Artificial Intelligence in Colorectal Cancer Diagnosis Using Clinical Data: Non-Invasive Approach
Abstract
1. Introduction
2. Proposed Methods
2.1. Machine Learning
Validation Techniques
2.2. Neural Networks
2.2.1. Feedforward Neural Networks
2.2.2. Training of a Neural Network
2.3. Performance Measurement
3. Experiments
3.1. The Data
3.2. CAD Systems Designed
4. Discussion
4.1. Classification Problem Solved with Traditional Machine Learning
4.1.1. Data Preprocessing and Labeling
4.1.2. Obtaining the Response Variable
Absolute Deviation
Weight of Each Predictor
Normalizing the Continuous Response Variable
4.1.3. Labeling the Response Variable
4.1.4. Developing the Machine Learning Model
4.1.5. K-Fold Cross Validation
5-Fold Cross Validation
25-Fold Cross-Validation
50-Fold Cross Validation
4.1.6. Holdout Validation
5% of Data Held out for Validation
15% of Data Held out for Validation
25% of Data Held out for Validation
4.2. Regression Problem Solved with Artificial Neural Networks
4.2.1. Reasoning of the Second Approach
4.2.2. The Architecture of the Network
4.2.3. Examination of the Networks
5. Results
5.1. Classification Problem
5.2. Regression Problem
6. Testing the System
7. Conclusions and Future Goals
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 12, 713–732. [Google Scholar] [CrossRef]
- Rwala, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef]
- Rampun, A.; Wang, H.; Scotney, B.; Morrow, P.; Zwiggelaar, R. Classification of mammographic microcalcification clusters with machine learning confidence levels. In Proceedings of the 14th International Workshop on Breast Imaging, Atlanta, GA, USA, 8–11 July 2018. [Google Scholar]
- Goel, N.; Yadav, A.; Singh, B.M. Medical image processing: A review. In Proceedings of the IEEE Second International Innovative Applications of Computational Intelligence on Power, Energy and Controls with Their Impact on Humanity (CIPECH), Ghaziabad, India, 18–19 November 2016. [Google Scholar]
- Ameling, S.; Wirth, S.; Paulus, D.; Lacey, G.; Vilarino, F. Texture-Based Polyp Detection in Colonoscopy. In Bildverarbeitung für die Medizin 2009: Informatik aktuell; Meinzer, H.P., Deserno, T.M., Handels, H., Tolxdorff, T., Eds.; Springer: Berlin, Heidelberg, 2009; pp. 346–350. [Google Scholar]
- Ali, H.; Sharif, M.; Yasmin, M.; Rehmani, M.H.; Riaz, F. A survey of feature extraction and fusion of deep learning for detection of abnormalities in video endoscopy of gastrointestinal-tract. Artif. Intell. Rev. 2020, 53, 2635–2707. [Google Scholar] [CrossRef]
- Alagappan, M.; Brown, J.R.G.; Mori, Y.; Berzin, T.M. Artificial intelligence in gastrointestinal endoscopy: The future is almost here. World J. Gastrointest. Endosc. 2018, 10, 239–249. [Google Scholar] [CrossRef]
- Gheorghe, G.; Bungau, S.; Ilie, M.; Behl, T.; Vesa, C.M.; Brisc, C.; Bacalbasa, N.; Turi, V.; Costache, R.S.; Diaconu, C.C. Early Diagnosis of Pancreatic Cancer: The Key for Survival. Diagnostics 2020, 10, 869. [Google Scholar] [CrossRef] [PubMed]
- Bel’skaya, L.V.; Sarf, E.A.; Shalygin, S.P.; Postnova, T.V.; Kosenok, V.K. Identification of salivary volatile organic compounds as potential markers of stomach and colorectal cancer: A pilot study. J. Oral Biosci. 2020, 62, 212–221. [Google Scholar] [CrossRef]
- Pang, S.-W.; Awi, N.J.; Armon, S.; Lim, W.-D.; Low, J.-H.; Peh, K.-B.; Peh, S.-C.; Teow, S.-Y. Current Update of Laboratory Molecular Diagnostics Advancement in Management of Colorectal Cancer (CRC). Diagnostics 2020, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsen, M.; Thorlacius-Ussing, L.; Vorum, H.; Moyer, M.P.; Stender, M.T.; Thorlacius-Ussing, O.; Honoré, B. Proteomic Characterization of Colorectal Cancer Cells versus Normal-Derived Colon Mucosa Cells: Approaching Identification of Novel Diagnostic Protein Biomarkers in Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 3466. [Google Scholar] [CrossRef]
- Jaberie, H.; Hosseini, S.V.; Naghibalhossaini, F. Evaluation of Alpha 1-Antitrypsin for the Early Diagnosis of Colorectal Cancer. Pathol. Oncol. Res. 2020, 26, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhou, G.; Wang, H.; Liu, Y.; Chen, B.; Chen, W.; Lin, C.; Wu, S.; Gong, A.; Xu, M. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int. J. Cancer 2019, 146, 2901–2912. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Cai, D.; Li, W.; Yu, T.; Mao, H.; Jiang, S.; Xiao, B. Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer. Clin. Biochem. 2019, 74, 60–68. [Google Scholar] [CrossRef]
- Toiyama, Y.; Okugawa, Y.; Goel, A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem. Biophys. Res. Commun. 2014, 455, 43–57. [Google Scholar] [CrossRef]
- Behrouz Sharif, S.; Hashemzadeh, S.; Mousavi Ardehaie, R.; Eftekharsadat, A.; Ghojazadeh, M.; Mehrtash, A.H.; Estiar, M.A.; Teimoori-Toolabi, L.; Sakhinia, E. Detection of aberrant methylated SEPT9 and NTRK3 genes in sporadic colorectal cancer patients as a potential diagnostic biomarker. Oncol. Lett. 2016, 12, 5335–5343. [Google Scholar] [CrossRef]
- Symonds, E.L.; Pedersen, S.K.; Murray, D.; Byrne, S.E.; Roy, A.; Karapetis, C.; Hollington, P.; Rabbitt, P.; Jones, F.S.; LaPointe, L.; et al. Circulating epigenetic biomarkers for detection of recurrent. Cancer 2020, 126, 1460–1469. [Google Scholar]
- Young, G.P.; Pedersen, S.K.; Mansfield, S.; Murray, D.H.; Baker, R.T.; Rabbitt, P.; Byrne, S.; Bambacas, L.; Hollington, P.; Symonds, E.L. A cross-sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor-derived DNA with CEA for detection of recurrent colorectal cancer. Cancer Med. 2016, 5, 2763–2772. [Google Scholar] [CrossRef]
- Liu, S.; Huang, Z.; Zhang, Q.; Fu, Y.; Cheng, L.; Liu, B.; Liu, X. Profiling of isomer-specific IgG N-glycosylation in cohort of Chinese colorectal cancer patients. BBA Gen. Subj. 2020, 1864, 129510. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Toyama, K.; Koide, Y.; Morise, Z.; Uyama, I.; Takahashi, H. Clinical Significance of Serum Carcinoembryonic Antigen and Carbohydrate Antigen 19-9 Levels Before Surgery and During Postoperative Follow-Up in Colorectal Cancer. Int. Surg. 2018, 103, 322–330. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Q.; Gao, Z.; Xu, X.; Lu, Q.; Wu, Y. Clinical value of detecting IQGAP3, B7-H4 and cyclooxygenase-2 in the diagnosis and prognostic evaluation of colorectal cancer. Cancer Cell Int. 2019, 19, 1–14. [Google Scholar] [CrossRef]
- Azer, S.A. Challenges Facing the Detection of Colonic Polyps: What Can Deep Learning Do? Medicina 2019, 55, 473. [Google Scholar] [CrossRef]
- Curado, M.; Escolano, F.; Lozano, M.A.; Hancock, E.R. Early Detection of Alzheimer’s Disease: Detecting Asymmetries with a Return Random Walk Link Predictor. Entropy 2020, 22, 465. [Google Scholar] [CrossRef] [PubMed]
- Dachena, C.; Casu, S.; Fanti, A.; Lodi, M.B.; Mazzarella, G. Combined Use of MRI, fMRIand Cognitive Data for Alzheimer’s Disease: Preliminary Results. Appl. Sci. 2019, 9, 3156. [Google Scholar] [CrossRef]
- Francisco, E.; Manuel, C.; Miguel, A.; Lozano, E.; Hancook, R. Dirichlet Graph Densifiers. In Structural, Syntactic, and Statistical Pattern Recognition; Robles-Kelly, A., Loog, M., Biggio, B., Escolano, F., Wilson, R., Eds.; S+SSPR 2016; Lecture Notes in Computer Science; Springer: Cham, Germany, 2016; Volume 10029. [Google Scholar] [CrossRef]
- Jianjia, W.; Richard, C.; Wilson, E.; Hancock, R. fMRI Activation Network Analysis Using Bose-Einstein Entropy. In Structural, Syntactic, and Statistical Pattern Recognition; Robles-Kelly, A., Loog, M., Biggio, B., Escolano, F., Wilson, R., Eds.; S+SSPR 2016; Lecture Notes in Computer Science; Springer: Cham, Germany, 2016; Volume 10029. [Google Scholar] [CrossRef]
- Manuel, C.; Francisco, E.; Miguel, A.; Lozano, E.; Hancock, R. Dirichlet densifiers for improved commute times estimation. Pattern Recognit. 2019, 91, 56–68. [Google Scholar]
- Park, H.-C.; Kim, Y.-J.; Lee, S.-W. Adenocarcinoma Recognition in Endoscopy Images Using Optimized Convolutional Neural Networks. Appl. Sci. 2020, 10, 1650. [Google Scholar] [CrossRef]
- Muhammad, W.; Hart, G.R.; Nartowt, B.; Farrell, J.J.; Johung, K.; Liang, Y.; Deng, J. Pancreatic Cancer Prediction through an Artificial Neural Network. Front. Artif. Intell. 2019, 5. [Google Scholar] [CrossRef]
- Sánchez-Peralta, L.F.; Pagador, J.B.; Picón, A.; Calderón, Á.J.; Polo, F.; Andraka, N.; Bilbao, R.; Glover, B.; Saratxaga, C.L.; Sánchez-Margallo, F.M. PICCOLO White-Light and Narrow-Band Imaging Colonoscopic Dataset: A Performance Comparative of Models and Datasets. Appl. Sci. 2020, 10, 8501. [Google Scholar] [CrossRef]
- Mohamad, N.; Zaini, F.; Johari, A.; Yassin, I.; Zabidi, A.; Hassan, H.A. Comparison between Levenberg-Marquardt and Scaled Conjugate Gradient training algorithms for Breast Cancer Diagnosis using MLP. In 6th International Colloquium on Signal Processing & Its Applications (CSPA); Faculty of Electrical Engineering, Universiti Teknologi MARA Malaysia: Malacca City, Malaysia, 2010. [Google Scholar]
- Nielsen, M. Neural Networks and Deep Learning. Available online: http://neuralnetworksanddeeplearning.com (accessed on 14 March 2021).
- Rampun, A.; Wang, H.; Scotney, B.; Morrow, P.; Zwiggelaar, R. Confidence Analysis for Breast Mass Image Classification. In Proceedings of the 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018; pp. 2072–2076. [Google Scholar]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef]
- Mathworks. Available online: https://www.mathworks.com/ (accessed on 10 July 2020).
- Chao, W.-L.; Manickavasagan, H.; Krishna, S.G. Application of Artificial Intelligence in the Detection and Differentiation of Colon Polyps: A Technical Review for Physicians. Diagnostics 2019, 9, 99. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Lee, M.B.; Pham, T.D.; Batchuluun, G.; Arsalan, M.; Park, K.R. Enhanced Image-Based Endoscopic Pathological Site Classification Using an Ensemble of Deep Learning Models. Sensors 2020, 20, 5982. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Tiwari, A. Breast cancer diagnosis using Genetically Optimized Neural Network model. Expert Syst. Appl. 2015, 42, 4611–4620. [Google Scholar] [CrossRef]
- Terrada, O.; Cherradi, B.; Raihani, A.; Bouattane, O. A novel medical diagnosis support system for predicting patients with atherosclerosis diseases. Inform. Med. Unlocked. 2020, 21, 100483. [Google Scholar] [CrossRef]













| K-Fold Cross Validation | Performance Measures of Models | ||||||
|---|---|---|---|---|---|---|---|
| Best Performing Model | Accuracy | Training Time | % of False Negatives | Sensitivity | Specificity | Precision | |
| 5 | Logistic regression | 71.1% | 7.56 s | 6.55% | 89.89% | 36.39% | 72.31% |
| 25 | Logistic Regression | 71.8% | 9 s | 6.22% | 90.41% | 37.34% | 72.72% |
| 50 | SVM | 71.3% | 30 s | 4% | 93.83% | 29.74% | 71.16% |
| % of Data Held out for Validation | Performance Measures of Models | ||||||
|---|---|---|---|---|---|---|---|
| Best Performing Model | Accuracy | Training Time | % of False Negatives | Sensitivity | Specificity | Precision | |
| 5 | Linear Discriminant | 77.8% | 9.7 s | 0% | 100% | 37.5% | 74.35% |
| 15 | SVM | 70.4% | 2.44 s | 3.7% | 94.25% | 25% | 70.08% |
| 25 | SVM | 70.2% | 1.53 s | 2.66% | 95.89% | 22.78% | 69.65% |
| Hidden Layers | Neurons on Each Hidden Layer | Performance Measures of Networks | |||
|---|---|---|---|---|---|
| Number of Epochs | Performance (MSE) | Training Time [s] | Gradient | ||
| 1 | 3 | 41 | 1.94 | 0 s | 6.8 |
| 10 | 26 | 1.2 | 0 s | 10.7 | |
| 20 | 32 | 3.49 | 0 s | 2.62 | |
| 5 | 3 | 39 | 33.4 | 0 s | 57.1 |
| 10 | 16 | 2.15 | 0 s | 6.07 | |
| 20 | 15 | 6.23 × 10−4 | 00:04 s | 6.45 | |
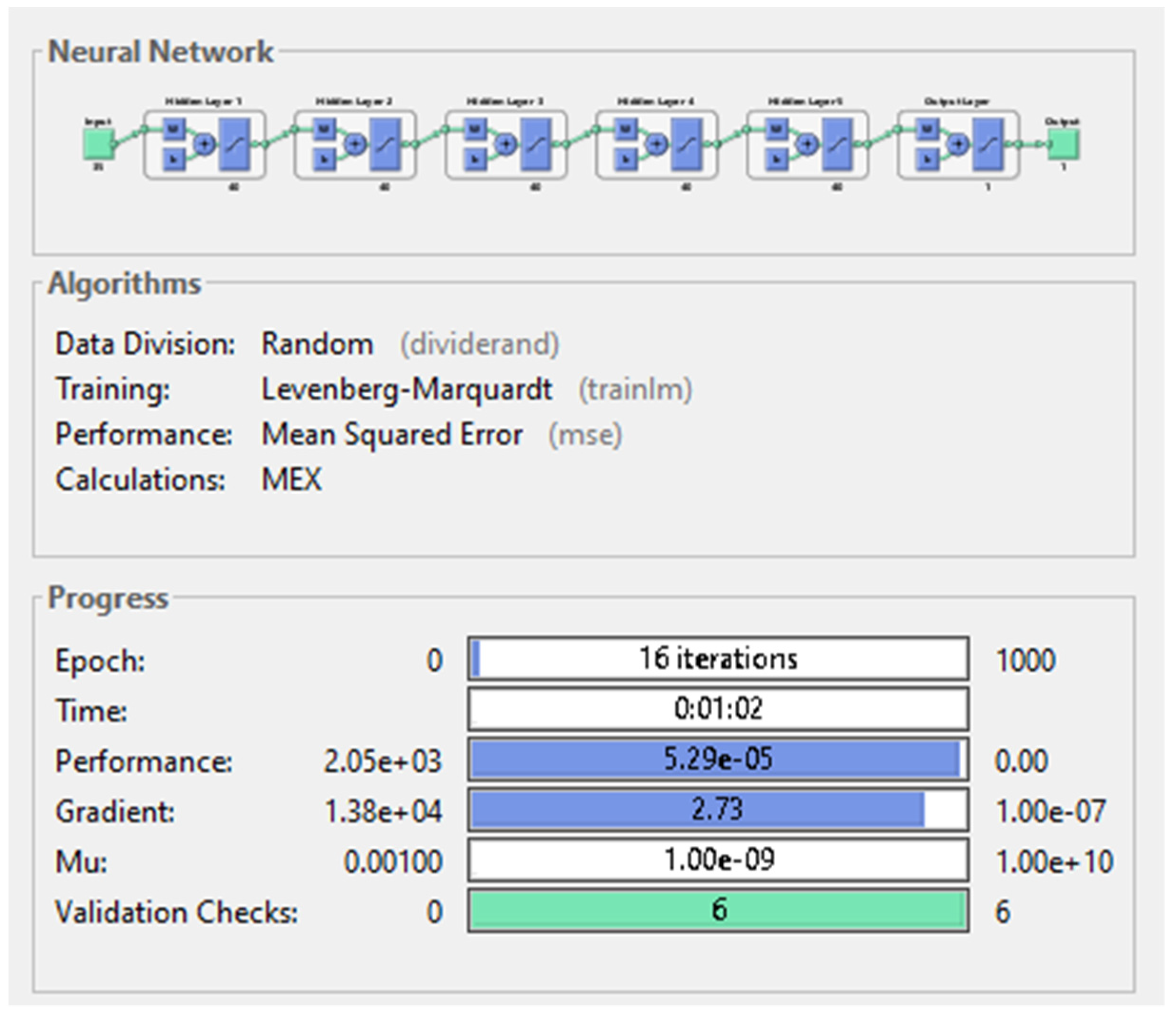
| 40 | 16 | 5.29 × 10−5 | 62 s | 2.73 | |
| 10 | 3 | 56 | 2.9 | 0 s | 25.5 |
| 10 | 20 | 0.508 | 00:02 s | 14.3 | |
| 20 | 14 | 5.9 × 10−3 | 00:17 s | 3.89 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lorenzovici, N.; Dulf, E.-H.; Mocan, T.; Mocan, L. Artificial Intelligence in Colorectal Cancer Diagnosis Using Clinical Data: Non-Invasive Approach. Diagnostics 2021, 11, 514. https://doi.org/10.3390/diagnostics11030514
Lorenzovici N, Dulf E-H, Mocan T, Mocan L. Artificial Intelligence in Colorectal Cancer Diagnosis Using Clinical Data: Non-Invasive Approach. Diagnostics. 2021; 11(3):514. https://doi.org/10.3390/diagnostics11030514
Chicago/Turabian StyleLorenzovici, Noémi, Eva-H. Dulf, Teodora Mocan, and Lucian Mocan. 2021. "Artificial Intelligence in Colorectal Cancer Diagnosis Using Clinical Data: Non-Invasive Approach" Diagnostics 11, no. 3: 514. https://doi.org/10.3390/diagnostics11030514
APA StyleLorenzovici, N., Dulf, E.-H., Mocan, T., & Mocan, L. (2021). Artificial Intelligence in Colorectal Cancer Diagnosis Using Clinical Data: Non-Invasive Approach. Diagnostics, 11(3), 514. https://doi.org/10.3390/diagnostics11030514







