Serum Extracellular Vesicle-Derived miRNAs in Patients with Non-Small Cell Lung Cancer—Search for Non-Invasive Diagnostic Biomarkers
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Ethics
2.2. Patient and Control Group Selection
2.3. Serum Collection
2.4. Extracellular Vesicles Isolation from Serum
2.5. RNA Isolation from EVs, Qualitative and Quantitative RNA Assessment
2.6. Reverse Transcription, Real-Time Quantitative PCR and miRNAs Level
2.7. Statistical Analysis
3. Results
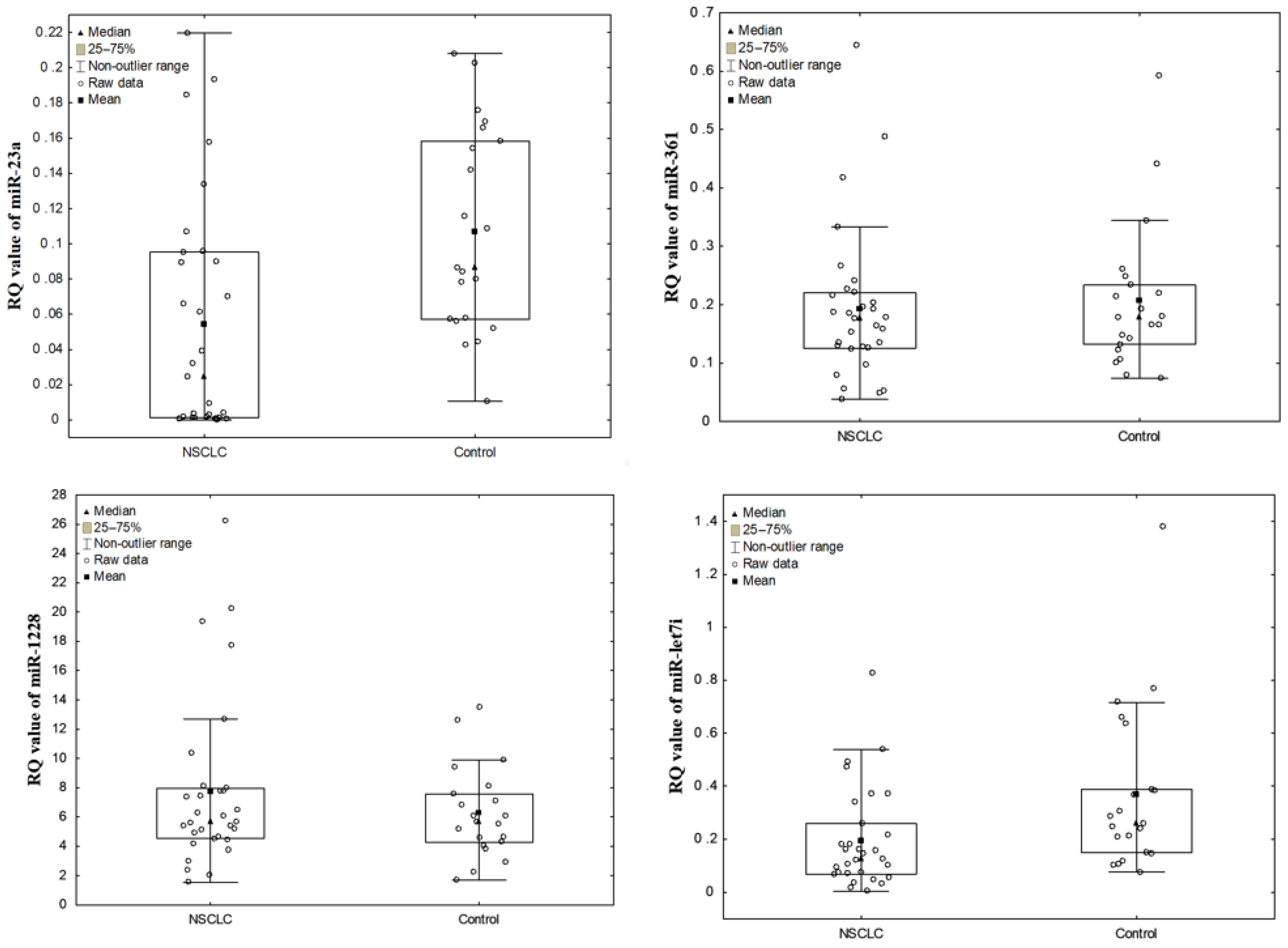
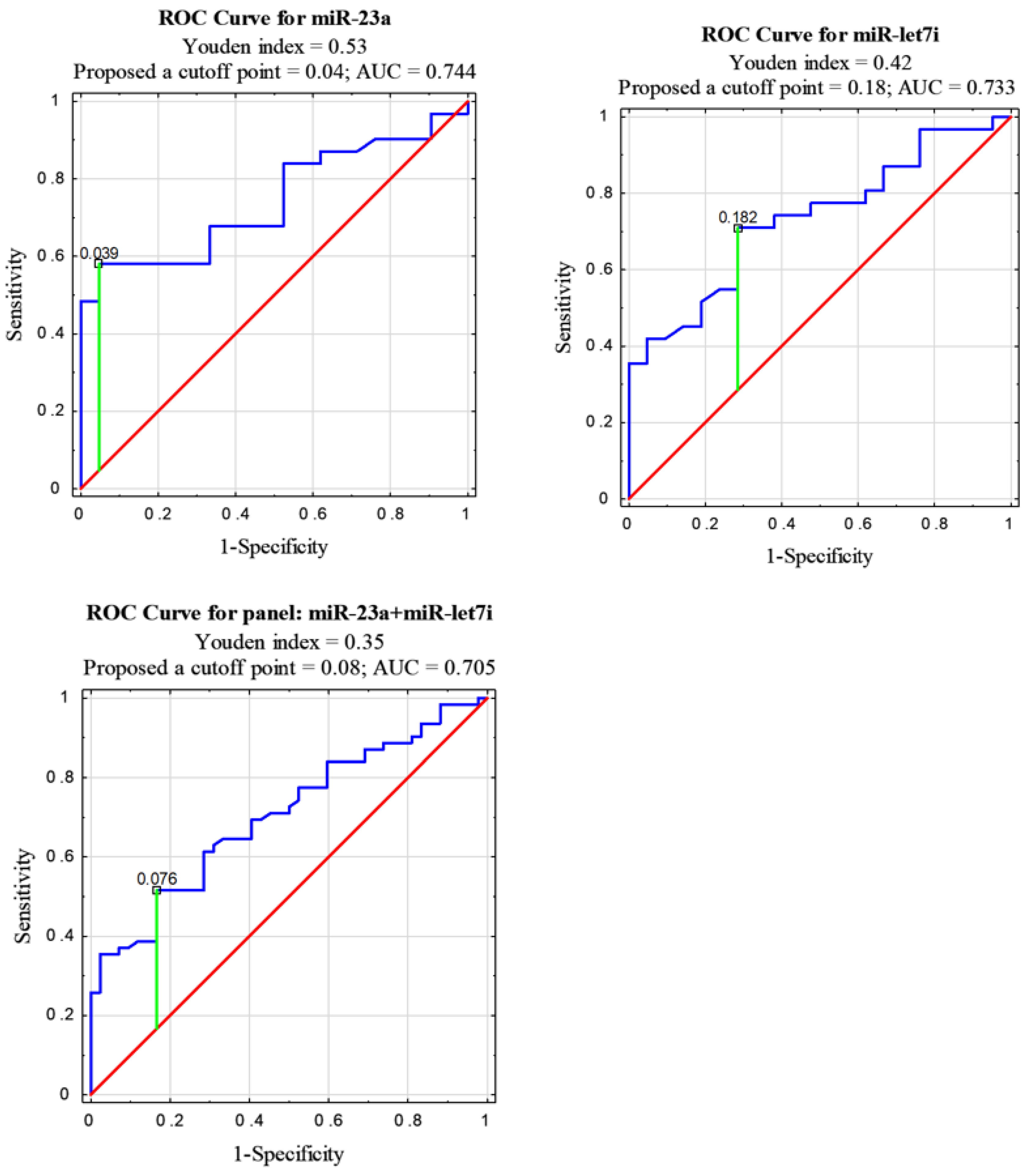
3.1. The Levels of NSCLC EV-Derived miRNAs vs. Control
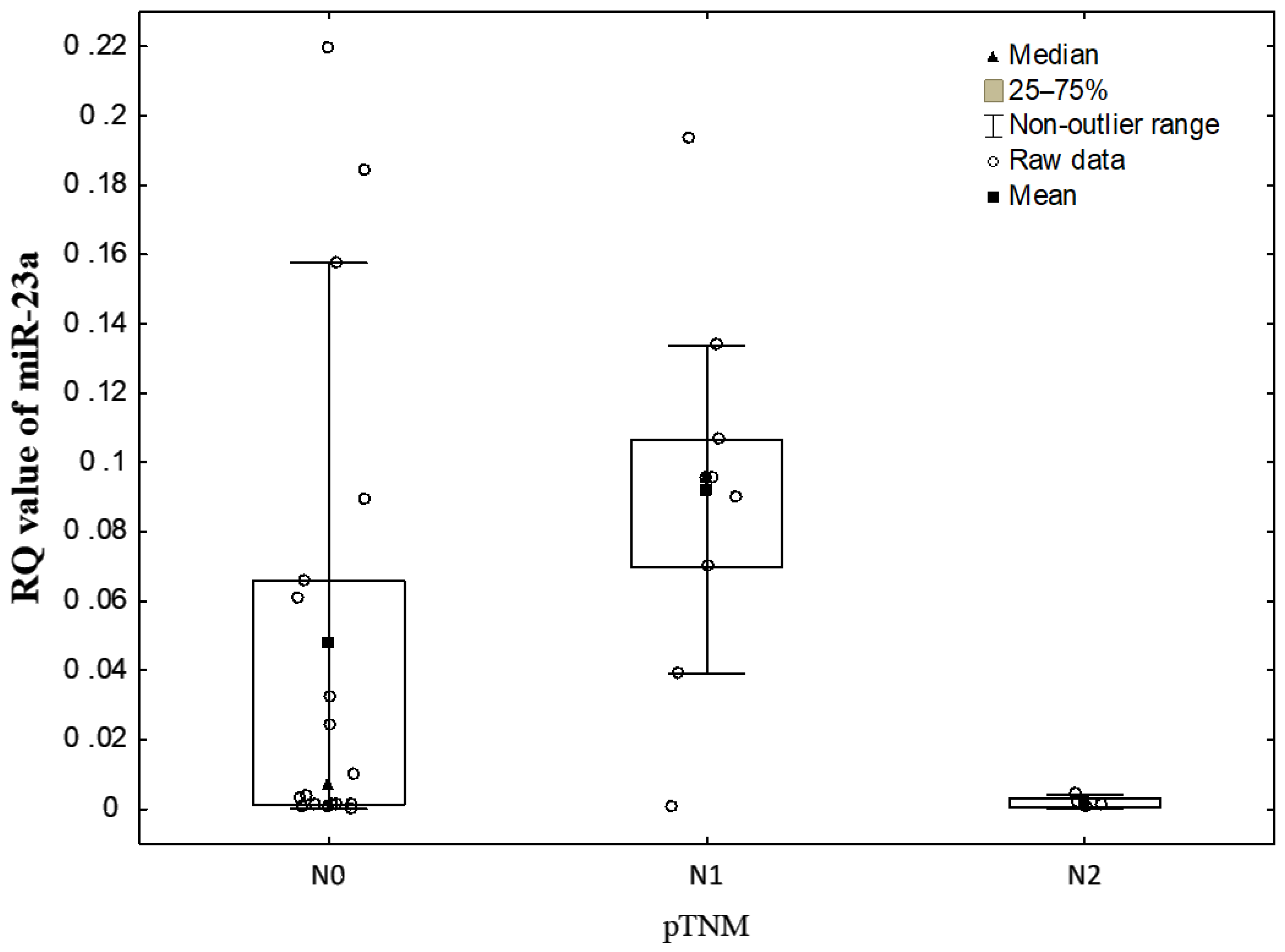
3.2. Correlations between EV-Derived miRNA Levels and Clinicopathological Characteristics of NSCLC Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, W.; Wang, J.; Liu, S.; Wang, S.; Cheng, Y.; Zhou, W.; Duan, C.; Zhang, C. MicroRNA-361-3p suppresses tumor cell proliferation and metastasis by directly targeting SH2B1 in NSCLC. J. Exp. Clin. Cancer Res. 2016, 35, 1–16. [Google Scholar] [CrossRef]
- Wang, P.; Lv, H.; Zhou, D.; Zhang, E. miR-204 suppresses non-small-cell lung carcinoma (NSCLC) invasion and migration by targeting JAK2. Genet. Mol. Res. 2016, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Arab, A.; Karimipoor, M.; Irani, S.; Kiani, A.; Zeinali, S.; Tafsiri, E.; Sheikhy, K. Potential circulating miRNA signature for early detection of NSCLC. Cancer Genet. 2017, 216, 150–158. [Google Scholar] [CrossRef]
- Qu, W.-Q.; Liu, L.; Yu, Z. Clinical value of microRNA-23a upregulation in non-small cell lung cancer. Int. J. Clin. Exp. Med. 2015, 8, 13598–13603. [Google Scholar]
- Yang, S.; Zhang, Y.; Zhao, X.; Wang, J.; Shang, J. microRNA-361 targets Wilms’ tumor 1 to inhibit the growth, migration and invasion of non-small-cell lung cancer cells. Mol. Med. Rep. 2016, 14, 5415–5421. [Google Scholar] [CrossRef] [PubMed]
- Soung, Y.H.; Ford, S.; Zhang, V.; Chung, J. Exosomes in Cancer Diagnostics. Cancers 2017, 9, 8. [Google Scholar] [CrossRef]
- Mathew, M.; Zade, M.; Mezghani, N.; Patel, R.; Wang, Y.; Momen-Heravi, F. Extracellular Vesicles as Biomarkers in Cancer Immunotherapy. Cancers 2020, 12, 2825. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Chen, Y.; Li, A.; Tan, C.; Liu, X. Exosomes play roles in sequential processes of tumor metastasis. Int. J. Cancer 2019, 144, 1486–1495. [Google Scholar] [CrossRef]
- Giallombardo, M.; Borrás, J.C.; Castiglia, M.; Van Der Steen, N.; Mertens, I.; Pauwels, P.; Peeters, M.; Rolfo, C. Exosomal miRNA Analysis in Non-small Cell Lung Cancer (NSCLC) Patients’ Plasma Through qPCR: A Feasible Liquid Biopsy Tool. J. Vis. Exp. 2016. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Yu, S.; Cao, H.; Shen, B.; Feng, J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget 2015, 6, 37151–37168. [Google Scholar] [CrossRef]
- Zhang, L.; Li, J.; Wang, Q.; Meng, G.; Lv, X.; Zhou, H.; Li, W.; Zhang, J. The relationship between microRNAs and the STAT3-related signaling pathway in cancer. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Cao, M.; Li, Y.; Lu, H.; Meng, Q.; Wang, L.; Cai, L.; Dong, X. miR-23a-mediated migration/invasion is rescued by its target, IRS-1, in non-small cell lung cancer cells. J. Cancer Res. Clin. Oncol. 2014, 140, 1661–1670. [Google Scholar] [CrossRef]
- An, Z.; Ren, J.; Yang, G.; Zhang, W.; Yu, C. MicroRNA let-7: Regulation, single nucleotide polymorphism, and therapy in lung cancer. J. Cancer Res. Ther. 2015, 11, 1. [Google Scholar] [CrossRef]
- Ma, Y.; Bao, C.; Kong, R.; Xing, X.; Zhang, Y.; Li, S.; Zhang, W.; Jiang, J.; Zhang, J.; Qiao, Z.; et al. MicroRNA-361-5p suppresses cancer progression by targeting signal transducer and activator of transcription 6 in non-small cell lung cancer. Mol. Med. Rep. 2015, 12, 7367–7373. [Google Scholar] [CrossRef]
- Roth, C.; Stückrath, I.; Pantel, K.; Izbicki, J.R.; Tachezy, M.; Schwarzenbach, H. Low Levels of Cell-Free Circulating miR-361-3p and miR-625 as Blood-Based Markers for Discriminating Malignant from Benign Lung Tumors. PLoS ONE 2012, 7, e38248. [Google Scholar] [CrossRef]
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non–Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.-X.; Zhang, M.-Y.; Li, R.; Liu, X.; Yin, Y.-H.; Qu, Y.-Q. Serum miR-1228-3p and miR-181a-5p as Noninvasive Biomarkers for Non-Small Cell Lung Cancer Diagnosis and Prognosis. BioMed Res. Int. 2020, 2020, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hetta, H.F.; Zahran, A.M.; Shafik, E.A.; El-Mahdy, R.I.; Mohamed, N.A.; Nabil, E.E.; Esmaeel, H.M.; Alkady, O.A.; Elkady, A.; Mohareb, D.A.; et al. Circulating miRNA-21 and miRNA-23a Expression Signature as Potential Biomarkers for Early Detection of Non-Small-Cell Lung Cancer. microRNA 2019, 8, 206–215. [Google Scholar] [CrossRef]
- Mei, Y.; Si, J.; Wang, Y.; Huang, Z.; Zhu, H.; Feng, S.; Wu, X.; Wu, L. Long Noncoding RNA GAS5 Suppresses Tumorigenesis by Inhibiting miR-23a Expression in Non-Small Cell Lung Cancer. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Zhou, X.; Li, S.; Qin, Y.; Chen, Y.; Liu, H. Inhibition of miR-23a increases the sensitivity of lung cancer stem cells to erlotinib through PTEN/PI3K/Akt pathway. Oncol. Rep. 2017, 38, 3064–3070. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Liu, S.; Dai, K.; Jin, L.; He, T.; Pan, X.; Lai, Y. MicroRNA-23a/24-2/27a as a potential diagnostic biomarker for cancer: A systematic review and meta-analysis. Mol. Clin. Oncol. 2017, 8, 159–169. [Google Scholar] [CrossRef]
- Fassina, A.; Cappellesso, R.; Fassan, M. Classification of Non-small Cell Lung Carcinoma in Transthoracic Needle Specimens Using MicroRNA Expression Profiling. Chest 2011, 140, 1305–1311. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ding, Z.; Zhang, C.; Zhang, X.; Meng, Q.; Wu, S.; Wang, S.; Yin, L.; Pu, Y.; Chen, R. MicroRNA-1228* inhibit apoptosis in A549 cells exposed to fine particulate matter. Environ. Sci. Pollut. Res. 2016, 23, 10103–10113. [Google Scholar] [CrossRef]
- Wu, X.; Xi, X.; Yan, Q.; Zhang, Z.; Cai, B.; Lu, W.; Wan, X. MicroRNA-361-5p facilitates cervical cancer progression through mediation of epithelial-to-mesenchymal transition. Med. Oncol. 2013, 30, 1–12. [Google Scholar] [CrossRef]
- Ma, R.; Wang, C.; Wang, J.; Wang, N.; Xu, J. miRNA–mRNA Interaction Network in Non-small Cell Lung Cancer. Interdiscip. Sci. Comput. Life Sci. 2015, 8, 209–219. [Google Scholar] [CrossRef]
- Geretto, M.; Pulliero, A.; Rosano, C.; Zhabayeva, D.; Bersimbaev, R.; Izzotti, A. Resistance to cancer chemotherapeutic drugs is determined by pivotal microRNA regulators. Am. J. Cancer Res. 2017, 7, 1350–1371. [Google Scholar] [PubMed]
- Cao, M.; Seike, M.; Soeno, C.; Mizutani, H.; Kitamura, K.; Minegishi, Y.; Noro, R.; Yoshimura, A.; Cai, L.; Gemma, A. MiR-23a regulates TGF-β-induced epithelial-mesenchymal transition by targeting E-cadherin in lung cancer cells. Int. J. Oncol. 2012, 41, 869–875. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Z.; Liao, B.-Y.; Yu, L.; Gao, X.; Lu, S.; Wang, S.; Dai, Z.; Zhang, X.; Chen, Q.; et al. Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. Int. J. Cancer 2014, 135, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Kanitz, A.; Imig, J.; Dziunycz, P.J.; Primorac, A.; Galgano, A.; Hofbauer, G.F.L.; Gerber, A.P.; Detmar, M. The Expression Levels of MicroRNA-361-5p and Its Target VEGFA Are Inversely Correlated in Human Cutaneous Squamous Cell Carcinoma. PLoS ONE 2012, 7, e49568. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Dong, P.; Xiong, Y.; Yue, J.; Ihira, K.; Konno, Y.; Kobayashi, N.; Todo, Y.; Watari, H. MicroRNA-361: A Multifaceted Player Regulating Tumor Aggressiveness and Tumor Microenvironment Formation. Cancers 2019, 11, 1130. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.-L.; Tian, F.-M.; Sun, C.-L. Downregulation of miR-361-5p associates with aggressive clinicopathological features and unfavorable prognosis in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 5132–5136. [Google Scholar] [PubMed]
- Akao, Y.; Nakagawa, Y.; Naoe, T. let-7 MicroRNA Functions as a Potential Growth Suppressor in Human Colon Cancer Cells. Biol. Pharm. Bull. 2006, 29, 903–906. [Google Scholar] [CrossRef]
- Thammaiah, C.K.; Jayaram, S. Role of let-7 family microRNA in breast cancer. Non-Coding RNA Res. 2016, 1, 77–82. [Google Scholar] [CrossRef]
- Wang, X.; Cao, L.; Wang, Y.; Wang, X.; Liu, N.; You, Y. Regulation of let-7 and its target oncogenes (Review). Oncol. Lett. 2012, 3, 955–960. [Google Scholar] [CrossRef]
- Trang, P.; Medina, P.P.; Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Homer, R.; Brown, D.; Bader, A.G.; Weidhaas, J.B.; et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene 2009, 29, 1580–1587. [Google Scholar] [CrossRef]
- Izzotti, A.; Calin, G.A.; Arrigo, P.; Steele, V.E.; Croce, C.M.; De Flora, S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2008, 23, 806–812. [Google Scholar] [CrossRef]
- Xia, X.-M.; Jin, W.-Y.; Shi, R.-Z.; Zhang, Y.-F.; Chen, J. Clinical significance and the correlation of expression between Let-7 and K-ras in non-small cell lung cancer. Oncol. Lett. 2010, 1, 1045–1047. [Google Scholar] [CrossRef] [PubMed]
- NCI Dictionary of Cancer Terms. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms (accessed on 13 January 2020).
- Lim, W.; Ridge, C.A.; Nicholson, A.G.; Mirsadraee, S. The 8th lung cancer TNM classification and clinical staging system: Review of the changes and clinical implications. Quant. Imaging Med. Surg. 2018, 8, 709–718. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, 1–10. [Google Scholar] [CrossRef]
- Akobeng, A.K. Understanding diagnostic tests 3: Receiver operating characteristic curves. Acta Paediatr. 2007, 96, 644–647. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Franklin, W.; Gazdar, A.F.; Bunn, P. Early detection of lung cancer: Clinical perspectives of recent advances in biology and radiology. Clin. Cancer Res. 2001, 7, 5–22. [Google Scholar] [PubMed]
- Pastuszak-Lewandoska, D.; Kordiak, J.; Czarnecka, K.H.; Migdalska-Sęk, M.; Nawrot, E.; Domańska-Senderowska, D.; Kiszałkiewicz, J.M.; Antczak, A.; Górski, P.; Brzeziańska-Lasota, E. Expression analysis of three miRNAs, miR-26a, miR-29b and miR-519d, in relation to MMP-2 expression level in non-small cell lung cancer patients: A pilot study. Med. Oncol. 2016, 33, 1–7. [Google Scholar] [CrossRef]
- Li, L.; Liu, H.; Liu, X.; Hu, H.; Liu, S. Clinical significance of exosomal miRNAs and proteins in three human cancers with high mortality in China (Review). Oncol. Lett. 2018, 17, 11–22. [Google Scholar] [CrossRef]
- Vanni, I.; Alama, A.; Grossi, F.; Bello, M.G.D.; Coco, S. Exosomes: A new horizon in lung cancer. Drug Discov. Today 2017, 22, 927–936. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Seremwe, M.; Dismuke, W.M.; Bieberich, E.; Stamer, W.D.; et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS ONE 2017, 12, e0170628. [Google Scholar] [CrossRef]
- Hsu, Y.-L.; Hung, J.-Y.; Chang, W.-A.; Lin, Y.-S.; Pan, Y.-C.; Tsai, P.-H.; Wu, C.-Y.; Kuo, P.-L. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene 2017, 36, 4929–4942. [Google Scholar] [CrossRef]
- He, Y.; Meng, C.; Shao, Z.; Wang, H.; Yang, S. MiR-23a Functions as a Tumor Suppressor in Osteosarcoma. Cell. Physiol. Biochem. 2014, 34, 1485–1496. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Tan, H.-Y.; Feng, Y.-G.; Zhang, C.; Chen, F.; Feng, Y. microRNA-23a in Human Cancer: Its Roles, Mechanisms and Therapeutic Relevance. Cancers 2018, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.D.; Esquela-Kerscher, A.; Stefani, G.; Byrom, M.; Kelnar, K.; Ovcharenko, D.; Wilson, M.; Wang, X.; Shelton, J.; Shingara, J.; et al. The let-7 MicroRNA Represses Cell Proliferation Pathways in Human Cells. Cancer Res. 2007, 67, 7713–7722. [Google Scholar] [CrossRef]
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef] [PubMed]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS Is Regulated by the let-7 MicroRNA Family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Cherni, I.; Weiss, G.J. miRNAs in lung cancer: Large roles for small players. Future Oncol. 2011, 7, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, S.R.; Thachil, T.; De Ieso, P.; Gee, H.; Moss, S.A.; Milic, N. Prognostic Role of MicroRNAs in Human Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis. Dis. Markers 2018, 2018, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Vucic, E.; Thu, K.L.; Pikor, L.; Enfield, K.S.S.; Yee, J.; English, J.C.; MacAulay, C.; Lam, S.; Jurisica, I.; Lam, W.L. Smoking status impacts microRNA mediated prognosis and lung adenocarcinoma biology. BMC Cancer 2014, 14, 778. [Google Scholar] [CrossRef]
- Izzotti, A.; Longobardi, M.; La Maestra, S.; Micale, R.T.; Pulliero, A.; Camoirano, A.; Geretto, M.; D’Agostini, F.; Balansky, R.; Miller, M.S.; et al. Release of MicroRNAs into Body Fluids from Ten Organs of Mice Exposed to Cigarette Smoke. Theranostics 2018, 8, 2147–2160. [Google Scholar] [CrossRef]
- Suzuki, K.; Yamada, H.; Nagura, A.; Ohashi, K.; Ishikawa, H.; Yamazaki, M.; Ando, Y.; Ichino, N.; Osakabe, K.; Sugimoto, K.; et al. Association of Cigarette Smoking with Serum MicroRNA Expression among Middle-Aged Japanese Adults. Fujita Med. J. 2016, 2, 1–5. [Google Scholar]
- Huang, J.; Wu, J.; Li, Y.; Li, X.; Yang, T.; Yang, Q.; Jiang, Y. Deregulation of Serum MicroRNA Expression Is Associated with Cigarette Smoking and Lung Cancer. BioMed Res. Int. 2014, 2014, 1–13. [Google Scholar] [CrossRef] [PubMed]
| Features | n (%) |
|---|---|
| Histopathological type | n = 31 |
| SCC | 15 (48.5) |
| ADC | 15 (48.5) |
| LCC | 1 (3) |
| Tumor size; T (pTNM staging system) | n = 31 |
| T1a + T1b + T1c | 10 (32) |
| T2a + T2b | 12 (39) |
| T3 + T4 | 8 (26) |
| Tx | 1 (3) |
| Intrathoracic lymph node involvement; N (pTNM staging system) | n = 31 |
| N0 | 18 (58) |
| N1 | 9 (29) |
| N2 | 4 (13) |
| Malignant stage (AJCC staging system) | n = 30 * |
| Stage I | 12 (40) |
| Stage II | 9 (30) |
| Stage III | 9 (30) |
| miRNA | Sample | RQ Value Mean ± SD (Median) | Range of RQ Value | Number (%) of Samples with | |
|---|---|---|---|---|---|
| RQ Value > 1 | RQ Value < 1 | ||||
| miR-23a | NSCLC | 0.054455 ± 0.066 (0.024) | 0.000100–0.219600 | 0 (0) | 31 (100) |
| Control | 0.107062 ± 0.058 (0.087) | 0.010600–0.208100 | 0 (0) | 21 (100) | |
| miR-361 | NSCLC | 0.193529 ± 0.130 (0.177) | 0.038100–0.644600 | 0 (0) | 31 (100) |
| Control | 0.206724 ± 0.124 (0.177) | 0.074100–0.592100 | 0 (0) | 21 (100) | |
| miR-1228 | NSCLC | 7.725716 ± 5.756 (5.661) | 1.551500–26.23720 | 31 (100) | 0 (0) |
| Control | 6.264948 ± 3.098 (5.637) | 1.689000–13.48830 | 21 (100) | 0 (0) | |
| miR-let7i | NSCLC | 0.192923 ± 0.187 (0.128) | 0.004200–0.826300 | 0 (0) | 31 (100) |
| Control | 0.369348 ± 0.312 (0.260) | 0.077400–1.378300 | 1 (95) | 20 (95) | |
| miRNA | AUC | 95% CI | p Value |
|---|---|---|---|
| miR-23a | 0.744 | 0.611–0.877 | 0.0003 |
| let7i | 0.733 | 0.598–0.867 | 0.0007 |
| let7i + miR-23a | 0.705 | 0.606–0.803 | 0.00001 |
| let7i + miR-361 | 0.651 | 0.546–0.756 | 0.0048 |
| let7i + miR-23a + miR-361 | 0.650 | 0.565–0.735 | 0.0005 |
| miR-23a + miR-361 | 0.612 | 0.505–0.718 | 0.0406 |
| let7i + miR-23a + miR-361 + miR-1228 | 0.582 | 0.505–0.659 | 0.0357 |
| miR-23a + miR-1228 | 0.551 | 0.441–0.662 | 0.364 |
| let7i + miR-1228 | 0.548 | 0.438–0.658 | 0.392 |
| miR-361 | 0.538 | 0.378–0.698 | 0.6379 |
| miR-361 + miR-1228 | 0.499 | 0.387–0.612 | 0.992 |
| miR-1228 | 0.459 | 0.3–0.619 | 0.619 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kryczka, J.; Migdalska-Sęk, M.; Kordiak, J.; Kiszałkiewicz, J.M.; Pastuszak-Lewandoska, D.; Antczak, A.; Brzeziańska-Lasota, E. Serum Extracellular Vesicle-Derived miRNAs in Patients with Non-Small Cell Lung Cancer—Search for Non-Invasive Diagnostic Biomarkers. Diagnostics 2021, 11, 425. https://doi.org/10.3390/diagnostics11030425
Kryczka J, Migdalska-Sęk M, Kordiak J, Kiszałkiewicz JM, Pastuszak-Lewandoska D, Antczak A, Brzeziańska-Lasota E. Serum Extracellular Vesicle-Derived miRNAs in Patients with Non-Small Cell Lung Cancer—Search for Non-Invasive Diagnostic Biomarkers. Diagnostics. 2021; 11(3):425. https://doi.org/10.3390/diagnostics11030425
Chicago/Turabian StyleKryczka, Jolanta, Monika Migdalska-Sęk, Jacek Kordiak, Justyna M. Kiszałkiewicz, Dorota Pastuszak-Lewandoska, Adam Antczak, and Ewa Brzeziańska-Lasota. 2021. "Serum Extracellular Vesicle-Derived miRNAs in Patients with Non-Small Cell Lung Cancer—Search for Non-Invasive Diagnostic Biomarkers" Diagnostics 11, no. 3: 425. https://doi.org/10.3390/diagnostics11030425
APA StyleKryczka, J., Migdalska-Sęk, M., Kordiak, J., Kiszałkiewicz, J. M., Pastuszak-Lewandoska, D., Antczak, A., & Brzeziańska-Lasota, E. (2021). Serum Extracellular Vesicle-Derived miRNAs in Patients with Non-Small Cell Lung Cancer—Search for Non-Invasive Diagnostic Biomarkers. Diagnostics, 11(3), 425. https://doi.org/10.3390/diagnostics11030425







