In-House Immunofluorescence Assay for Detection of SARS-CoV-2 Antigens in Cells from Nasopharyngeal Swabs as a Diagnostic Method for COVID-19
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Specimens
2.2. Test Evaluation
2.3. Cells from Viral Culture
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. COVID-19: Case Definitions. Updated in Public Health Surveillance for COVID-19. Published 16 December 2020. Available online: https://apps.who.int/iris/handle/10665/337834 (accessed on 23 July 2021).
- McCullough, P.A.; Kelly, R.J.; Ruocco, G.; Lerma, E.; Tumlin, J.; Wheelan, K.R.; Katz, N.; Lepor, N.E.; Vijay, K.; Carter, H.; et al. Pathophysiological Basis and Rationale for Early Outpatient Treatment of SARS-CoV-2 (COVID-19) Infection. Am. J. Med. 2021, 134, 16–22. [Google Scholar] [CrossRef]
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021, 20, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Feng, J.; Zhang, Q.; Guo, D.; Zhang, L.; Suo, T.; Hu, W.; Guo, M.; Wang, X.; Huang, Z.; et al. Analytical comparisons of SARS-CoV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerg. Microbes Infect. 2020, 9, 1175–1179. [Google Scholar] [CrossRef]
- Huang, C.G.; Lee, K.M.; Hsiao, M.J.; Yang, S.L.; Huang, P.N.; Gong, Y.N.; Hsieh, T.H.; Huang, P.W.; Lin, Y.J.; Liu, Y.C.; et al. Culture-Based Virus Isolation To Evaluate Potential Infectivity of Clinical Specimens Tested for COVID-19. J. Clin. Microbiol. 2020, 58, e01068-20. [Google Scholar] [CrossRef] [PubMed]
- Perera, R.; Tso, E.; Tsang, O.T.Y.; Tsang, D.N.C.; Fung, K.; Leung, Y.W.Y.; Chin, A.W.H.; Chu, D.K.W.; Cheng, S.M.S.; Poon, L.L.M.; et al. SARS-CoV-2 Virus Culture and Subgenomic RNA for Respiratory Specimens from Patients with Mild Coronavirus Disease. Emerg. Infect. Dis. 2020, 26, 2701–2704. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, T.; Spencer, E.A.; Brassey, J.; Heneghan, C. Viral cultures for COVID-19 infectious potential assessment—A systematic review. Clin. Infect. Dis. 2020, 73, e3884–e3899. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Q.; Sun, B.Q.; Fang, Z.F.; Zhao, J.C.; Liu, X.Y.; Li, Y.M.; Sun, X.Z.; Liang, H.F.; Zhong, B.; Huang, Z.F.; et al. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur. Respir. J. 2020, 56, 2001526. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2027–2034. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.X.; Liu, B.Z.; Deng, H.J.; Wu, G.C.; Deng, K.; Chen, Y.K.; Liao, P.; Qiu, J.F.; Lin, Y.; Cai, X.F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, M.; De Rosa, A.; Boccellino, M. Detection of SARS-CoV-2 Proteins Using an ELISA Test. Diagnostics 2021, 11, 698. [Google Scholar] [CrossRef] [PubMed]
- WHO. Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection Using Rapid Immunoassays: Interim Guidance; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Dinnes, J.; Deeks, J.J.; Adriano, A.; Berhane, S.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; Beese, S.; et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020, 8, Cd013705. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Skelly, D.; Goonawardane, N. A Novel Immunofluorescence Assay for the Rapid Serological Detection of SARS-CoV-2 Infection. Viruses 2021, 13, 747. [Google Scholar] [CrossRef] [PubMed]
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar] [CrossRef] [PubMed]
- Arya, R.; Kumari, S.; Pandey, B.; Mistry, H.; Bihani, S.C.; Das, A.; Prashar, V.; Gupta, G.D.; Panicker, L.; Kumar, M. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021, 433, 166725. [Google Scholar] [CrossRef]
- Millet, J.K.; Whittaker, G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015, 202, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Zeng, W.; Liu, G.; Ma, H.; Zhao, D.; Yang, Y.; Liu, M.; Mohammed, A.; Zhao, C.; Yang, Y.; Xie, J.; et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020, 527, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Tugaeva, K.V.; Hawkins, D.; Smith, J.L.R.; Bayfield, O.W.; Ker, D.S.; Sysoev, A.A.; Klychnikov, O.I.; Antson, A.A.; Sluchanko, N.N. The Mechanism of SARS-CoV-2 Nucleocapsid Protein Recognition by the Human 14-3-3 Proteins. J. Mol. Biol. 2021, 433, 166875. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef]
- Chan, K.H.; Sridhar, S.; Zhang, R.R.; Chu, H.; Fung, A.Y.; Chan, G.; Chan, J.F.; To, K.K.; Hung, I.F.; Cheng, V.C.; et al. Factors affecting stability and infectivity of SARS-CoV-2. J. Hosp. Infect. 2020, 106, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Yip, C.C.; Sridhar, S.; Cheng, A.K.; Leung, K.H.; Choi, G.K.; Chen, J.H.; Poon, R.W.; Chan, K.H.; Wu, A.K.; Chan, H.S.; et al. Evaluation of the commercially available LightMix® Modular E-gene kit using clinical and proficiency testing specimens for SARS-CoV-2 detection. J. Clin. Virol. 2020, 129, 104476. [Google Scholar] [CrossRef]
- Chan, J.F.; Yip, C.C.; To, K.K.; Tang, T.H.; Wong, S.C.; Leung, K.H.; Fung, A.Y.; Ng, A.C.; Zou, Z.; Tsoi, H.W.; et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J. Clin. Microbiol. 2020, 58, e00310-20. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Gao, Y.; Liu, X.; Nie, Z.; Sun, H.; Lin, K.; Peng, H.; Wang, S. Identification and functional analysis of the SARS-CoV-2 nucleocapsid protein. BMC Microbiol. 2021, 21, 58. [Google Scholar] [CrossRef] [PubMed]
- Che, X.-Y.; Qiu, L.-W.; Pan, Y.-X.; Wen, K.; Hao, W.; Zhang, L.-Y.; Wang, Y.-D.; Liao, Z.-Y.; Hua, X.; Cheng, V.C.C.; et al. Sensitive and specific monoclonal antibody-based capture enzyme immunoassay for detection of nucleocapsid antigen in sera from patients with severe acute respiratory syndrome. J. Clin. Microbiol. 2004, 42, 2629–2635. [Google Scholar] [CrossRef] [PubMed][Green Version]
- To, K.K.; Tsang, O.T.; Leung, W.S.; Tam, A.R.; Wu, T.C.; Lung, D.C.; Yip, C.C.; Cai, J.P.; Chan, J.M.; Chik, T.S.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef]
- Heimer, G.V.; Taylor, C.E. Improved mountant for immunofluorescence preparations. J. Clin. Pathol. 1974, 27, 254–256. [Google Scholar] [CrossRef] [PubMed]
- Chia, W.N.; Tan, C.W.; Foo, R.; Kang, A.E.Z.; Peng, Y.; Sivalingam, V.; Tiu, C.; Ong, X.M.; Zhu, F.; Young, B.E.; et al. Serological differentiation between COVID-19 and SARS infections. Emerg. Microbes Infect. 2020, 9, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
- Ascoli, C.A.; Aggeler, B. Overlooked benefits of using polyclonal antibodies. Biotechniques 2018, 65, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Lipman, N.S.; Jackson, L.R.; Trudel, L.J.; Weis-Garcia, F. Monoclonal versus polyclonal antibodies: Distinguishing characteristics, applications, and information resources. ILAR J. 2005, 46, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Tso, F.Y.; Lidenge, S.J.; Peña, P.B.; Clegg, A.A.; Ngowi, J.R.; Mwaiselage, J.; Ngalamika, O.; Julius, P.; West, J.T.; Wood, C. High prevalence of pre-existing serological cross-reactivity against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in sub-Saharan Africa. Int. J. Infect. Dis. 2021, 102, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Dobaño, C.; Santano, R.; Jiménez, A.; Vidal, M.; Chi, J.; Rodrigo Melero, N.; Popovic, M.; López-Aladid, R.; Fernández-Barat, L.; Tortajada, M.; et al. Immunogenicity and crossreactivity of antibodies to the nucleocapsid protein of SARS-CoV-2: Utility and limitations in seroprevalence and immunity studies. Transl. Res. 2021, 232, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Pyrc, K.; Sims, A.C.; Dijkman, R.; Jebbink, M.; Long, C.; Deming, D.; Donaldson, E.; Vabret, A.; Baric, R.; van der Hoek, L.; et al. Culturing the unculturable: Human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J. Virol. 2010, 84, 11255–11263. [Google Scholar] [CrossRef]
- Chan, J.F.; Chan, K.H.; Choi, G.K.; To, K.K.; Tse, H.; Cai, J.P.; Yeung, M.L.; Cheng, V.C.; Chen, H.; Che, X.Y.; et al. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: Implications for disease pathogenesis and clinical manifestation. J. Infect. Dis. 2013, 207, 1743–1752. [Google Scholar] [CrossRef] [PubMed]
- Kumavath, R.; Barh, D.; Andrade, B.S.; Imchen, M.; Aburjaile, F.F.; Ch, A.; Rodrigues, D.L.N.; Tiwari, S.; Alzahrani, K.J.; Góes-Neto, A.; et al. The Spike of SARS-CoV-2: Uniqueness and Applications. Front. Immunol. 2021, 12, 663912. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Z.; Yang, L.; Lian, X.; Xie, Y.; Li, S.; Xin, S.; Cao, P.; Lu, J. The MERS-CoV Receptor DPP4 as a Candidate Binding Target of the SARS-CoV-2 Spike. iScience 2020, 23, 101160. [Google Scholar] [CrossRef]
- Khandker, S.S.; Nik Hashim, N.H.H.; Deris, Z.Z.; Shueb, R.H.; Islam, M.A. Diagnostic Accuracy of Rapid Antigen Test Kits for Detecting SARS-CoV-2: A Systematic Review and Meta-Analysis of 17,171 Suspected COVID-19 Patients. J. Clin. Med. 2021, 10, 3493. [Google Scholar] [CrossRef]
- Jegerlehner, S.; Suter-Riniker, F.; Jent, P.; Bittel, P.; Nagler, M. Diagnostic accuracy of a SARS-CoV-2 rapid antigen test in real-life clinical settings. Int. J. Infect. Dis. 2021, 109, 118–122. [Google Scholar] [CrossRef]
- Candel, F.J.; Barreiro, P.; San Román, J.; Abanades, J.C.; Barba, R.; Barberán, J.; Bibiano, C.; Canora, J.; Cantón, R.; Calvo, C.; et al. Recommendations for use of antigenic tests in the diagnosis of acute SARS-CoV-2 infection in the second pandemic wave: Attitude in different clinical settings. Rev. Esp. Quimioter. 2020, 33, 466–484. [Google Scholar] [CrossRef]
- Larremore, D.B.; Wilder, B.; Lester, E.; Shehata, S.; Burke, J.M.; Hay, J.A.; Tambe, M.; Mina, M.J.; Parker, R. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci. Adv. 2021, 7, eabd5393. [Google Scholar] [CrossRef] [PubMed]
- Healy, B.; Khan, A.; Metezai, H.; Blyth, I.; Asad, H. The impact of false positive COVID-19 results in an area of low prevalence. Clin. Med. 2021, 21, e54–e56. [Google Scholar] [CrossRef] [PubMed]
- Surkova, E.; Nikolayevskyy, V.; Drobniewski, F. False-positive COVID-19 results: Hidden problems and costs. Lancet Respir. Med. 2020, 8, 1167–1168. [Google Scholar] [CrossRef]
- Braunstein, G.D.; Schwartz, L.; Hymel, P.; Fielding, J. False Positive Results With SARS-CoV-2 RT-PCR Tests and How to Evaluate a RT-PCR-Positive Test for the Possibility of a False Positive Result. J. Occup. Environ. Med. 2021, 63, e159–e162. [Google Scholar] [CrossRef] [PubMed]
- Madeley, C.R.; Peiris, J.S. Methods in virus diagnosis: Immunofluorescence revisited. J. Clin. Virol. 2002, 25, 121–134. [Google Scholar] [CrossRef]
- Liu, I.J.; Chen, P.J.; Yeh, S.H.; Chiang, Y.P.; Huang, L.M.; Chang, M.F.; Chen, S.Y.; Yang, P.C.; Chang, S.C.; Wang, W.K. Immunofluorescence assay for detection of the nucleocapsid antigen of the severe acute respiratory syndrome (SARS)-associated coronavirus in cells derived from throat wash samples of patients with SARS. J. Clin. Microbiol. 2005, 43, 2444–2448. [Google Scholar] [CrossRef]
- Monto, A.S.; Rhodes, L.M. Detection of coronavirus infection of man by immunofluorescence. Proc. Soc. Exp. Biol. Med. 1977, 155, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Orstavik, I.; Grandien, M.; Halonen, P.; Arstila, P.; Mordhorst, C.H.; Hornsleth, A.; Popow-Kraupp, T.; McQuillin, J.; Gardner, P.S.; Almeida, J.; et al. Viral diagnoses using the rapid immunofluorescence technique and epidemiological implications of acute respiratory infections among children in different European countries. Bull. World Health Organ. 1984, 62, 307–313. [Google Scholar] [PubMed]
- Sadeghi, C.D.; Aebi, C.; Gorgievski-Hrisoho, M.; Mühlemann, K.; Barbani, M.T. Twelve years’ detection of respiratory viruses by immunofluorescence in hospitalised children: Impact of the introduction of a new respiratory picornavirus assay. BMC Infect. Dis. 2011, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.H.; Yan, M.K.; To, K.K.; Lau, S.K.; Woo, P.C.; Cheng, V.C.; Li, W.S.; Chan, J.F.; Tse, H.; Yuen, K.Y. Use of the human colorectal adenocarcinoma (Caco-2) cell line for isolating respiratory viruses from nasopharyngeal aspirates. J. Med. Virol. 2013, 85, 874–879. [Google Scholar] [CrossRef]
- Faden, H. Comparison of midturbinate flocked-swab specimens with nasopharyngeal aspirates for detection of respiratory viruses in children by the direct fluorescent antibody technique. J. Clin. Microbiol. 2010, 48, 3742–3743. [Google Scholar] [CrossRef] [PubMed][Green Version]
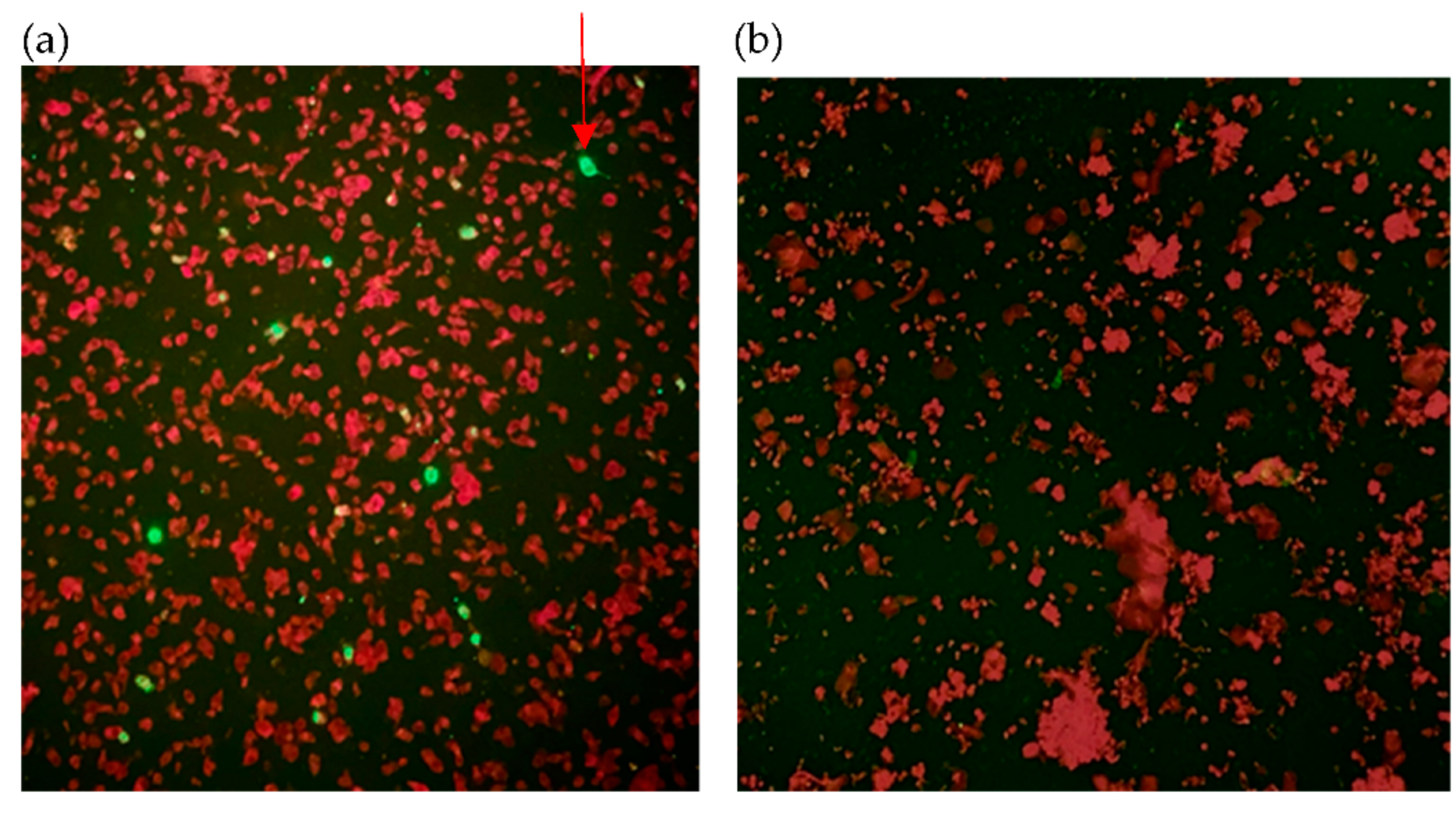
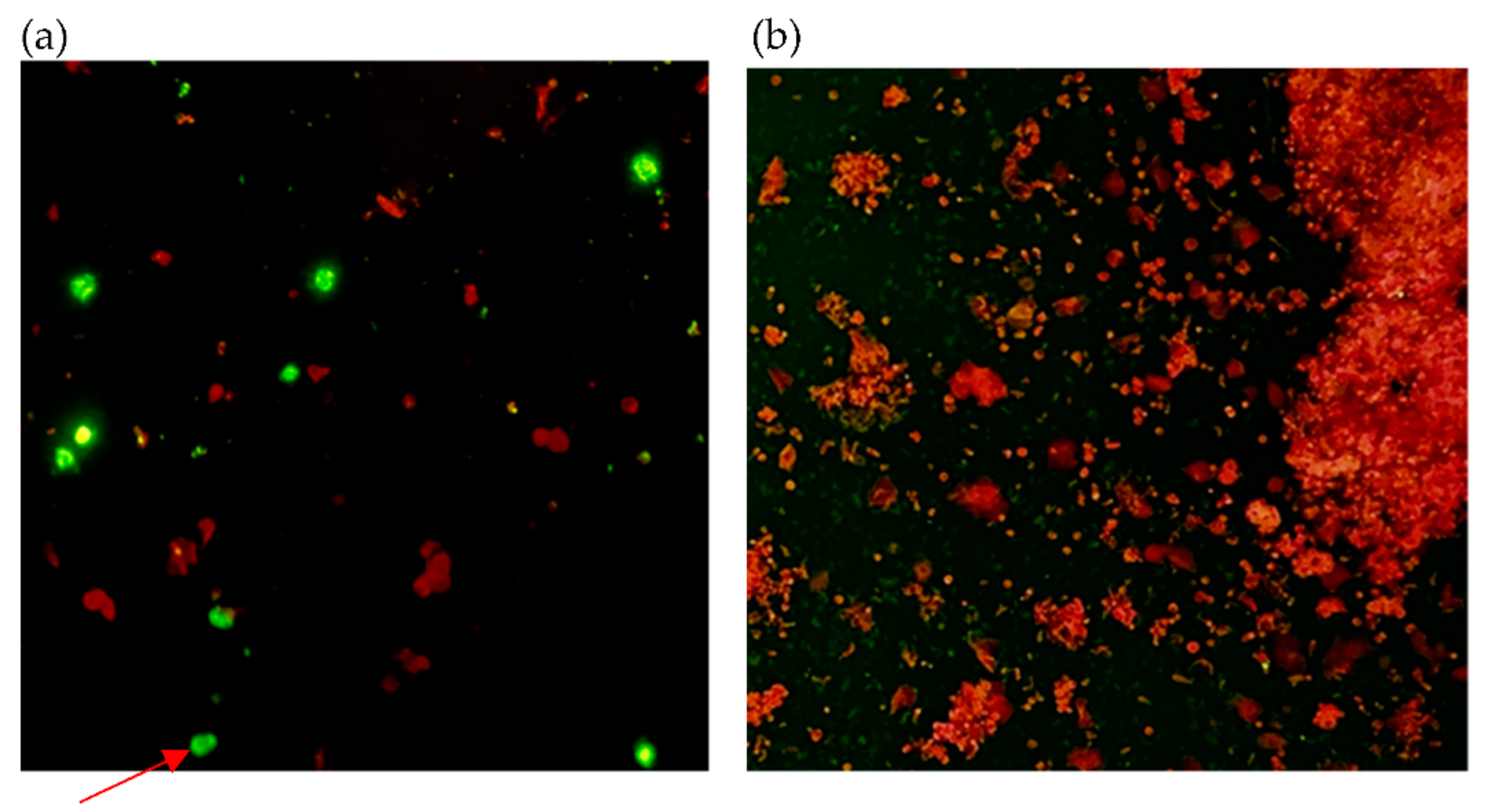
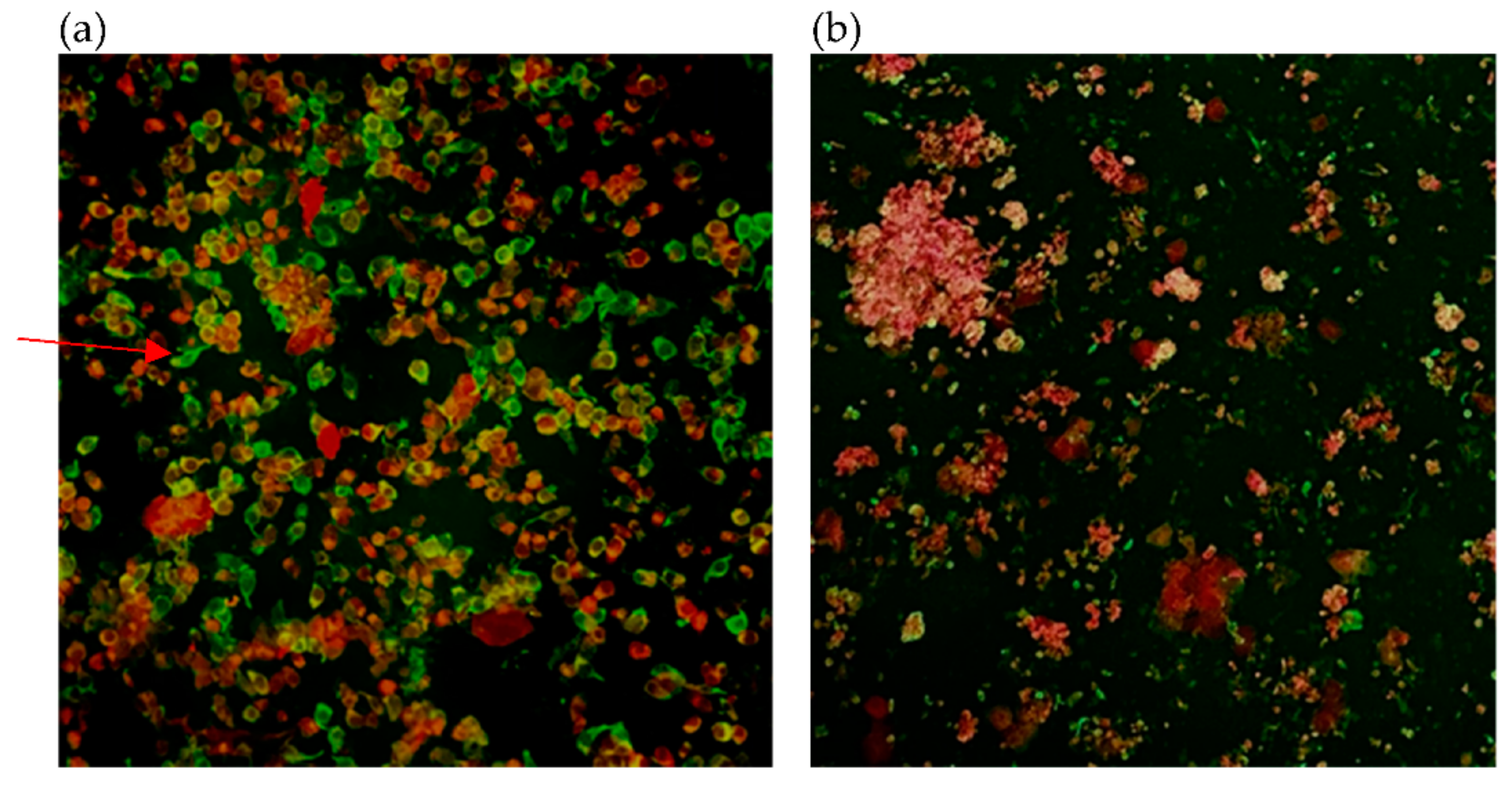
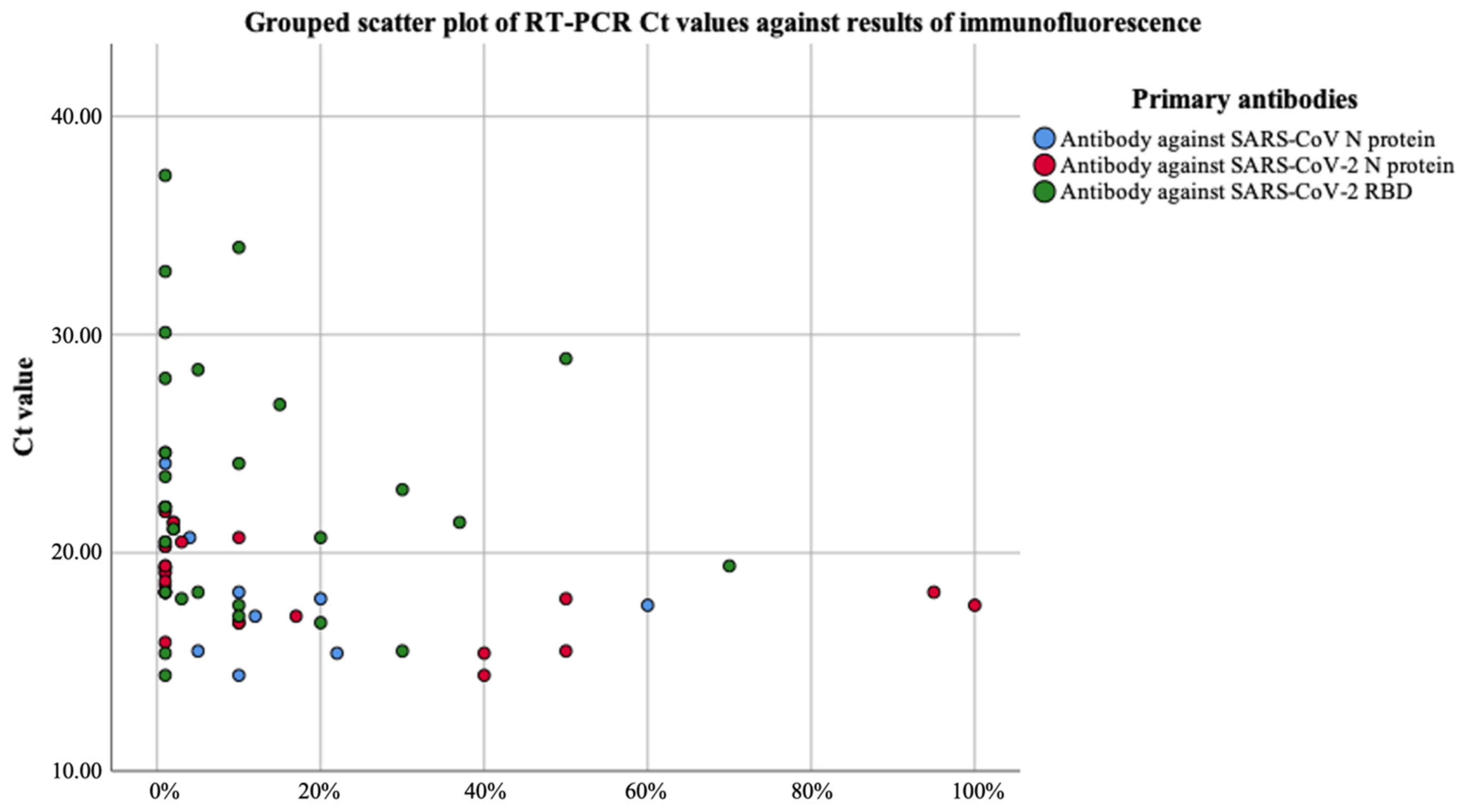
| Case Number | Collection Day | Result of Immunofluorescence | Result of RT-PCR | |||
|---|---|---|---|---|---|---|
| SARS-CoV N | SARS-CoV-2 N | SARS-CoV-2 RBD | Ct Value | Viral Load (copies/mL) | ||
| 1 | Day 1 | QI | 18.2 | 6.8 108 | ||
| 1 | Day 2 | 17.6 | 9.4 108 | |||
| 2 | Day 1 | 26.8 | 9.8 106 | |||
| 2 | Day 2 | 31.1 | 5.2 105 | |||
| 3 | Day 1 | 25.2 | 2.8 107 | |||
| 3 | Day 2 | QI | 19.3 | 1.5 109 | ||
| 4 | Day 1 | 30.1 | 9.8 105 | |||
| 4 | Day 2 | QI | QI | QI | 35.1 | 3.3 104 |
| 5 | Day 1 | 37.3 | 3.9 103 | |||
| 5 | Day 2 | 34.0 | 6.8 104 | |||
| 6 | Day 1 | 22.9 | 4.4 107 | |||
| 6 | Day 2 | 26.8 | 9.8 106 | |||
| 7 | Day 1 | 18.5 | 1.6 109 | |||
| 7 | Day 2 | QI | QI | QI | 18.7 | 1.2 109 |
| 8 | Day 1 | QI | 19.1 | 1.0 109 | ||
| 8 | Day 2 | 22.1 | 1.2 108 | |||
| 9 | Day 1 | 15.5 | 2.7 109 | |||
| 9 | Day 2 | 24.1 | 1.6 × 107 | |||
| 10 | Day 1 | 18.2 | 1.9 109 | |||
| 10 | Day 2 | 19.4 | 3.3 108 | |||
| 11 | Day 1 | 17.9 | 7.8 108 | |||
| 11 | Day 2 | QI | QI | QI | 23.0 | 4.1 107 |
| 12 | Day 1 | 24.6 | 7.0 107 | |||
| 12 | Day 2 | 28.0 | 8.1 106 | |||
| 13 | Day 1 | 20.5 | 1.0 109 | |||
| 13 | Day 2 | 20.7 | 2.9 109 | |||
| 14 | Day 1 | 14.39 | 2.3 1010 | |||
| 14 | Day 2 | QI | QI | QI | 22.44 | 1.7 108 |
| 15 | Day 1 | QI | QI | QI | 20.9 | 6.1 107 |
| 15 | Day 2 | QI | QI | QI | 30.9 | 1.3 105 |
| 16 | Day 1 | 18.2 | 3.0 108 | |||
| 16 | Day 2 | QI | QI | QI | 23.0 | 2.4 107 |
| 17 | Day 1 | QI | 18.7 | 9.6 108 | ||
| 17 | Day 2 | 20.3 | 1.7 108 | |||
| 18 | Day 1 | QI | 37.6 | 1.9 103 | ||
| 18 | Day 2 | 30.7 | 3.1 105 | |||
| 19 | Day 1 | 20.9 | 6.1 107 | |||
| 19 | Day 2 | 15.9 | 1.2 109 | |||
| 20 | Day 1 | NS | 37.8 | 2.4 103 | ||
| 20 | Day 2 | 33.9 | 1.4 105 | |||
| 21 | Day 1 | 16.8 | 3.8 109 | |||
| 21 | Day 2 | 21.4 | 3.1 108 | |||
| 22 | Day 1 | 32.9 | 4.9 105 | |||
| 22 | Day 2 | QI | QI | 27.4 | 7.0 106 | |
| 23 | Day 1 | 23.5 | 1.0 108 | |||
| 23 | Day 2 | 17.1 | 3.3 109 | |||
| 24 | Day 1 | QI | QI | QI | 28.9 | 2.8 106 |
| 24 | Day 2 | QI | QI | QI | 33.2 | 2.0 105 |
| 25 | Day 1 | 21.1 | 3.7 108 | |||
| 25 | Day 2 | 15.4 | 1.1 1010 | |||
| 26 | Day 1 | 25.2 | 2.8 107 | |||
| 26 | Day 2 | QI | QI | QI | 28.4 | 3.9 106 |
| 27 | Day 1 | 28.9 | 1.2 106 | |||
| 27 | Day 2 | 28.4 | 1.7 106 | |||
| 28 | Day 1 | 24.2 | 3.3 107 | |||
| 28 | Day 2 | 25.3 | 1.6 107 | |||
| 29 | Day 1 | 18.9 | 9.0 108 | |||
| 29 | Day 2 | QI | 21.9 | 1.1 108 | ||
| Parameters | SARS-CoV N | SARS-CoV-2 N | SARS-CoV-2 RBD |
|---|---|---|---|
| Sensitivity [95% confidence interval (CI)] | 48% (29–67%) | 59% (39–76%) | 59% (39–76%) |
| Specificity [95% confidence interval (CI)] | 98% (87–100%) | 98% (87–100%) | 80% (64–91%) |
| Positive predictive value (PPV) [95% confidence interval (CI)] | 93% (66–99%) | 94% (71–99%) | 68% (52–81%) |
| Negative predictive value (NPV) [95% confidence interval (CI)] | 72% (65–79%) | 76% (68–83%) | 73% (63–81%) |
| Youden (J) index | 0.46 | 0.56 | 0.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lam, A.H.-Y.; Cai, J.-P.; Leung, K.-Y.; Zhang, R.-R.; Liu, D.; Fan, Y.; Tam, A.R.; Cheng, V.C.-C.; To, K.K.-W.; Yuen, K.-Y.; et al. In-House Immunofluorescence Assay for Detection of SARS-CoV-2 Antigens in Cells from Nasopharyngeal Swabs as a Diagnostic Method for COVID-19. Diagnostics 2021, 11, 2346. https://doi.org/10.3390/diagnostics11122346
Lam AH-Y, Cai J-P, Leung K-Y, Zhang R-R, Liu D, Fan Y, Tam AR, Cheng VC-C, To KK-W, Yuen K-Y, et al. In-House Immunofluorescence Assay for Detection of SARS-CoV-2 Antigens in Cells from Nasopharyngeal Swabs as a Diagnostic Method for COVID-19. Diagnostics. 2021; 11(12):2346. https://doi.org/10.3390/diagnostics11122346
Chicago/Turabian StyleLam, Athene Hoi-Ying, Jian-Piao Cai, Ka-Yi Leung, Ricky-Ruiqi Zhang, Danlei Liu, Yujing Fan, Anthony Raymond Tam, Vincent Chi-Chung Cheng, Kelvin Kai-Wang To, Kwok-Yung Yuen, and et al. 2021. "In-House Immunofluorescence Assay for Detection of SARS-CoV-2 Antigens in Cells from Nasopharyngeal Swabs as a Diagnostic Method for COVID-19" Diagnostics 11, no. 12: 2346. https://doi.org/10.3390/diagnostics11122346
APA StyleLam, A. H.-Y., Cai, J.-P., Leung, K.-Y., Zhang, R.-R., Liu, D., Fan, Y., Tam, A. R., Cheng, V. C.-C., To, K. K.-W., Yuen, K.-Y., Hung, I. F.-N., & Chan, K.-H. (2021). In-House Immunofluorescence Assay for Detection of SARS-CoV-2 Antigens in Cells from Nasopharyngeal Swabs as a Diagnostic Method for COVID-19. Diagnostics, 11(12), 2346. https://doi.org/10.3390/diagnostics11122346








