations in Neuroblastoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
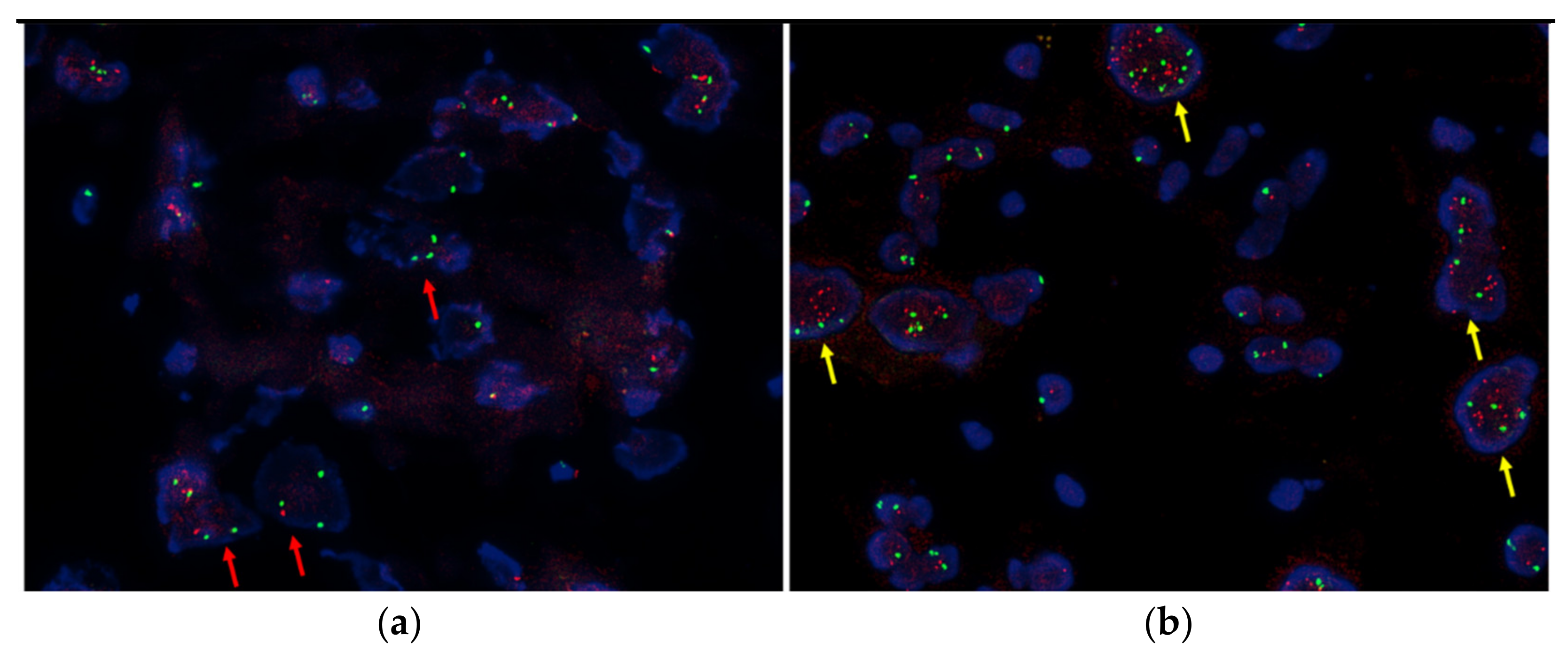
2.2. FISH
2.3. NGS
2.4. Statistical Analysis
3. Results
3.1. Clinicopathologic Features of the Patients
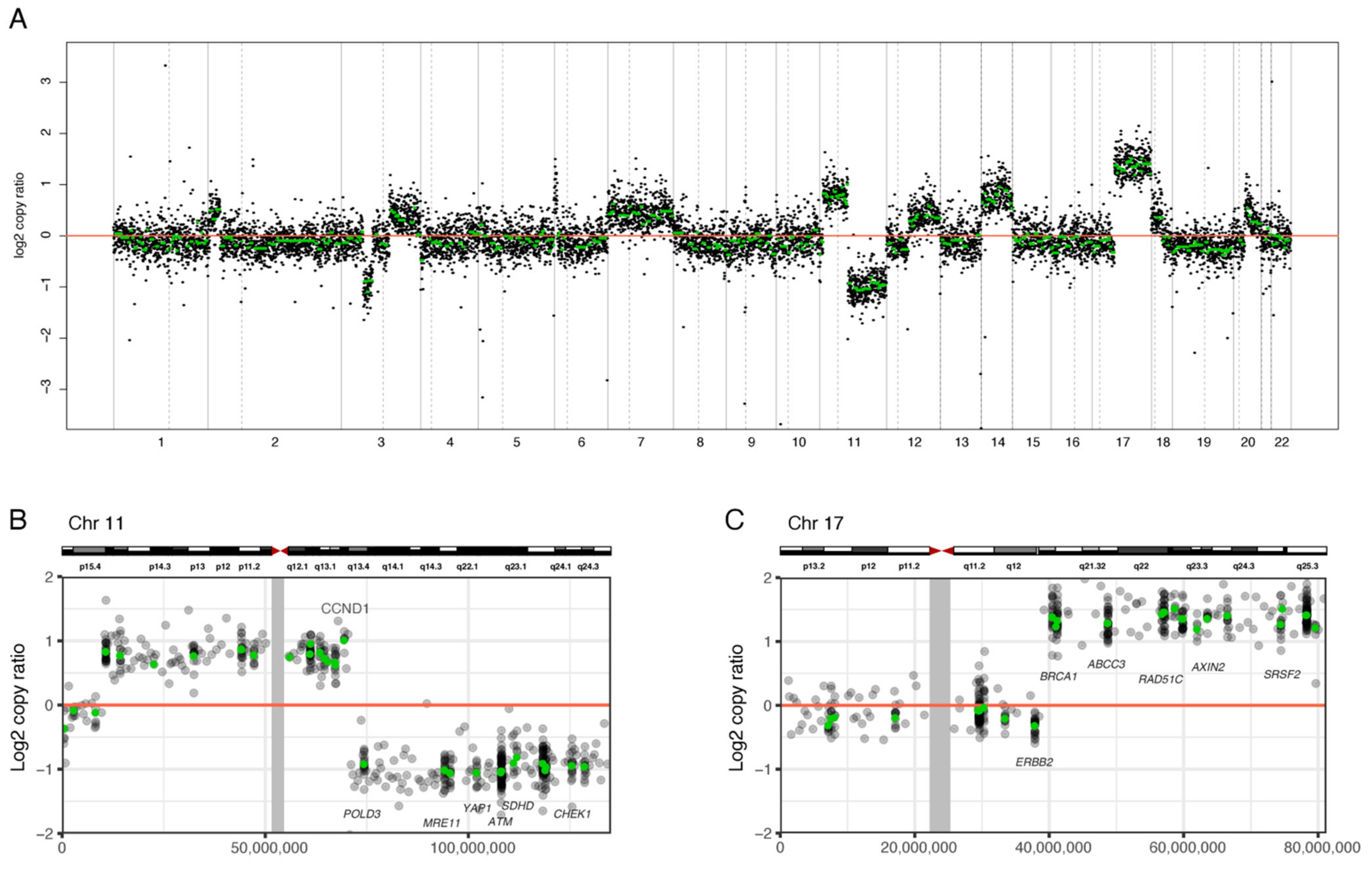
3.2. NGS Analysis of SCAs of the Whole Chromosome Set
3.3. Comparison of SCAs by FISH and NGS
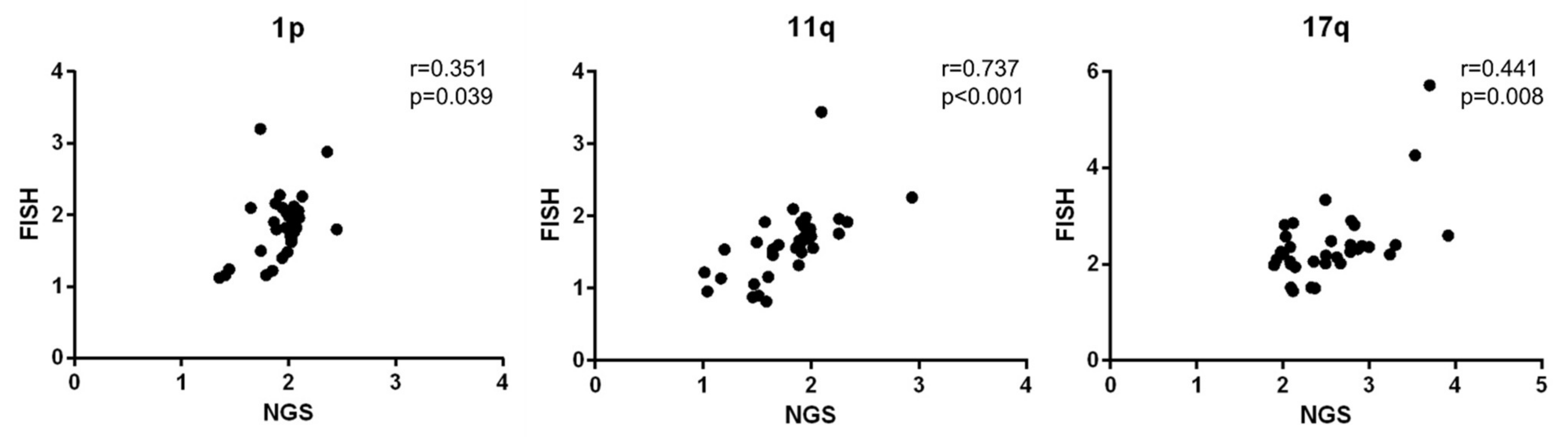
3.4. Copy Number Ratio of 1p, 11q, and 17q Measured by NGS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Mph, K.D.M.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Cohn, S.L.; Pearson, A.D.J.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D.; et al. The International Neuroblastoma Risk Group (INRG) Classification System: An INRG Task Force Report. J. Clin. Oncol. 2009, 27, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Deyell, R.; Attiyeh, E.F. Advances in the understanding of constitutional and somatic genomic alterations in neuroblastoma. Cancer Genet. 2011, 204, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Schleiermacher, G.; Mosseri, V.; London, W.B.; Maris, J.M.; Brodeur, G.M.; Attiyeh, E.; Haber, M.; Khan, J.; Nakagawara, A.; Speleman, F.; et al. Segmental chromosomal alterations have prognostic impact in neuroblastoma: A report from the INRG project. Br. J. Cancer 2012, 107, 1418–1422. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M. Recent Advances in Neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef]
- Janoueix-Lerosey, I.; Schleiermacher, G.; Michels, E.; Mosseri, V.; Ribeiro, A.; Lequin, D.; Vermeulen, J.; Couturier, J.; Peuchmaur, M.; Valent, A.; et al. Overall Genomic Pattern Is a Predictor of Outcome in Neuroblastoma. J. Clin. Oncol. 2009, 27, 1026–1033. [Google Scholar] [CrossRef]
- Gozzetti, A.; Le Beau, M.M. Fluorescence in situ hybridization: Uses and limitations. Semin. Hematol. 2000, 37, 320–333. [Google Scholar] [CrossRef]
- Meyerson, M.; Gabriel, S.; Getz, G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 2010, 11, 685–696. [Google Scholar] [CrossRef]
- Luthra, R.; Patel, K.P.; Routbort, M.J.; Broaddus, R.R.; Yau, J.; Simien, C.; Chen, W.; Hatfield, D.Z.; Medeiros, L.J.; Singh, R.R. A Targeted High-Throughput Next-Generation Sequencing Panel for Clinical Screening of Mutations, Gene Amplifications, and Fusions in Solid Tumors. J. Mol. Diagn. 2017, 19, 255–264. [Google Scholar] [CrossRef]
- Greengard, E.G. Molecularly Targeted Therapy for Neuroblastoma. Children 2018, 5, 142. [Google Scholar] [CrossRef]
- Lim, H.; Son, M.H.; Hyun, J.K.; Cho, H.W.; Ju, H.Y.; Lee, J.W.; Yoo, K.H.; Sung, K.W.; Koo, H.H. Clinical Significance of Segmental Chromosomal Aberrations in Patients with Neuroblastoma: First Report in Korean Population. J. Korean Med. Sci. 2020, 35, e82. [Google Scholar] [CrossRef]
- Choi, Y.B.; Bae, G.; Lee, N.H.; Kim, J.-S.; Lee, S.H.; Yoo, K.H.; Sung, K.W.; Koo, H.H. Clinical Significance of Persistent Tumor in Bone Marrow during Treatment of High-risk Neuroblastoma. J. Korean Med. Sci. 2015, 30, 1062–1067. [Google Scholar] [CrossRef] [PubMed]
- Shimada, H.; Ambros, I.M.; Dehner, L.P.; Hata, J.; Joshi, V.V.; Roald, B.; Stram, D.O.; Gerbing, R.B.; Lukens, J.N.; Matthay, K.K.; et al. The International Neuroblastoma Pathology Classification (the Shimada system). Cancer 1999, 86, 364–372. [Google Scholar] [CrossRef]
- Castel, V.; García-Miguel, P.; Cañete, A.; Melero, C.; Navajas, A.; Ruíz-Jiménez, J.; Navarro, S.; Badal, M. Prospective evaluation of the International Neuroblastoma Staging System (INSS) and the International Neuroblastoma Response Criteria (INRC) in a multicentre setting. Eur. J. Cancer 1999, 35, 606–611. [Google Scholar] [CrossRef]
- Ambros, P.F.; Ambros, I.M.; Brodeur, G.M.; Haber, M.; Khan, J.; Nakagawara, A.; Schleiermacher, G.; Speleman, F.; Spitz, R.; London, W.B.; et al. International consensus for neuroblastoma molecular diagnostics: Report from the International Neuroblastoma Risk Group (INRG) Biology Committee. Br. J. Cancer 2009, 100, 1471–1482. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Lee, J.W.; Shim, J.H.; Joung, J.-G.; Yun, J.W.; Bae, J.S.; Shin, H.-T.; Sung, K.W.; Park, W.-Y. Clinical Relevance of Genomic Changes in Recurrent Pediatric Solid Tumors. Transl. Oncol. 2018, 11, 1390–1397. [Google Scholar] [CrossRef]
- Schleiermacher, G.; Michon, J.; Huon, I.; D’Enghien, C.D.; Klijanienko, J.; Brisse, H.; Ribeiro, A.; Mosseri, V.; Rubie, H.; Munzer, C.; et al. Chromosomal CGH identifies patients with a higher risk of relapse in neuroblastoma without MYCN amplification. Br. J. Cancer 2007, 97, 238–246. [Google Scholar] [CrossRef]
- Fischer, M.; Bauer, T.; Oberthür, A.; Hero, B.; Theissen, J.; Ehrich, M.; Spitz, R.; Eils, R.; Westermann, F.; Brors, B.; et al. Integrated genomic profiling identifies two distinct molecular subtypes with divergent outcome in neuroblastoma with loss of chromosome 11q. Oncogene 2009, 29, 865–875. [Google Scholar] [CrossRef][Green Version]
- Bown, N.; Cotterill, S.; Lastowska, M.; O’Neill, S.; Pearson, A.D.J.; Plantaz, D.; Meddeb, M.; Danglot, G.; Brinkschmidt, C.; Christiansen, H.; et al. Gain of Chromosome Arm 17q and Adverse Outcome in Patients with Neuroblastoma. N. Engl. J. Med. 1999, 340, 1954–1961. [Google Scholar] [CrossRef] [PubMed]
- Attiyeh, E.F.; London, W.B.; Mossé, Y.P.; Wang, Q.; Winter, C.; Khazi, D.; McGrady, P.W.; Seeger, R.C.; Look, A.T.; Shimada, H.; et al. Chromosome 1p and 11q Deletions and Outcome in Neuroblastoma. N. Engl. J. Med. 2005, 353, 2243–2253. [Google Scholar] [CrossRef] [PubMed]
- Franke, F.; Christiansen, H.; Harbott, J.; Lampert, F. Tumour karyotype may be important in the prognosis of human neuroblastoma. J. Cancer Res. Clin. Oncol. 1986, 111, 266–272. [Google Scholar] [CrossRef]
- Maris, J.M.; White, P.S.; Beltinger, C.P.; Sulman, E.P.; Castleberry, R.P.; Shuster, J.J.; Look, A.T.; Brodeur, G.M. Significance of chromosome 1p loss of heterozygosity in neuroblastoma. Cancer Res. 1995, 55, 4664–4669. [Google Scholar]
- Carén, H.; Kryh, H.; Nethander, M.; Sjöberg, R.-M.; Träger, C.; Nilsson, S.; Abrahamsson, J.; Kogner, P.; Martinsson, T. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc. Natl. Acad. Sci. USA 2010, 107, 4323–4328. [Google Scholar] [CrossRef] [PubMed]
- Nakagawara, A.; Li, Y.; Izumi, H.; Muramori, K.; Inada, H.; Nishi, M. Neuroblastoma. Jpn. J. Clin. Oncol. 2018, 48, 214–241. [Google Scholar] [CrossRef] [PubMed]
- Fusco, P.; Esposito, M.R.; Tonini, G.P. Chromosome instability in neuroblastoma (Review). Oncol. Lett. 2018, 16, 6887–6894. [Google Scholar] [CrossRef] [PubMed]
- Cunsolo, C.L.; Bicocchi, M.P.; Petti, A.R.; Tonini, G.P. Numerical and structural aberrations in advanced neuroblastoma tumours by CGH analysis; survival correlates with chromosome 17 status. Br. J. Cancer 2000, 83, 1295–1300. [Google Scholar] [CrossRef][Green Version]
- Oostlander, A.; Meijer, G.; Ylstra, B. Microarray-based comparative genomic hybridization and its applications in human genetics. Clin. Genet. 2004, 66, 488–495. [Google Scholar] [CrossRef]
- Tan, D.S.P.; Lambros, M.B.K.; Natrajan, R.; Reis-Filho, J.S. Getting it right: Designing microarray (and not ‘microawry’) comparative genomic hybridization studies for cancer research. Lab. Investig. 2007, 87, 737–754. [Google Scholar] [CrossRef]
- Worst, B.C.; van Tilburg, C.M.; Balasubramanian, G.P.; Fiesel, P.; Witt, R.; Freitag, A.; Boudalil, M.; Previti, C.; Wolf, S.; Schmidt, S.; et al. Next-generation personalised medicine for high-risk paediatric cancer patients—The INFORM pilot study. Eur. J. Cancer 2016, 65, 91–101. [Google Scholar] [CrossRef]
- Spitz, R.; Oberthuer, A.; Zapatka, M.; Brors, B.; Hero, B.; Ernestus, K.; Oestreich, J.; Fischer, M.; Simon, T.; Berthold, F. Oligonucleotide array-based comparative genomic hybridization (aCGH) of 90 neuroblastomas reveals aberration patterns closely associated with relapse pattern and outcome. Genes Chromosom. Cancer 2006, 45, 1130–1142. [Google Scholar] [CrossRef]
- Theissen, J.; Oberthuer, A.; Hombach, A.; Volland, R.; Hertwig, F.; Fischer, M.; Spitz, R.; Zapatka, M.; Brors, B.; Ortmann, M.; et al. Chromosome 17/17q gain and unaltered profiles in high resolution array-CGH are prognostically informative in neuroblastoma. Genes Chromosom. Cancer 2014, 53, 639–649. [Google Scholar] [CrossRef] [PubMed]

| Number (n = 35) | Median (Range) | |
|---|---|---|
| Age (years) | 3.08 (11.2–0.2) | |
| Sex | ||
| Male | 21 | |
| Female | 14 | |
| Stage | ||
| 1 | 2 | |
| 2 | 6 | |
| 3 | 4 | |
| 4 | 23 | |
| MYCN status | ||
| Amplification | 3 | |
| No amplification | 32 | |
| Risk group | ||
| High risk | 22 | |
| Non high risk | 13 | |
| Primary site | ||
| Abdomen | 27 | |
| Mediastinum | 8 | |
| Diagnosis | ||
| Neuroblastoma, differentiating | 7 | |
| Neuroblastoma, poorly differentiated | 17 | |
| Neuroblastoma, undifferentiated | 1 | |
| Ganglioneuroblastoma | 6 | |
| Ganglioneuroblastoma, intermixed type (GNB-I) | 3 | |
| Ganglioneuroblastoma, nodular type (GNB-N) | 1 | |
| INPC | ||
| Favorable | 21 | |
| Non-favorable | 14 | |
| LDH (IU/L) | 590 (239–7789) | |
| Ferritin (ng/mL) | 93.3 (8.3–3771.8) | |
| NSE (ng/mL) | 46.8 (10.1–685) | |
| VMA (mg/day) | 6.9 (0.4–588.4) | |
| Relapse | ||
| Yes | 6 | |
| No | 29 | |
| Death | ||
| Yes | 1 | |
| No | 34 |
| 1p Deletion | FISH | Total | |||||
|---|---|---|---|---|---|---|---|
| Deletion | (%) | No Deletion | (%) | (%) | |||
| NGS | Deletion | 5 | (71.4) | 2 | (28.6) | 7 | |
| (%) | (100) | (6.7) | (20) | ||||
| No deletion | 0 | (0) | 28 | (100) | 28 | ||
| (%) | (0) | (93.3) | (80) | ||||
| Total (%) | 5 (14.3) | 30 (85.7) | 35 (100) | ||||
| 11q deletion | FISH | Total | |||||
| Deletion | (%) | No deletion | (%) | Fail | (%) | ||
| NGS | Deletion | 8 | (61.5) | 4 | (30.8) | 1 | 13 |
| (%) | (88.9) | (16) | (37.1) | ||||
| No deletion | 1 | (4.8) | 21 | (95.2) | 22 | ||
| (%) | (11.1) | (84) | (62.9) | ||||
| Total (%) | 9 (25.7) | 25 (71.4) | 1 (2.9) | 35 (100) | |||
| 17q gain | FISH | Total | |||||
| Gain | (%) | No gain | (%) | (%) | |||
| NGS | Gain | 6 | (40) | 9 | (60) | 15 | |
| (%) | (85.7) | (32.1) | (42.9) | ||||
| No gain | 1 | (5) | 19 | (95) | 20 | ||
| (%) | (14.3) | (67.9) | (57.1) | ||||
| Total (%) | 7 (20) | 28 (80) | 35 (100) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, E.; Lee, B.; Lee, J.W.; Sung, K.W.; Kim, J.-S. Comparison of Next-Generation Sequencing and Fluorescence In Situ Hybridization for Detection of Segmental Chromosomal Aberrations in Neuroblastoma. Diagnostics 2021, 11, 1702. https://doi.org/10.3390/diagnostics11091702
Kim E, Lee B, Lee JW, Sung KW, Kim J-S. Comparison of Next-Generation Sequencing and Fluorescence In Situ Hybridization for Detection of Segmental Chromosomal Aberrations in Neuroblastoma. Diagnostics. 2021; 11(9):1702. https://doi.org/10.3390/diagnostics11091702
Chicago/Turabian StyleKim, Eojin, Boram Lee, Ji Won Lee, Ki Woong Sung, and Jung-Sun Kim. 2021. "Comparison of Next-Generation Sequencing and Fluorescence In Situ Hybridization for Detection of Segmental Chromosomal Aberrations in Neuroblastoma" Diagnostics 11, no. 9: 1702. https://doi.org/10.3390/diagnostics11091702
APA StyleKim, E., Lee, B., Lee, J. W., Sung, K. W., & Kim, J.-S. (2021). Comparison of Next-Generation Sequencing and Fluorescence In Situ Hybridization for Detection of Segmental Chromosomal Aberrations in Neuroblastoma. Diagnostics, 11(9), 1702. https://doi.org/10.3390/diagnostics11091702






