Multi-Solvent Extraction Procedure for the Pioneer Fecal Metabolomic Analysis—Identification of Potential Biomarkers in Stable Kidney Transplant Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Volunteer Recruitment and Sample Collection
2.2. Fecal Metabolite Extraction
2.3. Sample Preparation
2.4. UHPLC Analysis
2.5. Peak Identification by GC/MS
2.6. GC-MS Analysis
2.7. Identification and Comparison of Volatile Compounds
2.8. Statistical Analysis Approaches for Metabolic Change Detection
3. Results
3.1. Patient Characteristics
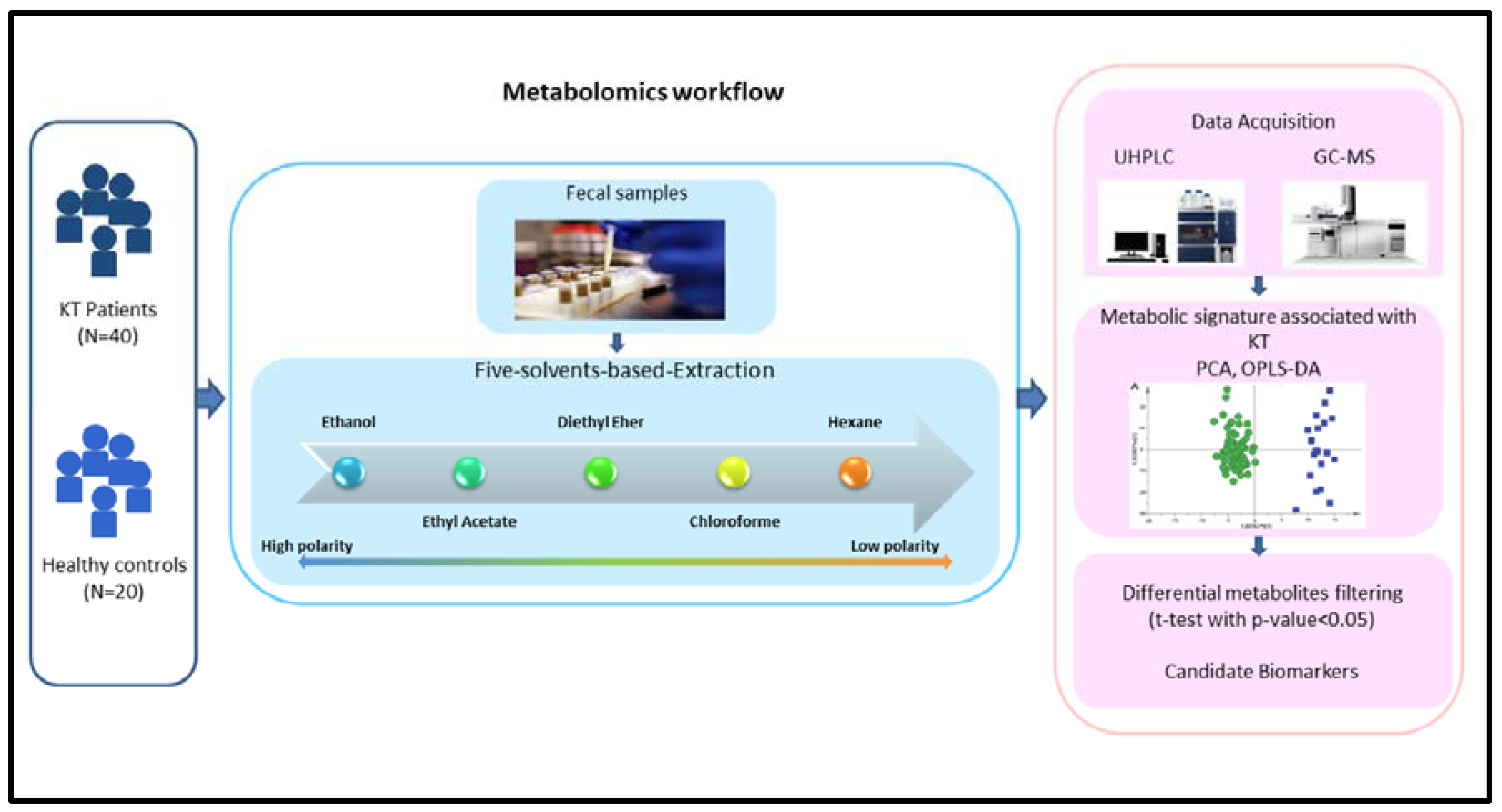
3.2. Metabolomics Workflow
3.3. Dynamic Changes in Unknown Fecal Metabolic Patterns in Kidney Transplant Patients
3.4. Fecal Metabolic Differences between Kidney Transplant Patients and Healthy Individuals and the Selection of Potential Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marcén, R. Immunosuppressive drugs in kidney transplantation: Impact on patient survival, and incidence of cardiovascular disease, malignancy and infection. Drugs 2009, 69, 2227–2243. [Google Scholar] [CrossRef]
- Kaczmarek, I.; Zaruba, M.-M.; Beiras-Fernandez, A.; Reimann, R.; Nickel, T.; Grinninger, C.; Sadoni, S.; Hagl, C.; Meiser, B. Tacrolimus with mycophenolate mofetil or sirolimus compared with calcineurin inhibitor-free immunosuppression (sirolimus/mycophenolate mofetil) after heart transplantation: 5-year results. J. Heart Lung Transplant. 2013, 32, 277–284. [Google Scholar] [CrossRef]
- Monteiro, M.S.; Carvalho, M.; Bastos, M.L.; Guedes de Pinho, P. Metabolomics analysis for biomarker discovery: Advances and challenges. Curr. Med. Chem. 2013, 20, 257–271. [Google Scholar] [CrossRef]
- Yin, P.; Xu, G. Metabolomics toward Biomarker Discovery. Methods Mol. Biol. 2017, 1619, 467–475. [Google Scholar] [CrossRef]
- Zhang, Q.; Yin, X.; Wang, H.; Wu, X.; Li, X.; Li, Y.; Zhang, X.; Fu, C.; Li, H.; Qiu, Y. Fecal Metabolomics and Potential Biomarkers for Systemic Lupus Erythematosus. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Loftfield, E.; Vogtmann, E.; Sampson, J.N.; Moore, S.C.; Nelson, H.; Knight, R.; Chia, N.; Sinha, R. Comparison of Collection Methods for Fecal Samples for Discovery Metabolomics in Epidemiologic Studies. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1483–1490. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Y.; Chen, D.-Q.; Chen, L.; Liu, J.-R.; Vaziri, N.D.; Guo, Y.; Zhao, Y.-Y. Microbiome–metabolome reveals the contribution of gut–kidney axis on kidney disease. J. Transl. Med. 2019, 17. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Montemurno, E.; Piccolo, M.; Vannini, L.; Lauriero, G.; Maranzano, V.; Gozzi, G.; Serrazanetti, D.; Dalfino, G.; Gobbetti, M.; et al. Microbiota and Metabolome Associated with Immunoglobulin A Nephropathy (IgAN). PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Yang, Y.; Yin, Y.; Chen, X.; Chen, C.; Xia, Y.; Qi, H.; Baker, P.N.; Zhang, H.; Han, T.-L. Evaluating different extraction solvents for GC-MS based metabolomic analysis of the fecal metabolome of adult and baby giant pandas. Sci. Rep. 2019, 9, 12017. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.F.; Zhu, Q.F.; Guo, N.; Zheng, S.J.; Wang, Y.L.; Wang, J.; Xu, J.; Liu, S.J.; He, K.; Hu, T.; et al. Comprehensive Profiling of Fecal Metabolome of Mice by Integrated Chemical Isotope Labeling-Mass Spectrometry Analysis. Anal Chem. 2018, 90, 3512–3520. [Google Scholar] [CrossRef]
- Sostare, J.; Di Guida, R.; Kirwan, J.; Chalal, K.; Palmer, E.; Dunn, W.B.; Viant, M.R. Comparison of modified Matyash method to conventional solvent systems for polar metabolite and lipid extractions. Anal. Chim. Acta. 2018, 1037, 301–315. [Google Scholar] [CrossRef]
- Castillo-Rodriguez, E.; Fernandez-Prado, R.; Esteras, R.; Perez-Gomez, M.V.; Gracia-Iguacel, C.; Fernandez-Fernandez, B.; Kanbay, M.; Tejedor, A.; Lazaro, A.; Ruiz-Ortega, M.; et al. Impact of Altered Intestinal Microbiota on Chronic Kidney Disease Progression. Toxins 2018, 10, 300. [Google Scholar] [CrossRef]
- Sampaio-Maia, B.; Simões-Silva, L.; Pestana, M.; Araujo, R.; Soares-Silva, I.J. Chapter Three—The Role of the Gut Microbiome on Chronic Kidney Disease. In Advances in Applied Microbiology; Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2016; Volume 96, pp. 65–94. [Google Scholar]
- Onal, E.M.; Afsar, B.; Covic, A.; Vaziri, N.D.; Kanbay, M. Gut microbiota and inflammation in chronic kidney disease and their roles in the development of cardiovascular disease. Hypertens. Res. 2019, 42, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Hecking, M.; Kainz, A.; Werzowa, J.; Haidinger, M.; Döller, D.; Tura, A.; Karaboyas, A.; Hörl, W.H.; Wolzt, M.; Sharif, A.; et al. Glucose metabolism after renal transplantation. Diabetes Care 2013, 36, 2763–2771. [Google Scholar] [CrossRef] [PubMed]
- Masson, P. Quality control techniques for routine analysis with liquid chromatography in laboratories. J. Chromatogr. A. 2007, 1158, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zheng, X.; Zhou, Q.; Villanueva, N.; Nian, W.; Liu, X.; Huan, T. Metabolomics-Based Discovery of Molecular Signatures for Triple Negative Breast Cancer in Asian Female Population. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Davies, R. The metabolomic quest for a biomarker in chronic kidney disease. Clin. Kidney J. 2018, 11, 694–703. [Google Scholar] [CrossRef]
- Kalim, S.; Rhee, E.P. An Overview of Renal Metabolomics. Kidney Int. 2017, 91, 61–69. [Google Scholar] [CrossRef]
- Posada-Ayala, M.; Zubiri, I.; Martin-Lorenzo, M.; Sanz-Maroto, A.; Molero, D.; Gonzalez-Calero, L.; Fernandez-Fernandez, B.; de la Cuesta, F.; Laborde, C.M.; Barderas, M.G.; et al. Identification of a urine metabolomic signature in patients with advanced-stage chronic kidney disease. Kidney Int. 2014, 85, 103–111. [Google Scholar] [CrossRef]
- Suhre, K.; Schwartz, J.E.; Sharma, V.K.; Chen, Q.; Lee, J.R.; Muthukumar, T.; Dadhania, D.M.; Ding, R.; Ikle, D.N.; Bridges, N.D.; et al. Urine Metabolite Profiles Predictive of Human Kidney Allograft Status. JASN 2016, 27, 626–636. [Google Scholar] [CrossRef]
- Chen, D.-Q.; Cao, G.; Chen, H.; Argyopoulos, C.P.; Yu, H.; Su, W.; Chen, L.; Samuels, D.C.; Zhuang, S.; Bayliss, G.P.; et al. Identification of serum metabolites associating with chronic kidney disease progression and anti-fibrotic effect of 5-methoxytryptophan. Nat. Commun. 2019, 10, 1476. [Google Scholar] [CrossRef] [PubMed]
- Deda, O.; Gika, H.G.; Wilson, I.D.; Theodoridis, G.A. An overview of fecal sample preparation for global metabolic profiling. J. Pharm. Biomed. Anal. 2015, 113, 137–150. [Google Scholar] [CrossRef]
- Chen, M.X.; Wang, S.-Y.; Kuo, C.-H.; Tsai, I.-L. Metabolome analysis for investigating host-gut microbiota interactions. J. Formos. Med. Assoc. 2019, 118, S10–S22. [Google Scholar] [CrossRef] [PubMed]
- Abbiss, H.; Maker, G.L.; Trengove, R.D. Metabolomics Approaches for the Diagnosis and Understanding of Kidney Diseases. Metabolites 2019, 9, 34. [Google Scholar] [CrossRef]
- Kouidhi, S.; Zidi, O.; Alhujaily, M.; Souai, N.; Mosbah, A.; Belali, T.M.; Ghedira, K.; El Kossai, I.; El Manaa, J.; Mnif, W.; et al. Fecal Metabolomics Reveals Distinct Profiles of Kidney Transplant Recipients and Healthy Controls. Diagnostics 2021, 11, 807. [Google Scholar] [CrossRef] [PubMed]
- Tambellini, N.P.; Zaremberg, V.; Turner, R.J.; Weljie, A.M. Evaluation of Extraction Protocols for Simultaneous Polar and Non-Polar Yeast Metabolite Analysis Using Multivariate Projection Methods. Metabolites 2013, 3, 592–605. [Google Scholar] [CrossRef]
- Ebshiana, A.A.; Snowden, S.G.; Thambisetty, M.; Parsons, R.; Hye, A.; Legido-Quigley, C. Metabolomic Method: UPLC-q-ToF Polar and Non-Polar Metabolites in the Healthy Rat Cerebellum Using an In-Vial Dual Extraction. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Deda, O.; Chatziioannou, A.C.; Fasoula, S.; Palachanis, D.; Raikos, Ν.; Theodoridis, G.A.; Gika, H.G. Sample preparation optimization in fecal metabolic profiling. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1047, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Moorhead, J.F.; Chan, M.K.; El-Nahas, M.; Varghese, Z. Lipid nephrotoxicity in chronic progressive glomerular and tubulo-interstitial disease. Lancet 1982, 2, 1309–1311. [Google Scholar] [CrossRef]
- Ruan, X.Z.; Varghese, Z.; Moorhead, J.F. An update on the lipid nephrotoxicity hypothesis. Nat. Rev. Nephrol. 2009, 5, 713–721. [Google Scholar] [CrossRef]
- Szczuko, M.; Kaczkan, M.; Drozd, A.; Maciejewska, D.; Palma, J.; Owczarzak, A.; Marczuk, N.; Rutkowski, P.; Małgorzewicz, S. Comparison of Fatty Acid Profiles in a Group of Female Patients with Chronic Kidney Diseases (CKD) and Metabolic Syndrome (MetS)–Similar Trends of Changes, Different Pathophysiology. Int. J. Mol. Sci. 2019, 20, 1719. [Google Scholar] [CrossRef] [PubMed]
- Gollasch, B.; Dogan, I.; Rothe, M.; Gollasch, M.; Luft, F.C. Effects of hemodialysis on blood fatty acids. Physiol. Rep. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-Q.; Chen, H.; Chen, L.; Vaziri, N.D.; Wang, M.; Li, X.-R.; Zhao, Y.-Y. The link between phenotype and fatty acid metabolism in advanced chronic kidney disease. Nephrol. Dial. Transpl. 2017, 32, 1154–1166. [Google Scholar] [CrossRef] [PubMed]
- Debnath, S.; Velagapudi, C.; Redus, L.; Thameem, F.; Kasinath, B.; Hura, C.E.; Lorenzo, C.; Abboud, H.E.; O’Connor, J.C. Tryptophan Metabolism in Patients with Chronic Kidney Disease Secondary to Type 2 Diabetes: Relationship to Inflammatory Markers. Int. J. Tryptophan. Res. 2017, 10. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, Q.; Xia, T.; Fu, S.; Tao, X.; Wen, Y.; Chan, S.; Gao, S.; Xiong, X.; Chen, W. Diagnostic value of plasma tryptophan and symmetric dimethylarginine levels for acute kidney injury among tacrolimus-treated kidney transplant patients by targeted metabolomics analysis. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Badawy, A.A.-B. Kynurenine Pathway of Tryptophan Metabolism: Regulatory and Functional Aspects. Int. J. Tryptophan Res. 2017, 10. [Google Scholar] [CrossRef]
- Aregger, F.; Uehlinger, D.E.; Fusch, G.; Bahonjic, A.; Pschowski, R.; Walter, M.; Schefold, J.C. Increased urinary excretion of kynurenic acid is associated with non-recovery from acute kidney injury in critically ill patients. BMC Nephrol. 2018, 19. [Google Scholar] [CrossRef]
- Zheng, L.; Wang, J.; Gao, W.; Hu, C.; Wang, S.; Rong, R.; Guo, Y.; Zhu, T.; Zhu, D. GC/MS-based urine metabolomics analysis of renal allograft recipients with acute rejection. J. Transl. Med. 2018, 16. [Google Scholar] [CrossRef]
- Liu, S.; Liang, S.; Liu, H.; Chen, L.; Sun, L.; Wei, M.; Jiang, H.; Wang, J. Metabolite Profiling of Feces and Serum in Hemodialysis Patients and the Effect of Medicinal Charcoal Tablets. Kidney Blood Press. Res. 2018, 43, 755–767. [Google Scholar] [CrossRef]
- Bassi, R.; Niewczas, M.A.; Biancone, L.; Bussolino, S.; Merugumala, S.; Tezza, S.; D’Addio, F.; Nasr, M.B.; Valderrama-Vasquez, A.; Usuelli, V.; et al. Metabolomic Profiling in Individuals with a Failing Kidney Allograft. PLoS ONE 2017, 12, e0169077. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Yu, J.; Wang, Y.; Lu, J.; Shang, E.-X.; Zhu, Z.; Guo, J.; Duan, J. Disorder of gut amino acids metabolism during CKD progression is related with gut microbiota dysbiosis and metagenome change. J. Pharm. Biomed. Anal. 2018, 149, 425–435. [Google Scholar] [CrossRef]
- Mishima, E.; Fukuda, S.; Mukawa, C.; Yuri, A.; Kanemitsu, Y.; Matsumoto, Y.; Akiyama, Y.; Fukuda, N.N.; Tsukamoto, H.; Asaji, K.; et al. Evaluation of the impact of gut microbiota on uremic solute accumulation by a CE-TOFMS-based metabolomics approach. Kidney Int. 2017, 92, 634–645. [Google Scholar] [CrossRef] [PubMed]
- Zeisel, S.H.; Warrier, M. Trimethylamine N-Oxide, the Microbiome, and Heart and Kidney Disease. Annu. Rev. Nutr. 2017, 37, 157–181. [Google Scholar] [CrossRef] [PubMed]
- Prokopienko, A.J.; Nolin, T.D. Microbiota-Derived Uremic Retention Solutes: Perpetrators of Altered Nonrenal Drug Clearance in Kidney Disease. Expert Rev. Clin. Pharm. 2018, 11, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhao, F.; Sun, J.; Lin, B.; Zhao, L.; Liu, Y.; Jin, Y.; Li, S.; Li, A.; Wei, Y. Alterations in the gut microbiota and metabolite profiles of thyroid carcinoma patients. Int. J. Cancer. 2019, 144, 2728–2745. [Google Scholar] [CrossRef]
- Guthrie, L.; Wolfson, S.; Kelly, L. The human gut chemical landscape predicts microbe-mediated biotransformation of foods and drugs. eLife 2019, 8, e42866. [Google Scholar] [CrossRef]
- Selma, M.V.; Espín, J.C.; Tomás-Barberán, F.A. Interaction between Phenolics and Gut Microbiota: Role in Human Health. J. Agric. Food Chem. 2009, 57, 6485–6501. [Google Scholar] [CrossRef]




| Subjects | Age (Y) Mean ± SD | Gender | Diet | BMI ± SD | Immunosuppressive Therapy | Period (Y) after Tx Mean ± SD |
|---|---|---|---|---|---|---|
| Patients | 42 ± 6 | 28 M/12 F | Low salt | 23.7 ± 5 | Str/Fk/MMF | 6 ± 5 |
| Controls | 44 ± 5 | 10 M/10 F | Balanced | 20 ± 4 | - | - |
| Extraction | RT | Metabolites | Sub Class | m/z | Chemical Structure | p-Value |
|---|---|---|---|---|---|---|
| Ethanol | 2,4 | Dodecanoic acid | Fatty acids | 103.1/89.1/117.1 | MF: C12H24O2 MW: 200.32 g/mol | <0.001 |
| 5,5 | Ethyl oleate | Dithianes | 43.05/55.12/69.06 | MF: C20H38O2 MW: 310.5 g/mol | <0.001 | |
| 8,9 | trans-O-Dithiane-4,5-diol | Fatty acid ester | 152.98/108.79/44.03 | MF: C4H8O2S2 MW: 152.2 g/mol | <0.001 | |
| 9,8 | Formic acid | Carboxylic acid | 29.02/46.04/45 | MF: CH2O2 MW: 46.025 g/mol | <0.001 | |
| 12,5 | l-Tryptophan | Essential amino acid | 130.07/159.09/232.06 | MF: C11H12N2O2 MW: 204.22 g/mol | <0.001 | |
| 16,08 | 4-Hydroxyphenylpyruvic acid | Benzenoids | 190.1/89.06/116.05 | MF: C9H8O4 MW: 180.16 g/mol | <0.001 | |
| 16,2 | 5alpha-Cholest-7-en-3beta-ol | Sterols | 43.05/386.33/255.22 | MF: C27H46O MW: 386.7 g/mol | <0.001 | |
| 17,6 | Valeric acid | Straight chain fatty acid | 60.02/27.03/29.05 | MF: C5H10O2 MW: 102.13 g/mol | <0.001 | |
| 18,02 | Erucic acid | Long-chain fatty acid | 321.2/303.3/255.1 | MF: C22H42O2 MW: 338.6 g/mol | <0.001 | |
| Ethyl Acetate | 4,1 | Palmitic acid | Long-chain fatty acid | 74/87/143 | MF: C16H32O2 MW: 256.42 g/mol | <0.001 |
| 10,5 | Octadecanoic acid | Long-chain fatty acid | 265.4/283.4/266.5 | MF: C18H36O2 MW: 284.5 g/mol | <0.001 | |
| 13,48 | Isostearic acid | Long-chain fatty acid | 74.04/87.04/255.2 | MF: C18H36O2 MW: 284.5 g/mol | <0.001 | |
| 13,9 | Stigmastanol | Sterols | 43.99/107.71/215.7 | MF: C29H52O MW: 416.7 g/mol | <0.001 | |
| Diethyl Ether | 3,8 | 5beta-Coprostanol | Cholestane steroids | 81.09/95.02/67.1 | MF: C27H48O MW: 388.7 g/mol | <0.001 |
| 10,2 | p-Anisic acid | Benzenoids | 135.04/209.07/165.03 | MF: C8H8O3 MW: 152.15 g/mol | <0.001 | |
| 11,5 | Hentriacontane | Alkanes | 71/85/99 | MF: C31H64 MW: 436.8 g/mol | <0.001 | |
| 16,9 | 13-Methylmyristic acid | Long-chain fatty acid | 73.05/89.04/43.05 | MF: C15H30O2 MW: 242.4 g/mol | <0.001 | |
| Chloroform | 4,03 | 2,4-Di-tert-butylphenol | Benzenoids | 191.99/57.23/163.15 | MF: C14H22O MW: 206.32g/mol | <0.001 |
| 14,56 | Androst-5-ene-3,17-dione | Androstane steroids | 286/177/91 | MF: C19H26O2 MW: 286.4 g/mol | <0.001 | |
| Hexane | 8,8 | Oleic Acid | Long-chain fatty acid | 41/55/43 | MF: C18H34O2 MW: 282.5 g/mol | <0.001 |
| 12,6 | Aspartylglycine ethyl ester | Alkyl-phenylketones | 88.1/70.09/43.12 | MF: C8H14N2O5 MW: 218.21 g/mol | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kouidhi, S.; Souai, N.; Alhujaily, M.; Zidi, O.; Kochbati, A.; Redissi, A.; Belali, T.M.; Kossai, I.E.; El Manaa, J.; Cherif, A.; et al. Multi-Solvent Extraction Procedure for the Pioneer Fecal Metabolomic Analysis—Identification of Potential Biomarkers in Stable Kidney Transplant Patients. Diagnostics 2021, 11, 962. https://doi.org/10.3390/diagnostics11060962
Kouidhi S, Souai N, Alhujaily M, Zidi O, Kochbati A, Redissi A, Belali TM, Kossai IE, El Manaa J, Cherif A, et al. Multi-Solvent Extraction Procedure for the Pioneer Fecal Metabolomic Analysis—Identification of Potential Biomarkers in Stable Kidney Transplant Patients. Diagnostics. 2021; 11(6):962. https://doi.org/10.3390/diagnostics11060962
Chicago/Turabian StyleKouidhi, Soumaya, Nessrine Souai, Muhanad Alhujaily, Oumaima Zidi, Ameni Kochbati, Alaeddine Redissi, Tareg M. Belali, Imene El Kossai, Jamelddine El Manaa, Ameur Cherif, and et al. 2021. "Multi-Solvent Extraction Procedure for the Pioneer Fecal Metabolomic Analysis—Identification of Potential Biomarkers in Stable Kidney Transplant Patients" Diagnostics 11, no. 6: 962. https://doi.org/10.3390/diagnostics11060962
APA StyleKouidhi, S., Souai, N., Alhujaily, M., Zidi, O., Kochbati, A., Redissi, A., Belali, T. M., Kossai, I. E., El Manaa, J., Cherif, A., Mnif, W., & Mosbah, A. (2021). Multi-Solvent Extraction Procedure for the Pioneer Fecal Metabolomic Analysis—Identification of Potential Biomarkers in Stable Kidney Transplant Patients. Diagnostics, 11(6), 962. https://doi.org/10.3390/diagnostics11060962









