Kynurenine and Hemoglobin as Sex-Specific Variables in COVID-19 Patients: A Machine Learning and Genetic Algorithms Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients, Sample Collection, and Processing
2.1.1. Patients Enrollment and Sample Collection
2.1.2. Metabolomics Profile of Plasma Samples
2.1.3. Sample Preparation
2.1.4. LC-MS/MS Method
2.1.5. DI-MS/MS Method
2.1.6. Quantification
2.1.7. Cytokine and Chemokine Quantification in Plasma Samples
2.2. Statistical Analysis
2.2.1. Descriptive Statistics
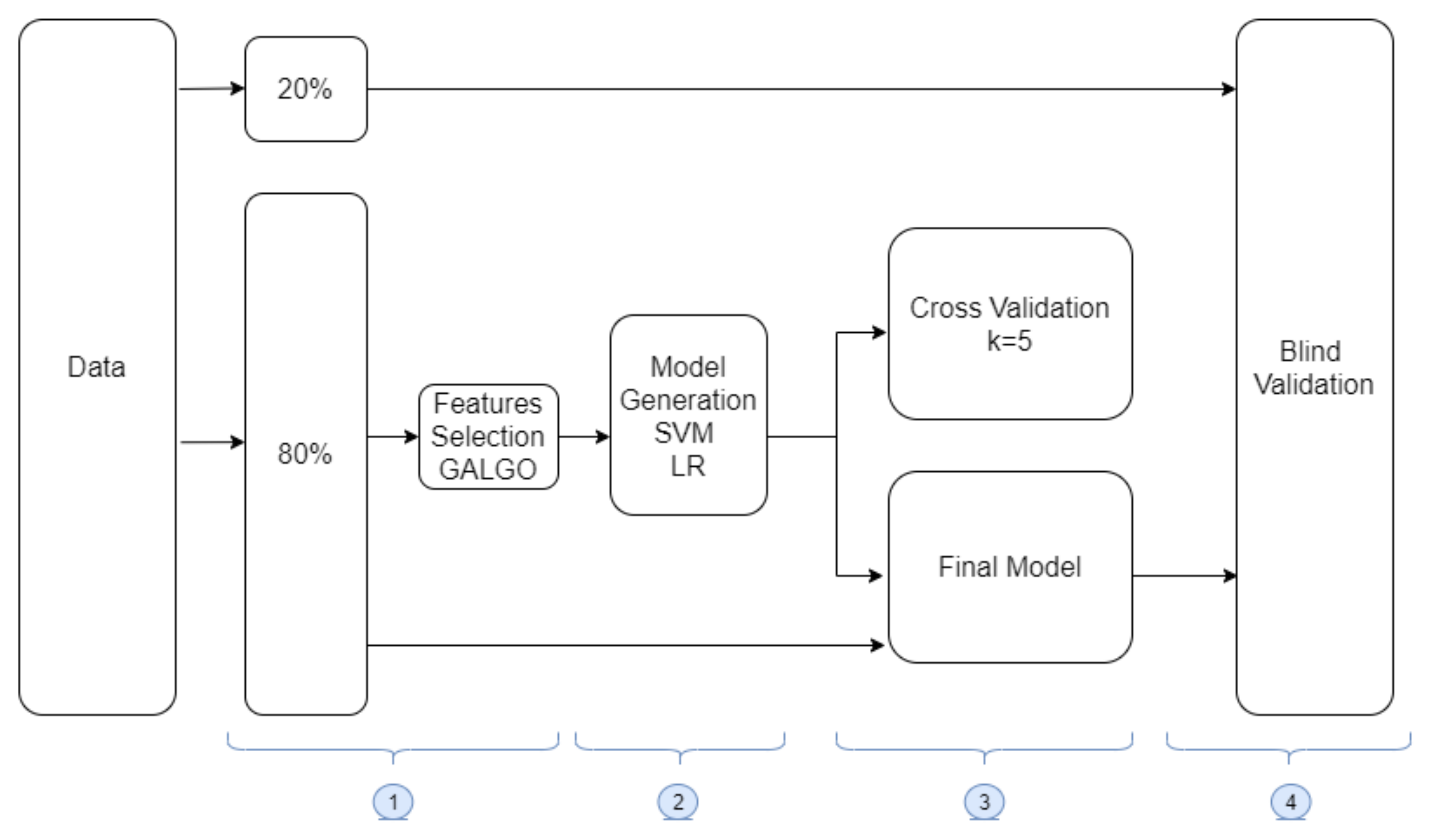
2.2.2. Machine Learning Methodology
2.2.3. Data Preparation and Feature Selection
2.2.4. Model Generation
2.2.5. Model Training and Validation
2.2.6. Blind Testing
2.2.7. Models Evaluation Metrics
3. Results
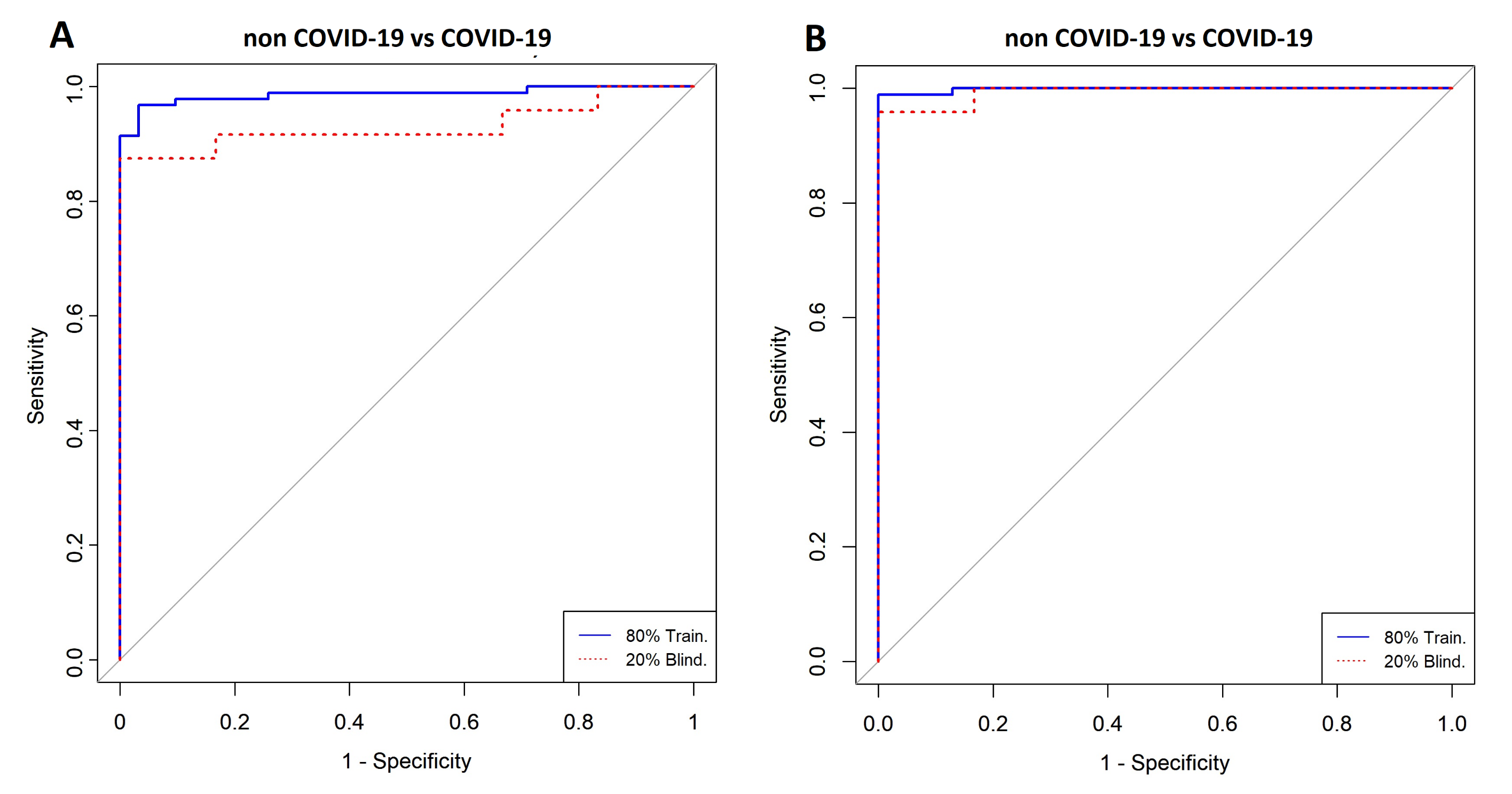
3.1. Comparison between COVID-19 Patients and Non-COVID-19 (Negative Controls)
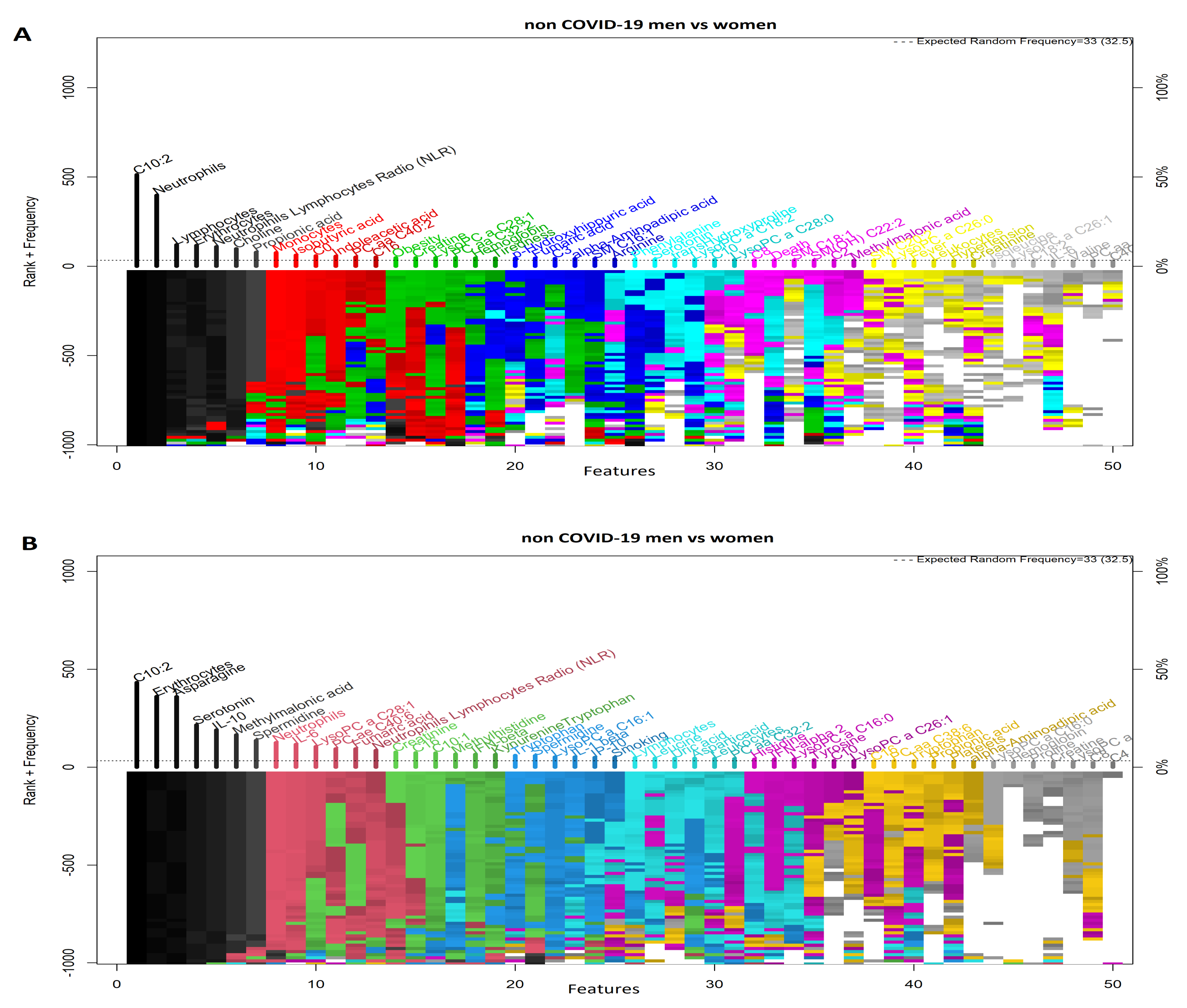
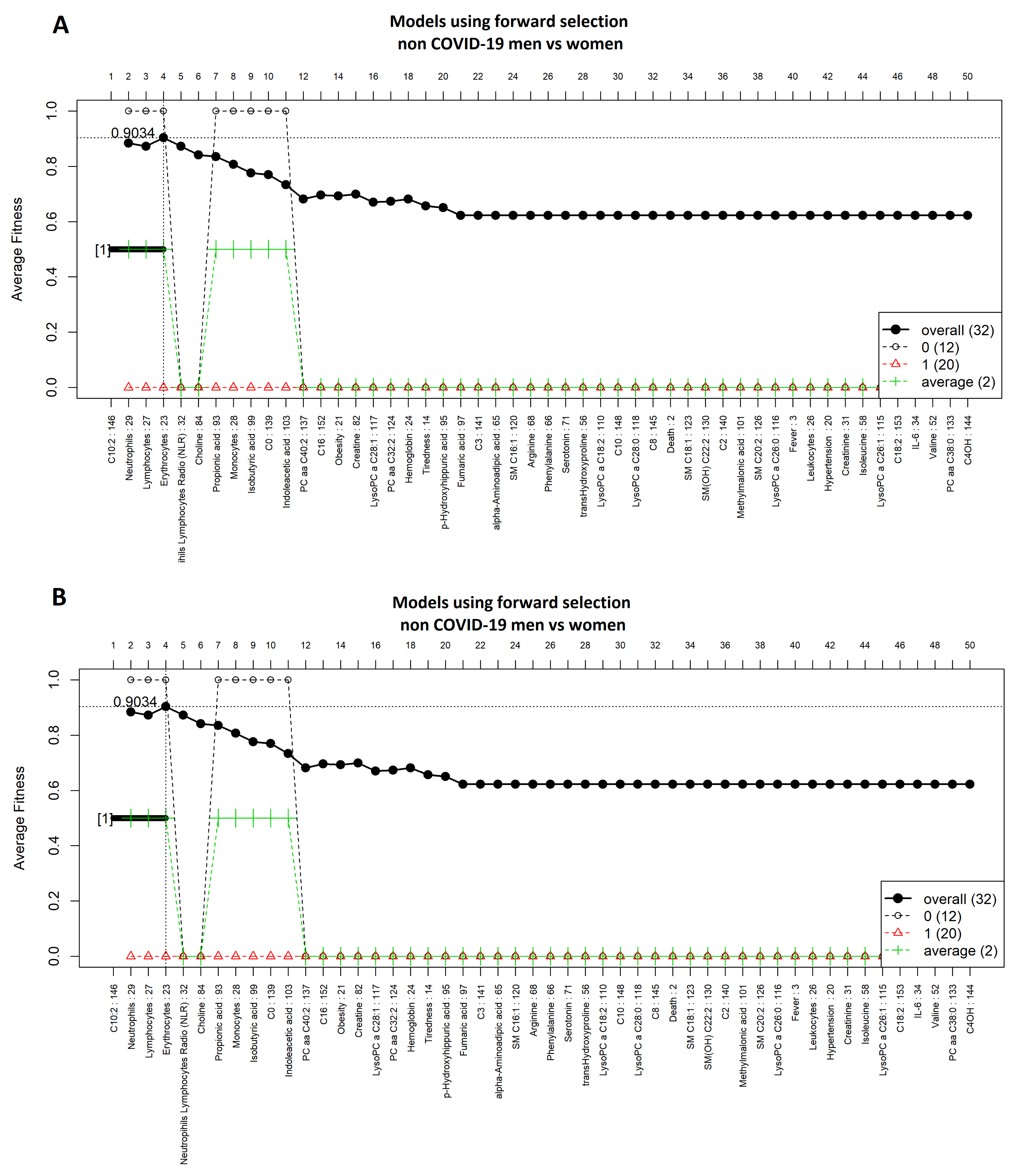
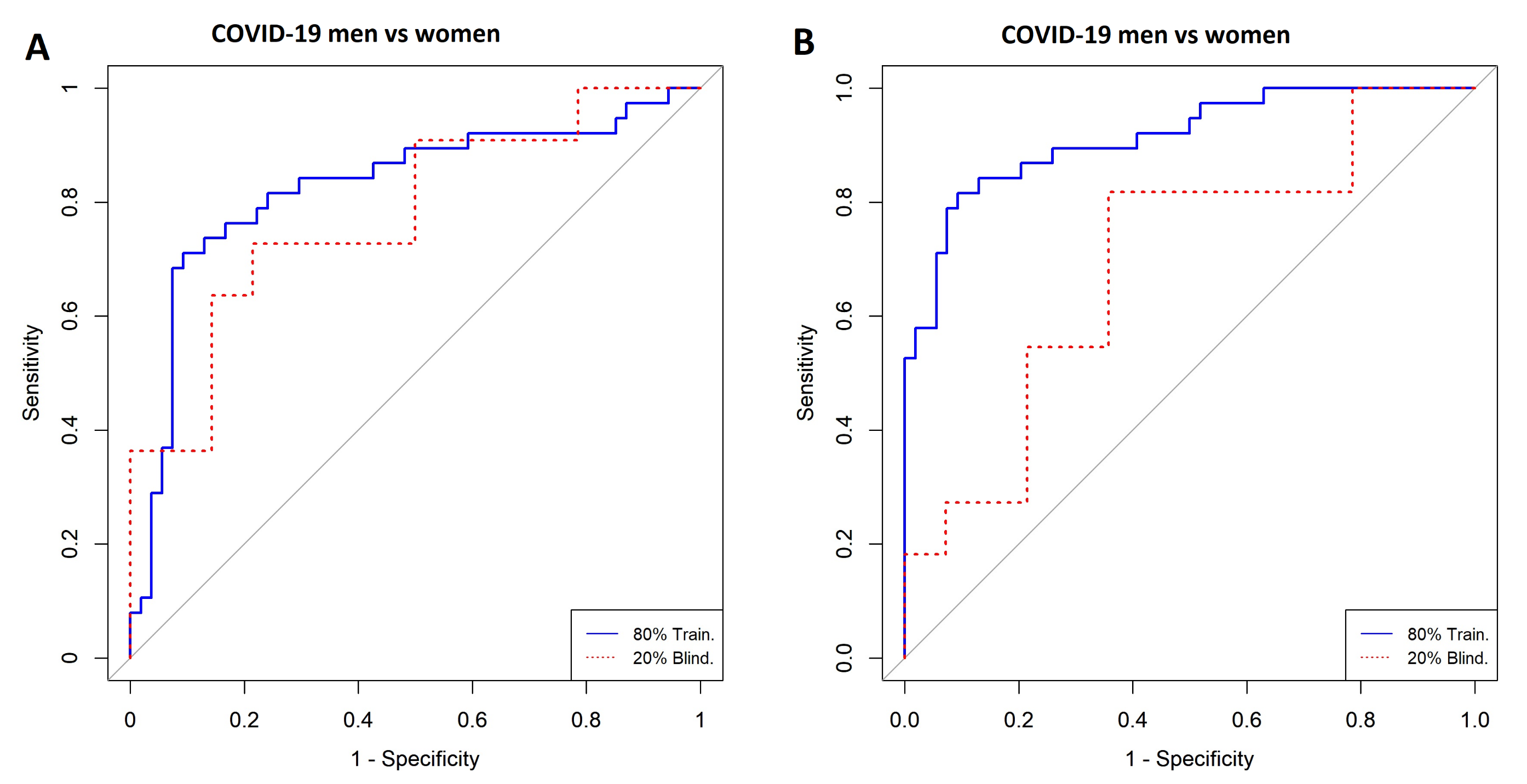
3.2. Comparison of COVID-19 Status by Sex
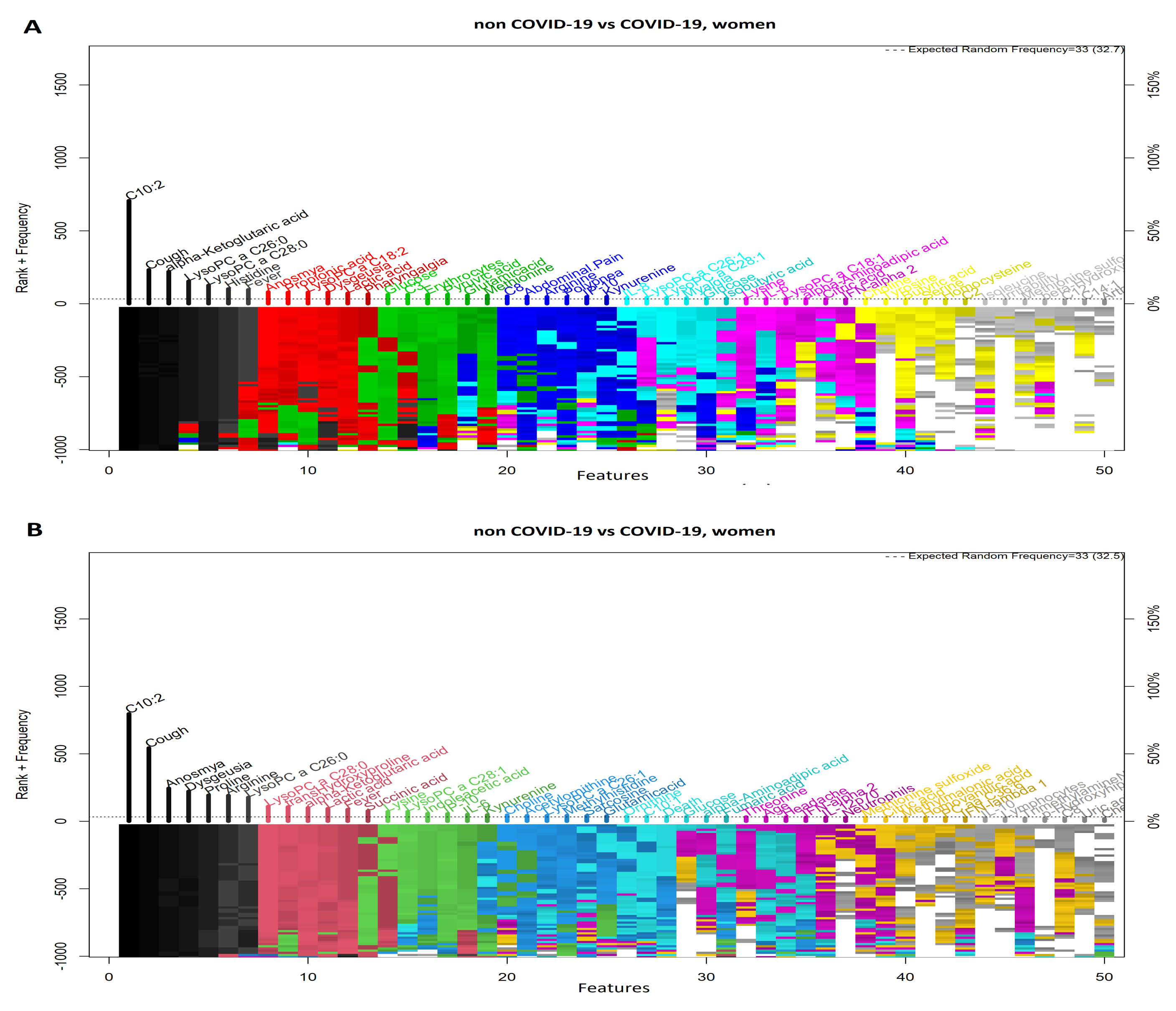
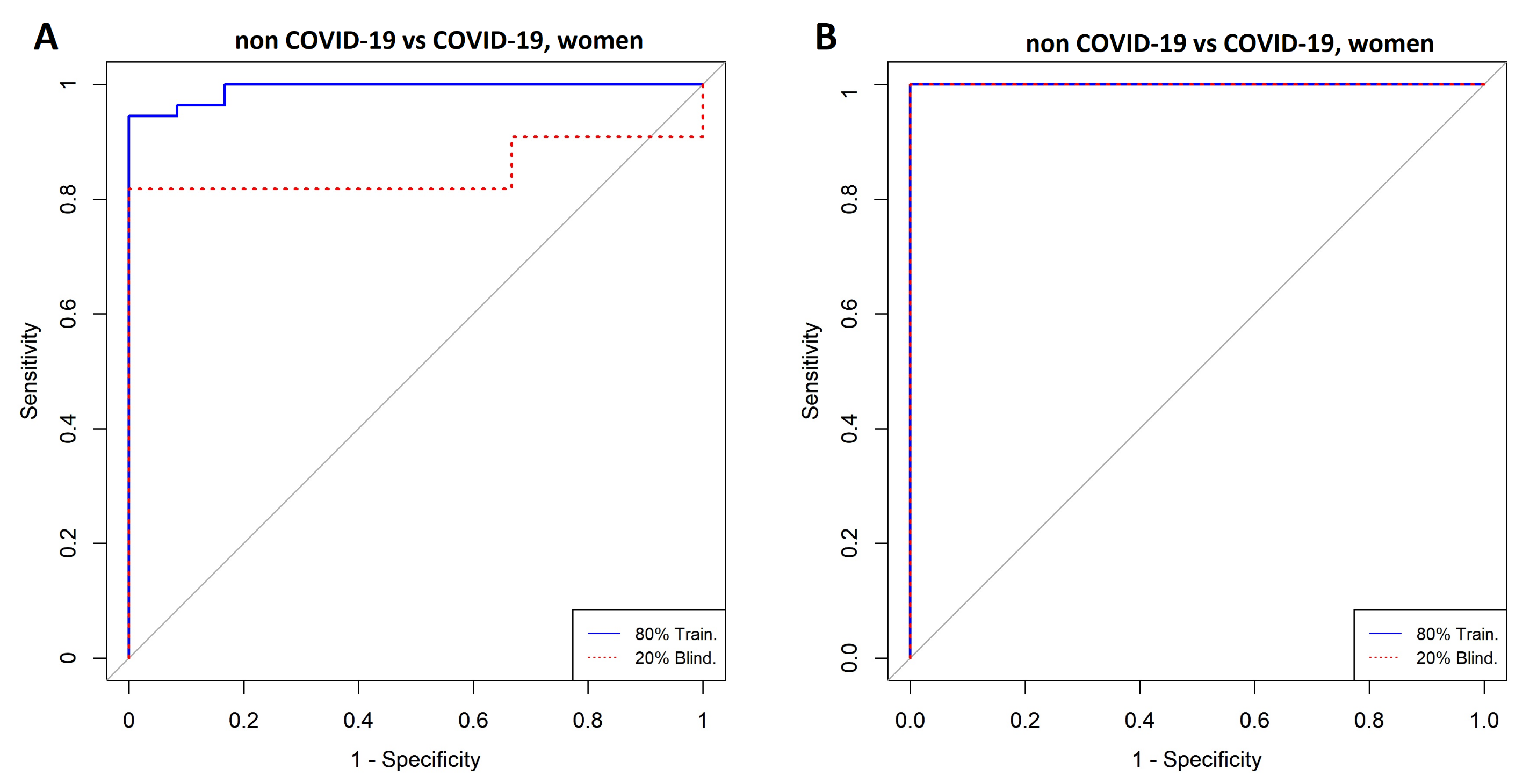
3.3. Comparison between Women with COVID-19 and Women without COVID-19
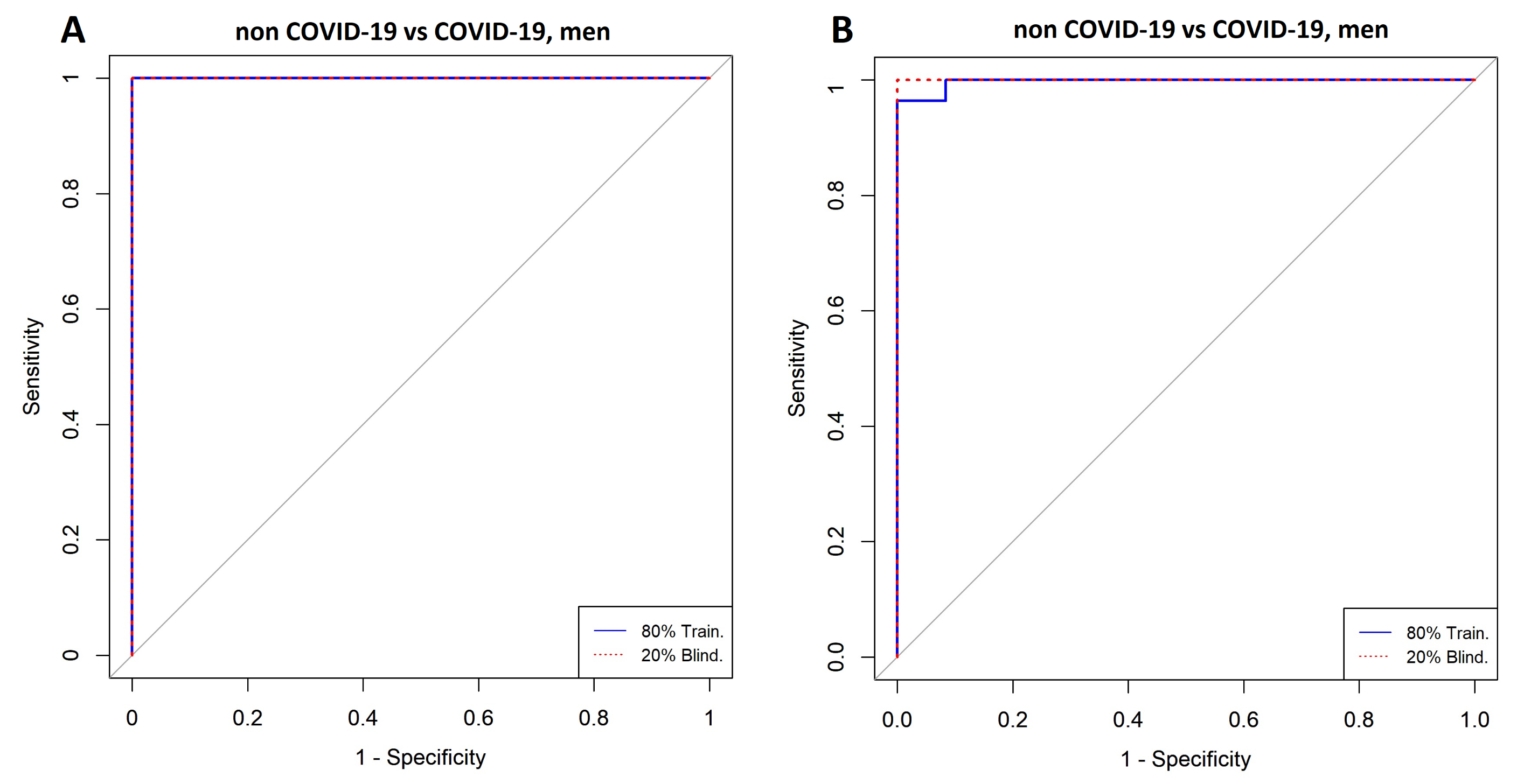
3.4. Comparison between Men with COVID-19 and Men without COVID-19
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1
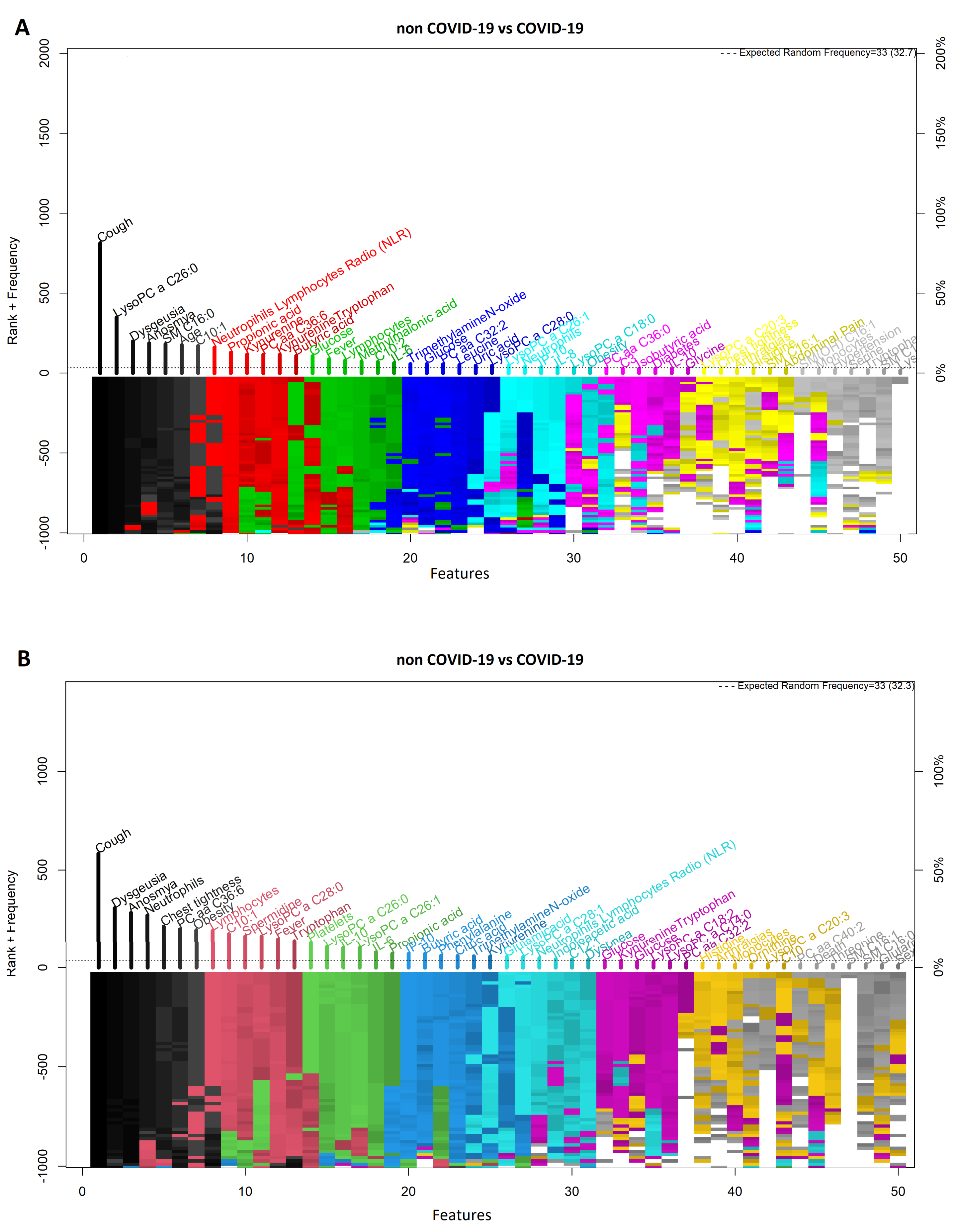
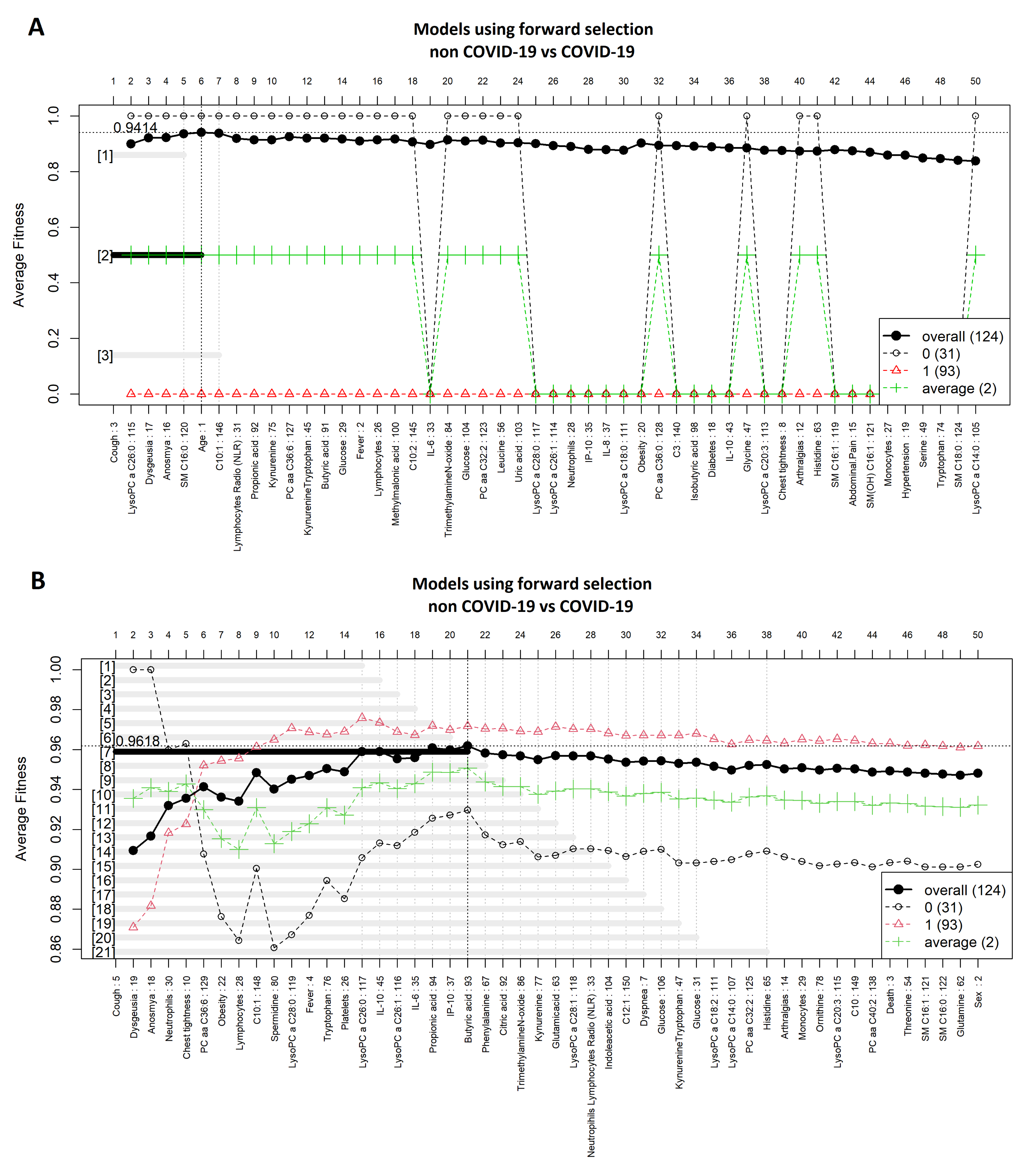
| COVID-19 vs. Non COVID-19 | Model | ||
|---|---|---|---|
| SVM | LR | ||
| Included Variables | 21 | 6 | |
| Cross-Validation (k = 5) | AUC | 0.99 | 0.98 |
| CI 95% | 0.99–1 | 0.96–1 | |
| Specificity | 1 | 0.97 | |
| Sensitivity | 0.99 | 0.95 | |
| Accuracy | 0.99 | 0.96 | |
| Training (80%) | AUC | 0.99 | 0.98 |
| CI 95% | 0.99–1 | 0.96-1 | |
| Specificity | 1 | 0.96 | |
| Sensitivity | 0.98 | 0.96 | |
| Accuracy | 0.99 | 0.96 | |
| Blind (20%) | AUC | 0.99 | 0.93 |
| CI 95% | 0.97–1 | 0.93-1 | |
| Specificity | 1 | 1 | |
| Sensitivity | 0.95 | 0.87 | |
| Accuracy | 0.96 | 0.9 | |

| Non COVID-19 Men vs. COVID-19 Women | Model | ||
|---|---|---|---|
| SVM | LR | ||
| Included Variables | 16 | 4 | |
| Cross Validation (k = 5) | AUC | 1 | 1 |
| CI 95% | 1–1 | 1–1 | |
| Specificity | 1 | 1 | |
| Sensitivity | 1 | 1 | |
| Accuracy | 1 | 1 | |
| Training (80%) | AUC | 1 | 1 |
| CI 95% | 1–1 | 1–1 | |
| Specificity | 1 | 1 | |
| Sensitivity | 1 | 1 | |
| Accuracy | 1 | 1 | |
| Blind (20%) | AUC | 0.83 | 1 |
| CI 95% | 0.76–0.90 | 1–1 | |
| Specificity | 0.66 | 1 | |
| Sensitivity | 1 | 1 | |
| Accuracy | 0.8 | 1 | |
References
- Ghosh, S.; Klein, R.S. Sex Drives Dimorphic Immune Responses to Viral Infections. J. Immunol. 2017, 198, 1782–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Karlberg, J.; Chong, D.S.; Lai, W.Y. Do men have a higher case fatality rate of severe acute respiratory syndrome than women do? Am. J. Epidemiol. 2004, 159, 229–231. [Google Scholar] [CrossRef] [Green Version]
- Matsuyama, R.; Nishiura, H.; Kutsuna, S.; Hayakawa, K.; Ohmagari, N. Clinical determinants of the severity of Middle East respiratory syndrome (MERS): A systematic review and meta-analysis. BMC Public Health 2016, 16, 1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eshima, N.; Tokumaru, O.; Hara, S.; Bacal, K.; Korematsu, S.; Tabata, M.; Karukaya, S.; Yasui, Y.; Okabe, N.; Matsuishi, T. Sex- and age-related differences in morbidity rates of 2009 pandemic influenza A H1N1 virus of swine origin in Japan. PLoS ONE 2011, 6, e19409. [Google Scholar] [CrossRef]
- Peckham, H.; de Gruijter, N.M.; Raine, C.; Radziszewska, A.; Ciurtin, C.; Wedderburn, L.R.; Rosser, E.C.; Webb, K.; Deakin, C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020, 11, 6317. [Google Scholar] [CrossRef] [PubMed]
- Ten-Caten, F.; Gonzalez-Dias, P.; Castro, I.; Ogava, R.L.T.; Giddaluru, J.; Silva, J.C.S.; Martins, F.; Goncalves, A.N.A.; Costa-Martins, A.G.; Araujo, J.D.; et al. In-depth analysis of laboratory parameters reveals the interplay between sex, age, and systemic inflammation in individuals with COVID-19. Int. J. Infect. Dis. 2021, 105, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Zhang, J.; Wang, T.; Cui, P.; Chen, Z.; Jiang, J.; Zhou, S.; Dai, J.; Wang, B.; Yuan, S.; et al. Potential Influence of Menstrual Status and Sex Hormones on Female Severe Acute Respiratory Syndrome Coronavirus 2 Infection: A Cross-sectional Multicenter Study in Wuhan, China. Clin. Infect. Dis. 2021, 72, e240–e248. [Google Scholar] [CrossRef] [PubMed]
- Rastrelli, G.; Di Stasi, V.; Inglese, F.; Beccaria, M.; Garuti, M.; Di Costanzo, D.; Spreafico, F.; Greco, G.F.; Cervi, G.; Pecoriello, A.; et al. Low testosterone levels predict clinical adverse outcomes in SARS-CoV-2 pneumonia patients. Andrology 2021, 9, 88–98. [Google Scholar] [CrossRef]
- Chang, E.; Varghese, M.; Singer, K. Gender and Sex Differences in Adipose Tissue. Curr. Diab. Rep. 2018, 18, 69. [Google Scholar] [CrossRef]
- Karastergiou, K.; Fried, S.K. Cellular Mechanisms Driving Sex Differences in Adipose Tissue Biology and Body Shape in Humans and Mouse Models. Adv. Exp. Med. Biol. 2017, 1043, 29–51. [Google Scholar] [CrossRef]
- Cai, Y.; Kim Daniel, J.; Takahashi, T.; Broadhurst David, I.; Yan, H.; Ma, S.; Rattray Nicholas, J.W.; Casanovas-Massana, A.; Israelow, B.; Klein, J.; et al. Kynurenic acid may underlie sex-specific immune responses to COVID-19. Sci. Signal. 2021, 14, eabf8483. [Google Scholar] [CrossRef]
- Dix, A.; Vlaic, S.; Guthke, R.; Linde, J. Use of systems biology to decipher host–pathogen interaction networks and predict biomarkers. Clin. Microbiol. Infect. 2016, 22, 600–606. [Google Scholar] [CrossRef] [Green Version]
- Hou, Q.; Bing, Z.T.; Hu, C.; Li, M.Y.; Yang, K.H.; Mo, Z.; Xie, X.W.; Liao, J.L.; Lu, Y.; Horie, S.; et al. RankProd Combined with Genetic Algorithm Optimized Artificial Neural Network Establishes a Diagnostic and Prognostic Prediction Model that Revealed C1QTNF3 as a Biomarker for Prostate Cancer. EBioMedicine 2018, 32, 234–244. [Google Scholar] [CrossRef]
- Xie, Y.; Meng, W.Y.; Li, R.Z.; Wang, Y.W.; Qian, X.; Chan, C.; Yu, Z.F.; Fan, X.X.; Pan, H.D.; Xie, C.; et al. Early lung cancer diagnostic biomarker discovery by machine learning methods. Transl. Oncol. 2021, 14, 100907. [Google Scholar] [CrossRef]
- Cristianini, N.; Ricci, E. Support Vector Machines. In Encyclopedia of Algorithms; Springer: Boston, MA, USA, 2008; pp. 928–932. [Google Scholar] [CrossRef]
- Velazquez-Pupo, R.; Sierra-Romero, A.; Torres-Roman, D.; Shkvarko, Y.V.; Santiago-Paz, J.; Gomez-Gutierrez, D.; Robles-Valdez, D.; Hermosillo-Reynoso, F.; Romero-Delgado, M. Vehicle Detection with Occlusion Handling, Tracking, and OC-SVM Classification: A High Performance Vision-Based System. Sensors 2018, 18, 374. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Ren, Z.; Tang, W.; Wang, H.; Chen, P. Intelligent gearbox diagnosis methods based on SVM, wavelet lifting and RBR. Sensors 2010, 10, 4602–4621. [Google Scholar] [CrossRef]
- Ruiz-Gonzalez, R.; Gomez-Gil, J.; Gomez-Gil, F.J.; Martinez-Martinez, V. An SVM-based classifier for estimating the state of various rotating components in agro-industrial machinery with a vibration signal acquired from a single point on the machine chassis. Sensors 2014, 14, 20713–20735. [Google Scholar] [CrossRef]
- Men, H.; Fu, S.; Yang, J.; Cheng, M.; Shi, Y.; Liu, J. Comparison of SVM, RF and ELM on an Electronic Nose for the Intelligent Evaluation of Paraffin Samples. Sensors 2018, 18, 285. [Google Scholar] [CrossRef] [Green Version]
- Dreiseitl, S.; Ohno–Machado, L. Logistic regression and artificial neural network classification models: A methodology review. J. Biomed. Inform. 2002, 35, 352–359. [Google Scholar] [CrossRef] [Green Version]
- Lessmann, S.; Stahlbock, R.; Crone, S.F. Genetic Algorithms for Support Vector Machine Model Selection. In Proceedings of the 2006 International Joint Conference on Neural Networks, Vancouver, BC, Canada, 16–21 July 2006. [Google Scholar] [CrossRef]
- Dalal, V.; Carmicheal, J.; Dhaliwal, A.; Jain, M.; Kaur, S.; Batra, S.K. Radiomics in stratification of pancreatic cystic lesions: Machine learning in action. Cancer Lett. 2020, 469, 228–237. [Google Scholar] [CrossRef]
- Xu, W.; Xu, M.; Wang, L.; Zhou, W.; Xiang, R.; Shi, Y.; Zhang, Y.; Piao, Y. Integrative analysis of DNA methylation and gene expression identified cervical cancer-specific diagnostic biomarkers. Signal Transduct. Target Ther. 2019, 4, 55. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.H.; Lin, C.H.; Lane, H.Y. Machine Learning and Novel Biomarkers for the Diagnosis of Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 2761. [Google Scholar] [CrossRef]
- Manoochehri, Z.; Salari, N.; Rezaei, M.; Khazaie, H.; Manoochehri, S.; Pavah, B.K. Comparison of support vector machine based on genetic algorithm with logistic regression to diagnose obstructive sleep apnea. J. Res. Med. Sci. 2018, 23, 65. [Google Scholar] [CrossRef]
- Guhathakurata, S.; Kundu, S.; Chakraborty, A.; Banerjee, J.S. A novel approach to predict COVID-19 using support vector machine. In Data Science for COVID-19; Academic Press: Cambridge, MA, USA, 2021; pp. 351–364. [Google Scholar] [CrossRef]
- Jiang, X.; Coffee, M.; Bari, A.; Wang, J.; Jiang, X.; Huang, J.; Shi, J.; Dai, J.; Cai, J.; Zhang, T.; et al. Towards an Artificial Intelligence Framework for Data-Driven Prediction of Coronavirus Clinical Severity. Comput. Mater. Contin. 2020, 63, 537–551. [Google Scholar] [CrossRef]
- Xu, W.; Sun, N.N.; Gao, H.N.; Chen, Z.Y.; Yang, Y.; Ju, B.; Tang, L.L. Risk factors analysis of COVID-19 patients with ARDS and prediction based on machine learning. Sci. Rep. 2021, 11, 2933. [Google Scholar] [CrossRef]
- Lu, J.Q.; Musheyev, B.; Peng, Q.; Duong, T.Q. Neural network analysis of clinical variables predicts escalated care in COVID-19 patients: A retrospective study. PeerJ. 2021, 9, e11205. [Google Scholar] [CrossRef]
- Li, X.; Ge, P.; Zhu, J.; Li, H.; Graham, J.; Singer, A.; Richman, P.S.; Duong, T.Q. Deep learning prediction of likelihood of ICU admission and mortality in COVID-19 patients using clinical variables. PeerJ 2020, 8, e10337. [Google Scholar] [CrossRef]
- Hou, W.; Zhao, Z.; Chen, A.; Li, H.; Duong, T.Q. Machining learning predicts the need for escalated care and mortality in COVID-19 patients from clinical variables. Int. J. Med Sci. 2021, 18, 1739–1745. [Google Scholar] [CrossRef]
- Ancochea, J.; Izquierdo, J.L.; Soriano, J.B. Evidence of Gender Differences in the Diagnosis and Management of Coronavirus Disease 2019 Patients: An Analysis of Electronic Health Records Using Natural Language Processing and Machine Learning. J. Women’s Health 2021, 30, 393–404. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, L.; Johnson, M.; Mandal, R.; Wishart, D.S. Comprehensive Targeted Metabolomic Assay for Urine Analysis. Anal. Chem. 2020, 92, 10627–10634. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.E.; Smith, T.A.; Ziganshin, B.A.; Elefteriades, J.A. The mystery of the Z-score. Aorta 2016, 4, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Trevino, V.; Falciani, F. GALGO: An R package for multivariate variable selection using genetic algorithms. Bioinformatics 2006, 22, 1154–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couronné, R.; Probst, P.; Boulesteix, A.L. Random forest versus logistic regression: A large-scale benchmark experiment. BMC Bioinform. 2018, 19, 270. [Google Scholar] [CrossRef]
- Chakravarthi, B.R.; Priyadharshini, R.; Muralidaran, V.; Jose, N.; Suryawanshi, S.; Sherly, E.; McCrae, J.P. DravidianCodeMix: Sentiment Analysis and Offensive Language Identification Dataset for Dravidian Languages in Code-Mixed Text. arXiv 2021, arXiv:2106.09460. [Google Scholar]
- Kleinbaum, D.G.; Klein, M. Introduction to logistic regression. In Logistic Regression; Springer: New York, NY, USA, 2010; pp. 1–39. [Google Scholar]
- Zou, X.; Hu, Y.; Tian, Z.; Shen, K. Logistic regression model optimization and case analysis. In Proceedings of the 2019 IEEE 7th International Conference on Computer Science and Network Technology (ICCSNT), Dalian, China, 19–20 October 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 135–139. [Google Scholar]
- Suthaharan, S. Machine learning models and algorithms for big data classification. Integr. Ser. Inf. Syst 2016, 36, 1–12. [Google Scholar]
- Miller, A. Subset Selection in Regression; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Rakotomamonjy, A. Optimizing Area Under Roc Curve with SVMs. In Proceedings of the Conference: ROC Analysis in Artificial Intelligence, 1st International Workshop, ROCAI-2004, Valencia, Spain, 22 August 2004; pp. 71–80. [Google Scholar]
- Yin, M.; Wortman Vaughan, J.; Wallach, H. Understanding the effect of accuracy on trust in machine learning models. In Proceedings of the 2019 CHI Conference on Human Factors in computing Systems, Glasgow, UK, 4–9 May 2019; pp. 1–12. [Google Scholar]
- Wang, M.; Jiang, N.; Li, C.; Wang, J.; Yang, H.; Liu, L.; Tan, X.; Chen, Z.; Gong, Y.; Yin, X.; et al. Sex-Disaggregated Data on Clinical Characteristics and Outcomes of Hospitalized Patients with COVID-19: A Retrospective Study. Front. Cell. Infect. Microbiol. 2021, 11, 467. [Google Scholar] [CrossRef]
- Feng, J.z.; Wang, Y.; Peng, J.; Sun, M.w.; Zeng, J.; Jiang, H. Comparison between logistic regression and machine learning algorithms on survival prediction of traumatic brain injuries. J. Crit. Care 2019, 54, 110–116. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z. Performance evaluation of machine learning methods for breast cancer prediction. Appl. Comput. Math. 2018, 7, 212–216. [Google Scholar] [CrossRef]
- Kang, J.; Schwartz, R.; Flickinger, J.; Beriwal, S. Machine learning approaches for predicting radiation therapy outcomes: A clinician’s perspective. Int. J. Radiat. Oncol. Biol. Phys. 2015, 93, 1127–1135. [Google Scholar] [CrossRef]
- Ayon, S.I.; Islam, M.M.; Hossain, M.R. Coronary artery heart disease prediction: A comparative study of computational intelligence techniques. IETE J. Res. 2020, 1–20. [Google Scholar] [CrossRef]
- Liang, M.; Cai, Z.; Zhang, H.; Huang, C.; Meng, Y.; Zhao, L.; Li, D.; Ma, X.; Zhao, X. Machine learning-based analysis of rectal cancer MRI radiomics for prediction of metachronous liver metastasis. Acad. Radiol. 2019, 26, 1495–1504. [Google Scholar] [CrossRef]
- Zoabi, Y.; Deri-Rozov, S.; Shomron, N. Machine learning-based prediction of COVID-19 diagnosis based on symptoms. Npj Digit. Med. 2021, 4, 3. [Google Scholar] [CrossRef]
- Tandan, M.; Acharya, Y.; Pokharel, S.; Timilsina, M. Discovering symptom patterns of COVID-19 patients using association rule mining. Comput. Biol. Med. 2021, 131, 104249. [Google Scholar] [CrossRef]
- Fraser, D.D.; Slessarev, M.; Martin, C.M.; Daley, M.; Patel, M.A.; Miller, M.R.; Patterson, E.K.; O’Gorman, D.B.; Gill, S.E.; Wishart, D.S.; et al. Metabolomics profiling of critically ill coronavirus disease 2019 patients: Identification of diagnostic and prognostic biomarkers. Crit. Care Explor. 2020, 2, e0272. [Google Scholar] [CrossRef]
- Delafiori, J.; Navarro, L.C.; Siciliano, R.F.; De Melo, G.C.; Busanello, E.N.B.; Nicolau, J.C.; Sales, G.M.; De Oliveira, A.N.; Val, F.F.A.; De Oliveira, D.N.; et al. COVID-19 automated diagnosis and risk assessment through metabolomics and machine learning. Anal. Chem. 2021, 93, 2471–2479. [Google Scholar] [CrossRef]
- Sindelar, M.; Stancliffe, E.; Schwaiger-Haber, M.; Anbukumar, D.S.; Albrecht, R.A.; Adkins-Travis, K.; Garcia-Sastre, A.; Shriver, L.P.; Patti, G.J. Longitudinal metabolomics of human plasma reveals robust prognostic markers of COVID-19 disease severity. medRxiv 2021. [Google Scholar] [CrossRef]
- Dana, P.M.; Sadoughi, F.; Hallajzadeh, J.; Asemi, Z.; Mansournia, M.A.; Yousefi, B.; Momen-Heravi, M. An insight into the sex differences in COVID-19 patients: What are the possible causes? Prehosp. Disaster Med. 2020, 35, 438–441. [Google Scholar] [CrossRef]
- Anai, M.; Akaike, K.; Iwagoe, H.; Akasaka, T.; Higuchi, T.; Miyazaki, A.; Naito, D.; Tajima, Y.; Takahashi, H.; Komatsu, T.; et al. Decrease in hemoglobin level predicts increased risk for severe respiratory failure in COVID-19 patients with pneumonia. Respir. Investig. 2021, 59, 187–193. [Google Scholar] [CrossRef]
- Hopp, M.T.; Domingo-Fernández, D.; Gadiya, Y.; Detzel, M.S.; Graf, R.; Schmalohr, B.F.; Kodamullil, A.T.; Imhof, D.; Hofmann-Apitius, M. Linking COVID-19 and Heme-Driven Pathophysiologies: A Combined Computational–Experimental Approach. Biomolecules 2021, 11, 644. [Google Scholar] [CrossRef]
- Cavezzi, A.; Troiani, E.; Corrao, S. COVID-19: Hemoglobin, iron, and hypoxia beyond inflammation. A narrative review. Clin. Pract. 2020, 10, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Eleftheriadis, T.; Pissas, G.; Antoniadi, G.; Liakopoulos, V.; Stefanidis, I. Kynurenine, by activating aryl hydrocarbon receptor, decreases erythropoietin and increases hepcidin production in HepG2 cells: A new mechanism for anemia of inflammation. Exp. Hematol. 2016, 44, 60–67.e1. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.; Schroecksnadel, K.; Mattle, V.; Winkler, C.; Konwalinka, G.; Fuchs, D. Possible role of cytokine-induced tryptophan degradation in anaemia of inflammation. Eur. J. Haematol. 2004, 72, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Ellingson, M.K.; Wong, P.; Israelow, B.; Lucas, C.; Klein, J.; Silva, J.; Mao, T.; Oh, J.E.; Tokuyama, M.; et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 2020, 588, 315–320. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef]
- Webb, K.; Peckham, H.; Radziszewska, A.; Menon, M.; Oliveri, P.; Simpson, F.; Deakin, C.T.; Lee, S.; Ciurtin, C.; Butler, G.; et al. Sex and Pubertal Differences in the Type 1 Interferon Pathway Associate With Both X Chromosome Number and Serum Sex Hormone Concentration. Front. Immunol. 2019, 9, 3167. [Google Scholar] [CrossRef]
- Drobnik, W.; Liebisch, G.; Audebert, F.X.; Frohlich, D.; Gluck, T.; Vogel, P.; Rothe, G.; Schmitz, G. Plasma ceramide and lysophosphatidylcholine inversely correlate with mortality in sepsis patients. J. Lipid Res. 2003, 44, 754–761. [Google Scholar] [CrossRef] [Green Version]
- Park, D.W.; Kwak, D.S.; Park, Y.Y.; Chang, Y.; Huh, J.W.; Lim, C.M.; Koh, Y.; Song, D.K.; Hong, S.B. Impact of serial measurements of lysophosphatidylcholine on 28-day mortality prediction in patients admitted to the intensive care unit with severe sepsis or septic shock. J. Crit. Care 2014, 29, 882.e5–882.e11. [Google Scholar] [CrossRef]
- Knuplez, E.; Marsche, G. An Updated Review of Pro- and Anti-Inflammatory Properties of Plasma Lysophosphatidylcholines in the Vascular System. Int. J. Mol. Sci. 2020, 21, 4501. [Google Scholar] [CrossRef]
- Bienvenu, L.A.; Noonan, J.; Wang, X.; Peter, K. Higher mortality of COVID-19 in males: Sex differences in immune response and cardiovascular comorbidities. Cardiovasc. Res. 2020, 116, 2197–2206. [Google Scholar] [CrossRef]
- Biswas, M.; Rahaman, S.; Biswas, T.K.; Haque, Z.; Ibrahim, B. Association of Sex, Age, and Comorbidities with Mortality in COVID-19 Patients: A Systematic Review and Meta-Analysis. Intervirology 2020, 64, 36–47. [Google Scholar] [CrossRef]






| Non-COVID-19 | COVID-19 | |||||
|---|---|---|---|---|---|---|
| Variable | Men | Women | Value | Men | Women | Value |
| Age | 40 (37–53) | 40 (37–46) | 0.59 | 53 (42–63) | 58 (52–62) | 0.11 |
| Lab Data | ||||||
| Erythrocytes (million/mL) | 5.5 ± 0.4 | 4.9 ± 0.3 | <0.0001 | 5.4 (5.1–5.7) | 4.9 ± 0.6 | <0.0001 |
| Hemoglobin (g/dL) | 16.3 (15.5–17) | 14.7 ± 0.9 | <0.0001 | 16.1 (15.3–16.7) | 13.9 ± 1.9 | <0.0001 |
| Platelets (thousands/mL) | 260.7 ± 58.5 | 297.2 ± 65.9 | 0.03 | 262.6 ± 94.9 | 227.4 ± 74.7 | 0.07 |
| Leukocytes (×10) | 7.3 ± 2.1 | 7.3 ± 2.3 | 0.84 | 8.4 (5.7–10) | 7.4 (5.3–10.6) | 0.67 |
| Lymphocytes (%) | 33 ± 9.9 | 28.1 ± 8 | 0.05 | 9.7 (6–16.3) | 15.1 (10.3–26.4) | 0.0006 |
| Monocytes (%) | 7.5 ± 2.3 | 6.2 ± 2.5 | 0.2 | 4.8 (2.9–7.2) | 4.5 (3–7.6) | 0.92 |
| Neutrophils (%) | 55.4 (50.5–61) | 63.2 ± 9 | 0.04 | 84.8 (72.9–89.5) | 78.5 (66.7–84.8) | 0.02 |
| Glucose | 93.2 ± 13.6 | 93.5 (84.3–106.8) | 0.79 | 119 (96.5–140.5) | 121 (97–264) | 0.27 |
| Creatinine | 1 ± 0.16 | 0.8 (0.7–0.9) | 0.0009 | 0.95 (0.8–1.2) | 0.8 (0.7–1) | 0.003 |
| Urea | 32.8 (27.9–39.9) | 29.1 ± 9.7 | 0.12 | 33.6 (25.4–42.5) | 49.6 (32–76.7) | 0.008 |
| Variable | Men | Women | Value | Men | Women | Value |
| Symptomatology, n (%) | ||||||
| Fever | 0 (0) | 0 (0) | >0.9 | 42 (61.8) | 27(55.1) | 0.57 |
| Cough | 0 (0) | 0 (0) | >0.9 | 57 (83.8) | 38 (77.6) | 0.47 |
| Headache | 12 (66.7) | 17 (77) | 0.49 | 41 (60.3 | 33 (67.4) | 0.56 |
| Dyspnoea | 2 (11.1) | 3 (13.6) | >0.9 | 47 (69.1) | 29 (59.2) | 0.33 |
| Irritability | 1 (5.6) | 2 (9) | >0.9 | 4 (5.9) | 3 (6.1) | >0.9 |
| Diarrhea | 1 (6.6) | 1 (4.6) | >0.9 | 10 (14.7) | 8 (16.3) | 0.8 |
| Chest tightness | 0 (0) | 2 (9.1) | 0.49 | 18 (26.5) | 15 (30.6) | 0.68 |
| Chills | 2 (11.1) | 3 (12.6) | >0.9 | 28 (41.2) | 18 (36.7) | 0.7 |
| Pharyngalgia | 7 (38.9) | 10 (45.95 | 0.75 | 30 (44.1) | 16 (32.7) | 0.25 |
| Myalgia | 7 (38.9) | 8 (36.4) | >0.9 | 33 (48.5) | 33 (67.4) | 0.06 |
| Arthralgias | 4 (22.2) | 7 (31.8) | 0.72 | 32 (47.1) | 32 (65.3) | 0.06 |
| Rhinorrhea | 1 (5.6) | 5 (22.7) | 0.19 | 12 (17.9) | 9 (18.4) | >0.9 |
| Polypnea | 0 (0) | 1 (4.6) | >0.9 | 6 (8.8) | 7 (14.3) | 0.38 |
| Vomiting | 0 (0) | 0 (0) | >0.9 | 4 (5.9) | 5 (10.2) | 0.49 |
| Abdominal pain | 2 (11.1) | 2 (9.1) | >0.9 | 8 (10.5) | 5 (10.2) | >0.9 |
| Conjunctivitis | 0 (0) | 1 (4.6) | >0.9 | 2 (2.9) | 1 (2) | >0.9 |
| Cyanosis | 0 (0) | 1 (4.6) | >0.9 | 0 (0) | 1 (2) | 0.42 |
| Anosmya | 0 (0) | 0 (0) | >0.9 | 10 (14.7) | 13 (26.5) | 0.16 |
| Dysgeusia | 0 (0) | 0 (0) | >0.9 | 10 (14.7) | 14 (28.6) | 0.1 |
| Comorbidities (self-reported), n (%) | ||||||
| Diabetes | 1 (5.6) | 2 (9.1) | >0.9 | 15 (22.1) | 18 (36.7) | 0.09 |
| Hypertension | 5 (27.8) | 4 (18.2) | 0.71 | 26 (38.8) | 18 (36.7) | 0.85 |
| COPD | 0 (0) | 0 (0) | >0.9 | 1 (1.5) | 0 (0) | >0.9 |
| Asthma | 2 (11.1) | 0 (0) | 0.19 | 0 (0) | 2 (4.1) | 0.17 |
| Immunosuppression | 0 (0) | 2 (9.1) | 0.49 | 1 (1.5) | 0 (0) | >0.9 |
| HIV/AIDS | 1 (5.6) | 0 (0) | 0.45 | 0 (0) | 0 (0) | >0.9 |
| Cardiovascular disease | 0 (0) | 0 (0) | >0.9 | 2 (2.9) | 1 (2) | >0.9 |
| Obesity (>30 kg/m2) | 1 (5.6) | 2 (9.1) | >0.9 | 17 (25) | 14 (28.6) | 0.68 |
| Chronic renal insufficiency | 1 (5.6) | 0 (0) | 0.45 | 3 (4.4) | 1 (2) | 0.64 |
| Smoking | 2 (11.1) | 2 (9.1) | >0.9 | 6 (8.8) | 1 (2) | 0.24 |
| Treatment (self-reported), n (%) | ||||||
| Antipyretics | 3 (16.7) | 1 (4.6) | 0.31 | 20 (30.3) | 14 (28.6) | >0.9 |
| Men with COVID-19 vs. Women with COVID-19 | Model | ||
|---|---|---|---|
| SVM | LR | ||
| Included Variables | 3 | 2 | |
| Cross-validation (k = 5) | AUC | 0.91 | 0.82 |
| CI 95% | 0.86–0.98 | 0.71–0.92 | |
| Specificity | 0.94 | 0.90 | |
| Sensitivity | 0.80 | 0.73 | |
| Accuracy | 0.88 | 0.83 | |
| Training (80%) | AUC | 0.92 | 0.82 |
| CI 95% | 0.85–0.98 | 0.73–0.92 | |
| Specificity | 0.92 | 0.90 | |
| Sensitivity | 0.84 | 0.71 | |
| Accuracy | 0.89 | 0.82 | |
| Blind (20%) | AUC | 0.66 | 0.77 |
| CI 95% | 0.47–0.91 | 0.58–0.97 | |
| Specificity | 0.71 | 0.78 | |
| Sensitivity | 0.72 | 0.72 | |
| Accuracy | 0.72 | 0.76 | |
| Women with COVID-19 vs. Women without COVID-19 | Model | ||
|---|---|---|---|
| SVM | LR | ||
| Included Variables | 29 | 12 | |
| Cross-validation (k = 5) | AUC | 1 | 0.99 |
| CI 95% | 1–1 | 0.96–1 | |
| Specificity | 1 | 1 | |
| Sensitivity | 1 | 1 | |
| Accuracy | 1 | 1 | |
| Training (80%) | AUC | 1 | 1 |
| CI 95% | 1–1 | 0.97–1 | |
| Specificity | 1 | 1 | |
| Sensitivity | 1 | 1 | |
| Accuracy | 1 | 1 | |
| Blind (20%) | AUC | 1 | 0.85 |
| CI 95% | 1–1 | 0.64–1 | |
| Specificity | 1 | 1 | |
| Sensitivity | 1 | 1 | |
| Accuracy | 1 | 1 | |
| Men with COVID-19 vs. Men without COVID-19 | Model | ||
|---|---|---|---|
| SVM | LR | ||
| Included Variables | 4 | 8 | |
| Cross-validation (k = 5) | AUC | 0.99 | 1 |
| CI 95% | 0.99–1 | 1–1 | |
| Specificity | 1 | 1 | |
| Sensitivity | 0.97 | 1 | |
| Accuracy | 0.97 | 1 | |
| Training (80%) | AUC | 0.99 | 1 |
| CI 95% | 0.98–1 | 1–1 | |
| Specificity | 1 | 1 | |
| Sensitivity | 0.96 | 1 | |
| Accuracy | 0.97 | 1 | |
| Blind (20%) | AUC | 1 | 1 |
| CI 95% | 1–1 | 1–1 | |
| Specificity | 1 | 1 | |
| Sensitivity | 1 | 1 | |
| Accuracy | 1 | 1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Celaya-Padilla, J.M.; Villagrana-Bañuelos, K.E.; Oropeza-Valdez, J.J.; Monárrez-Espino, J.; Castañeda-Delgado, J.E.; Oostdam, A.S.H.-V.; Fernández-Ruiz, J.C.; Ochoa-González, F.; Borrego, J.C.; Enciso-Moreno, J.A.; et al. Kynurenine and Hemoglobin as Sex-Specific Variables in COVID-19 Patients: A Machine Learning and Genetic Algorithms Approach. Diagnostics 2021, 11, 2197. https://doi.org/10.3390/diagnostics11122197
Celaya-Padilla JM, Villagrana-Bañuelos KE, Oropeza-Valdez JJ, Monárrez-Espino J, Castañeda-Delgado JE, Oostdam ASH-V, Fernández-Ruiz JC, Ochoa-González F, Borrego JC, Enciso-Moreno JA, et al. Kynurenine and Hemoglobin as Sex-Specific Variables in COVID-19 Patients: A Machine Learning and Genetic Algorithms Approach. Diagnostics. 2021; 11(12):2197. https://doi.org/10.3390/diagnostics11122197
Chicago/Turabian StyleCelaya-Padilla, Jose M., Karen E. Villagrana-Bañuelos, Juan José Oropeza-Valdez, Joel Monárrez-Espino, Julio E. Castañeda-Delgado, Ana Sofía Herrera-Van Oostdam, Julio César Fernández-Ruiz, Fátima Ochoa-González, Juan Carlos Borrego, Jose Antonio Enciso-Moreno, and et al. 2021. "Kynurenine and Hemoglobin as Sex-Specific Variables in COVID-19 Patients: A Machine Learning and Genetic Algorithms Approach" Diagnostics 11, no. 12: 2197. https://doi.org/10.3390/diagnostics11122197
APA StyleCelaya-Padilla, J. M., Villagrana-Bañuelos, K. E., Oropeza-Valdez, J. J., Monárrez-Espino, J., Castañeda-Delgado, J. E., Oostdam, A. S. H.-V., Fernández-Ruiz, J. C., Ochoa-González, F., Borrego, J. C., Enciso-Moreno, J. A., López, J. A., López-Hernández, Y., & Galván-Tejada, C. E. (2021). Kynurenine and Hemoglobin as Sex-Specific Variables in COVID-19 Patients: A Machine Learning and Genetic Algorithms Approach. Diagnostics, 11(12), 2197. https://doi.org/10.3390/diagnostics11122197







