Development of an Enzyme-Linked Immunosorbent Assay (ELISA) for Accurate and Prompt Coronavirus Disease 2019 (COVID-19) Diagnosis Using the Rational Selection of Serological Biomarkers
Abstract
:1. Introduction
2. Study Design
2.1. Study Population
2.2. Proteins
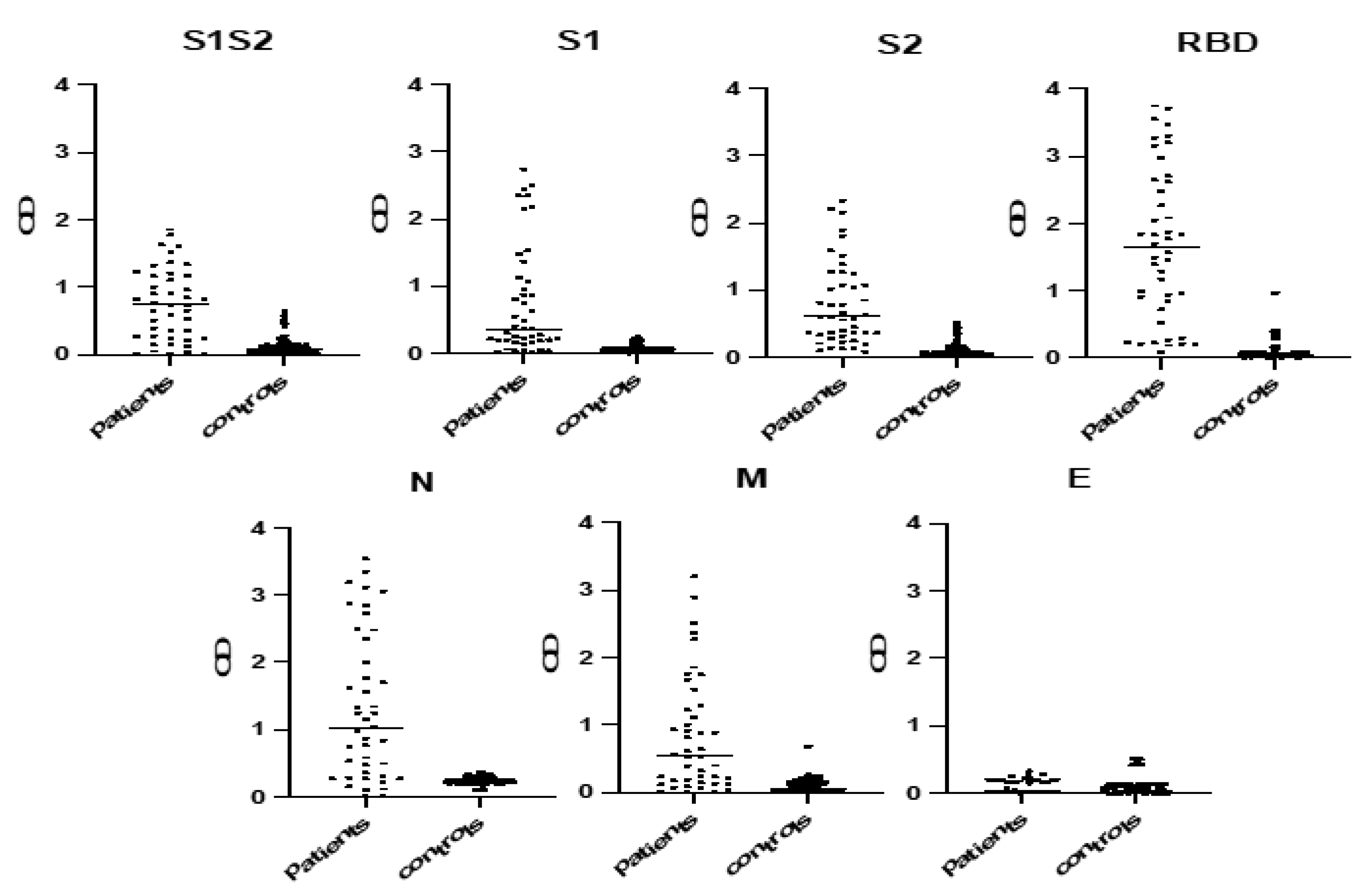
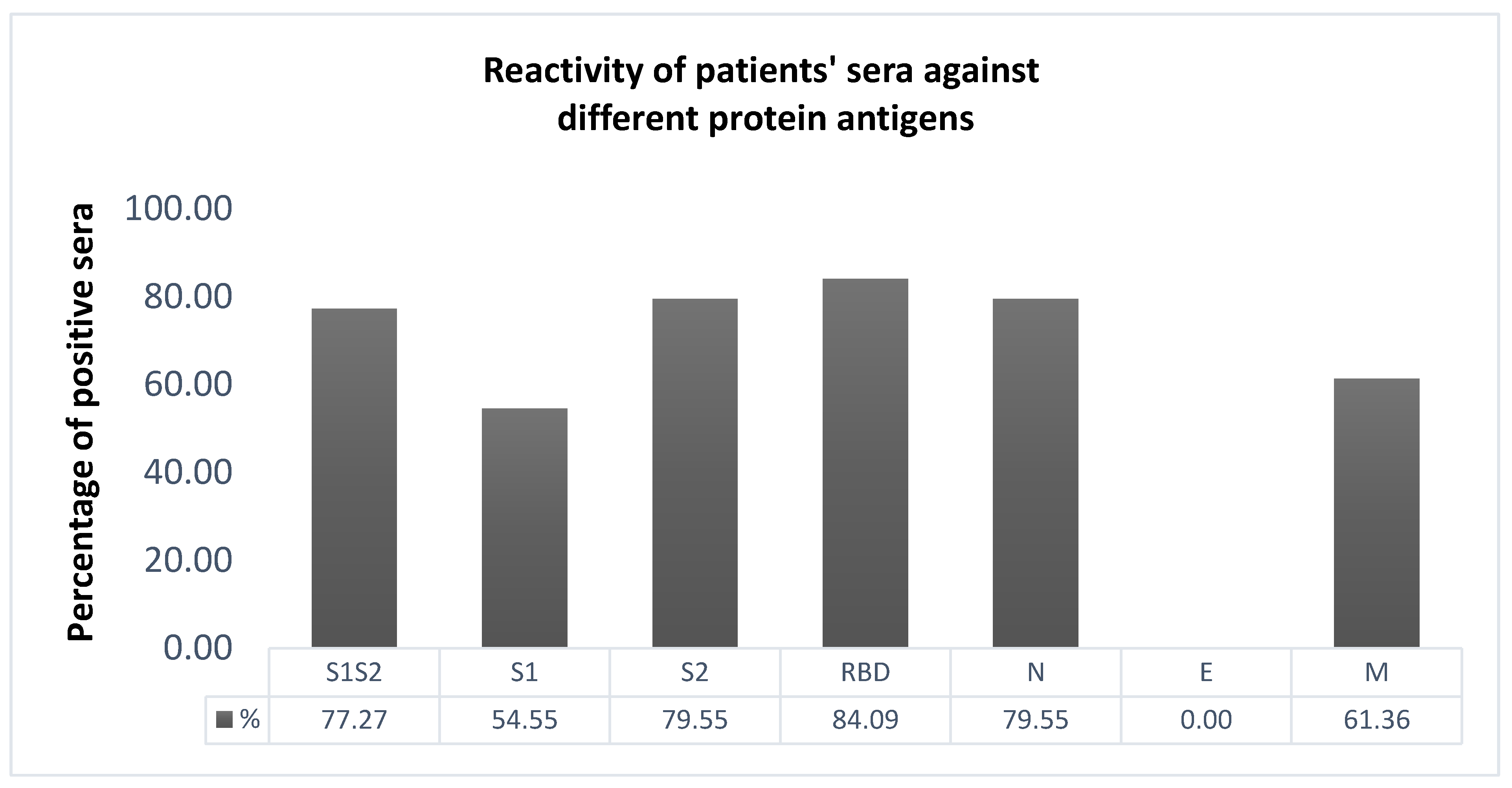
2.3. Selection of the Most Immunoreactive Protein Antigens
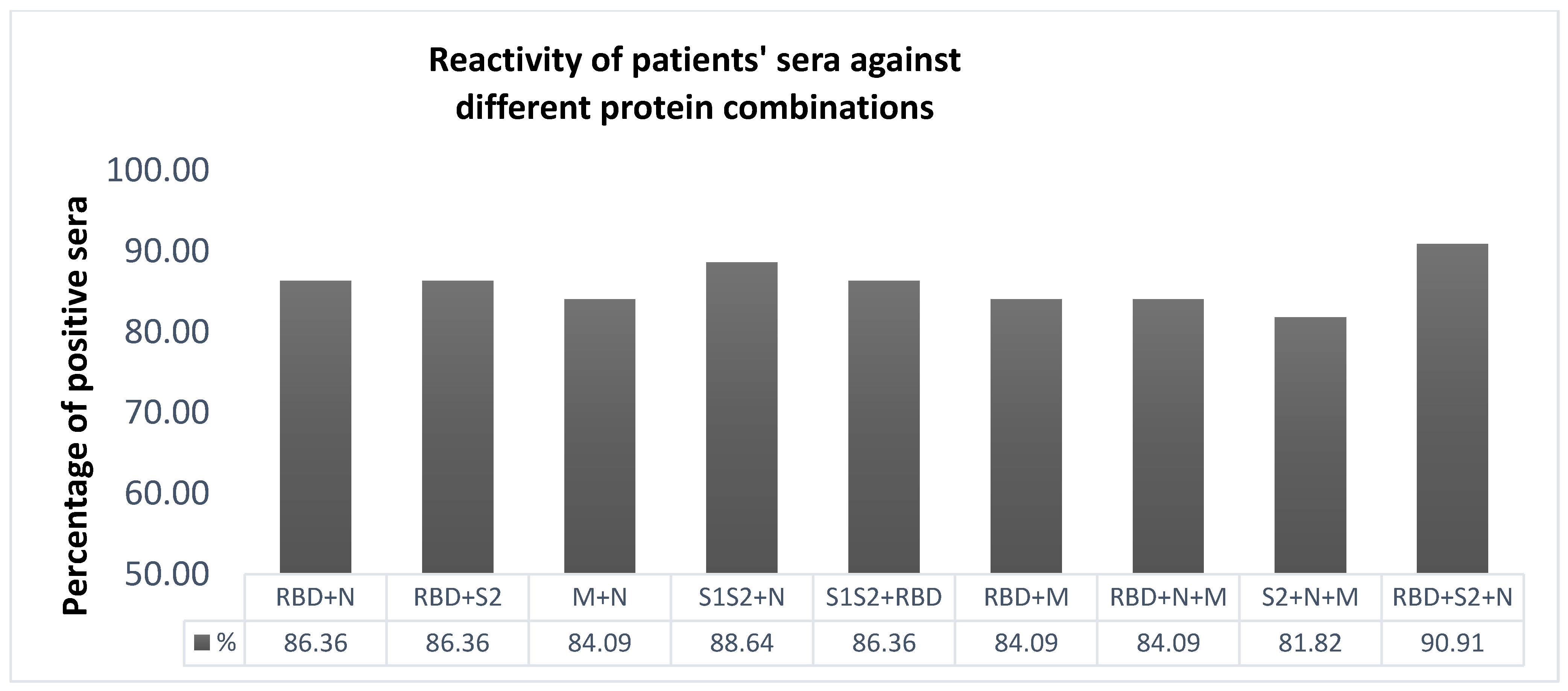
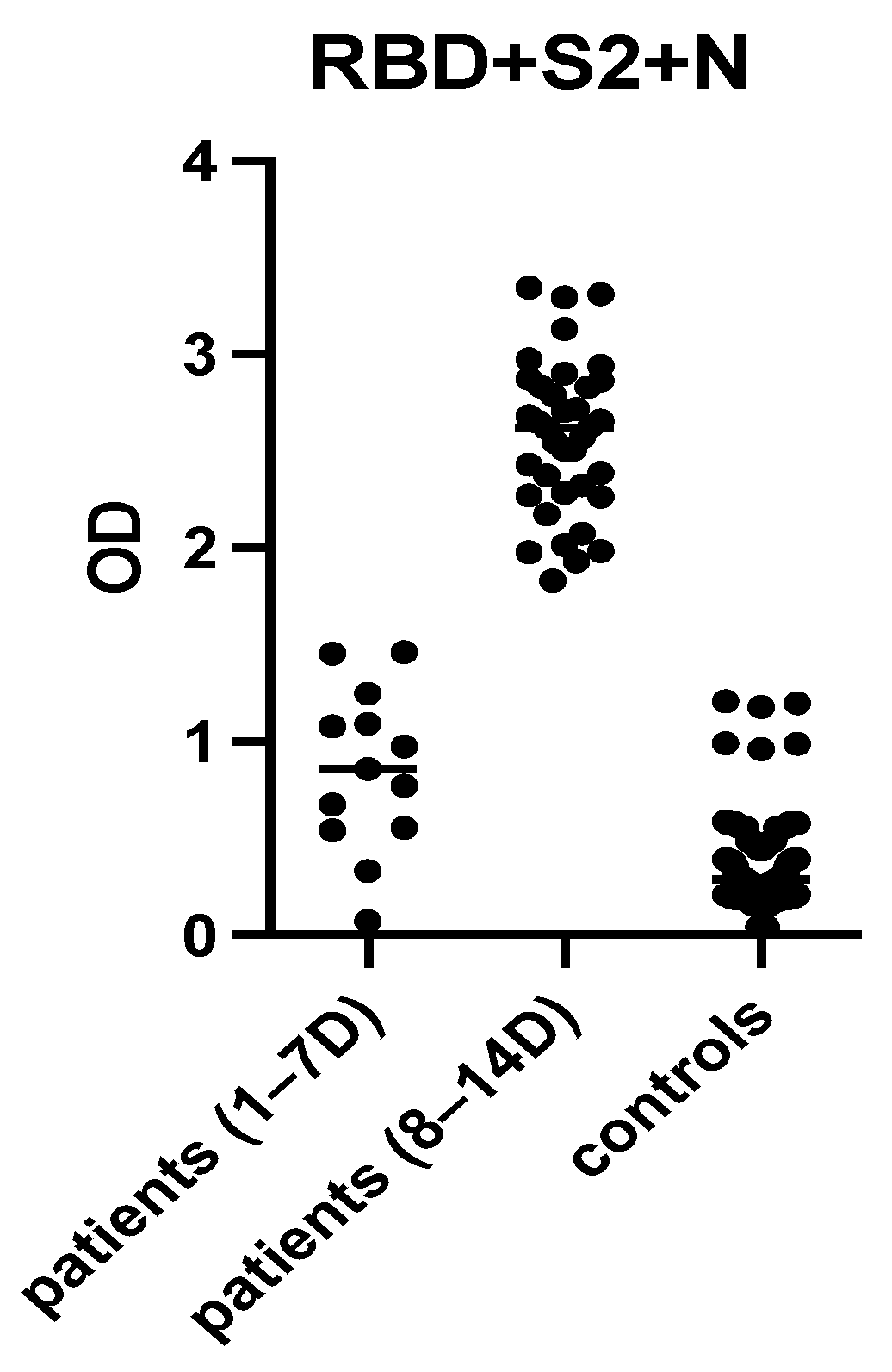
2.4. Selection of the Most Immunoreactive Combination of Protein Antigens
2.5. IgA and IgM Assessment
2.6. Determining Inter-Assay Variability and Repeatability
2.7. Sera Heat Treatment for Viral Inactivation
2.8. Inhibition of IgG Antibodies against Three Antigens
2.9. Comparing the “In-House” SARS-CoV-2 ELISA to Commercially Available Assays
2.10. Implementation of the Developed ELISA in PCR-Positive Children
2.11. Statistical Analysis
3. Results
3.1. ELISA Development
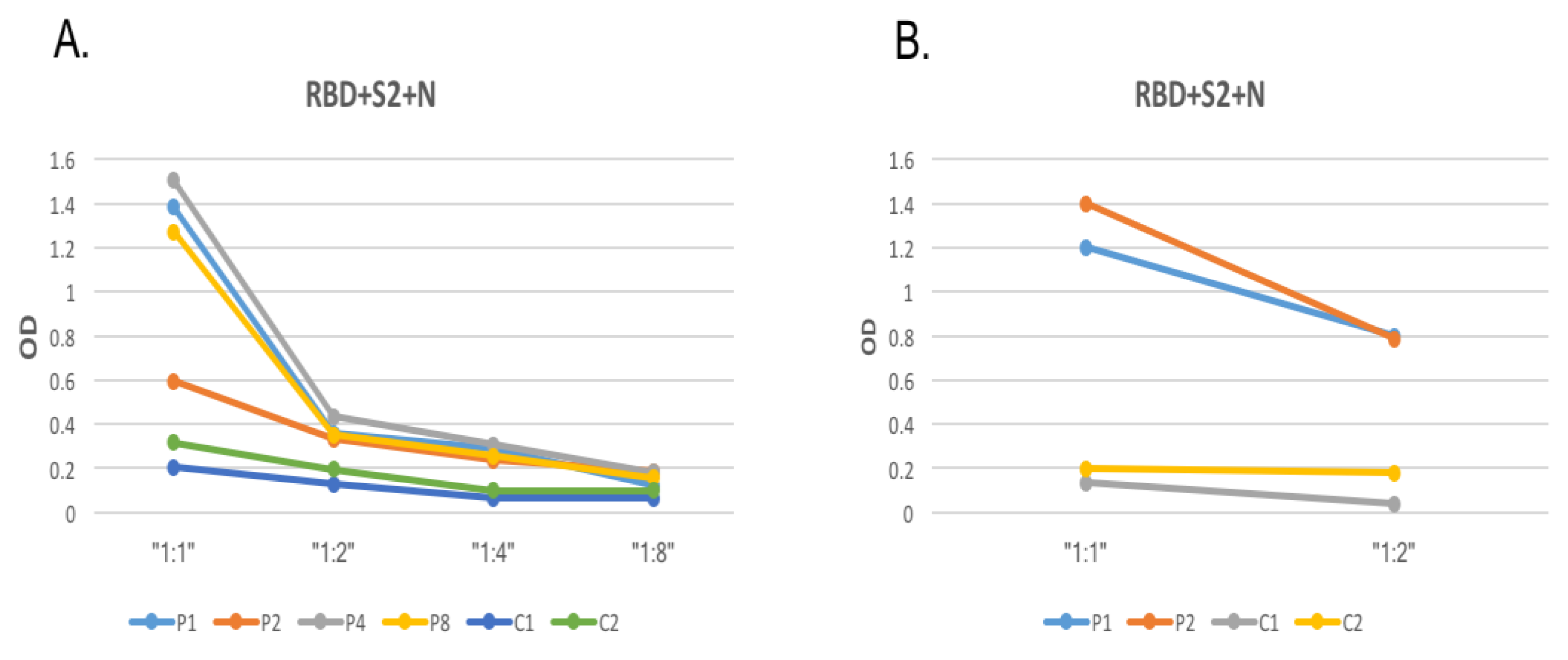
3.2. Optimization of SARS-CoV-2 Protein Coating Concentration and Serum Dilution
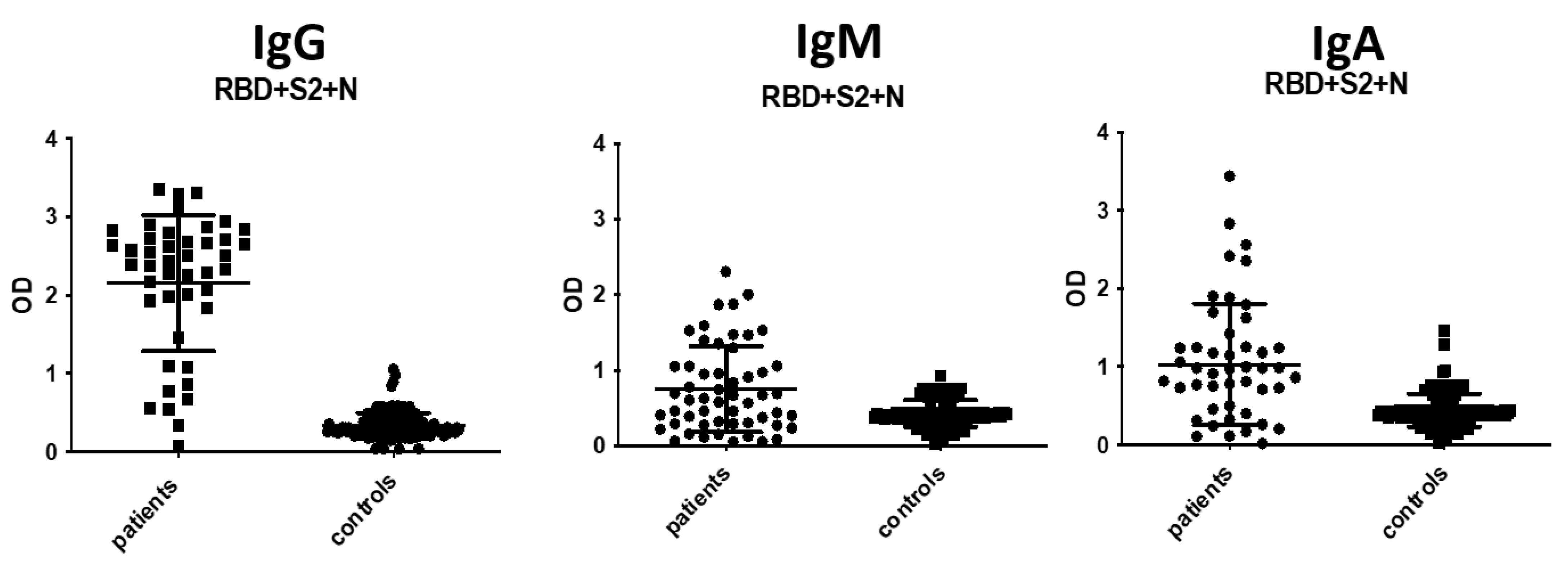
3.3. IgA and IgM Antibody Detection
3.4. Determining Inter-Assay and Intra-Assay Variability and Repeatability
3.5. Inhibition of IgG Antibody Binding
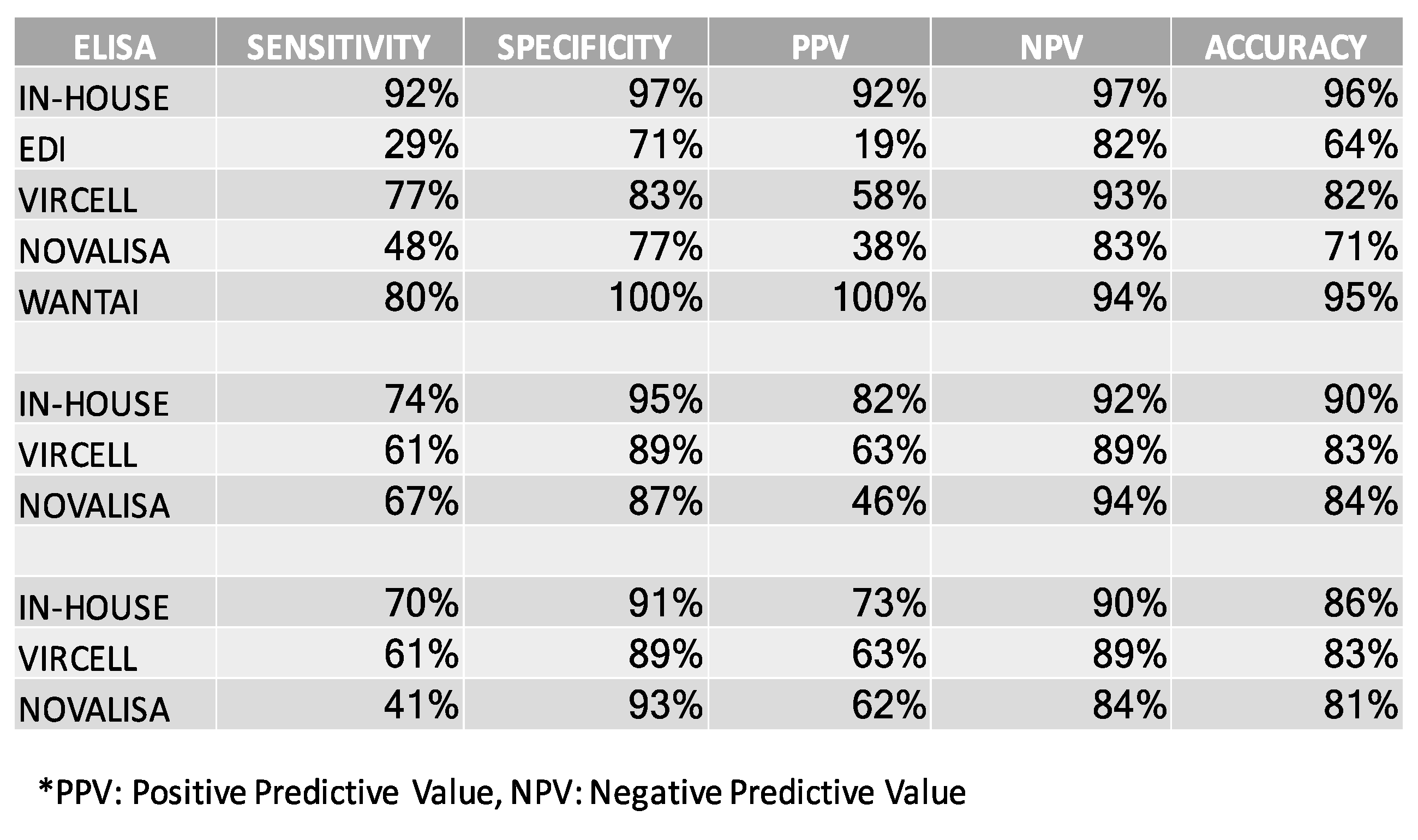
3.6. Comparing the In-House SARS-CoV-2 ELISA to Commercially Available Assays
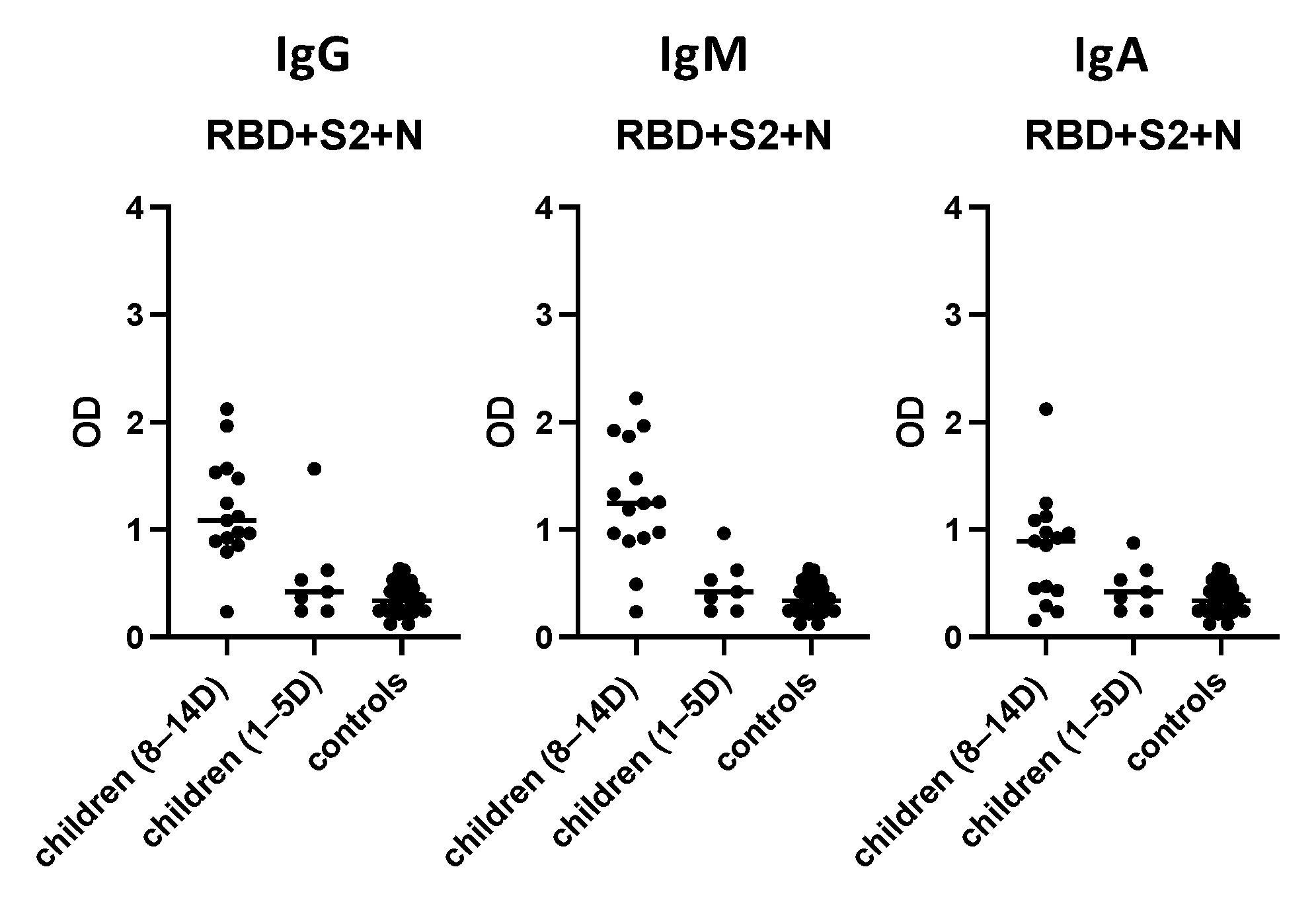
3.7. Evaluation of the “In-House” ELISA in Children
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hui, D.S.; Esam, I.A.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.T.; Leung, K.; Leung, G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. Lancet 2020, 395, 689–697. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Coronavirus Disease (COVID-19) Outbreak Situation. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 16 September 2021).
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2021, 325, 1113. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Lau, E.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020, 26, 1491–1493. [Google Scholar] [CrossRef]
- Tabata, S.; Imai, K.; Kawano, S.; Ikeda, M.; Kodama, T.; Miyoshi, K.; Obinata, H.; Mimura, S.; Kodera, T.; Kitagaki, M.; et al. Clinical characteristics of COVID-19 in 104 people with SARS-CoV-2 infection on the Diamond Princess cruise ship: A retrospective analysis. Lancet Infect. Dis. 2020, 20, 1043–1050. [Google Scholar] [CrossRef]
- Verity, R.; Okell, L.C.; Dorigatti, I.; Winskill, P.; Whittaker, C.; Imai, N.; Cuomo-Dannenburg, G.; Thompson, H.; Walker, P.; Fu, H.; et al. Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect. Dis. 2020, 20, 669–677. [Google Scholar] [CrossRef]
- Long, Q.X.; Tang, X.J.; Shi, Q.L.; Li, Q.; Deng, H.J.; Yuan, J.; Hu, J.L.; Xu, W.; Zhang, Y.; Lv, F.J.; et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020, 26, 1200–1204. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Føns, S.; Krogfelt, K.A. How can we interpret SARS-CoV-2 antibody test results? Pathog. Dis. 2021, 79, ftaa069. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef] [Green Version]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef] [Green Version]
- DeDiego, M.L.; Alvarez, E.; Almazán, F.; Rejas, M.T.; Lamirande, E.; Roberts, A.; Shieh, W.J.; Zaki, S.R.; Subbarao, K.; Enjuanes, L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007, 81, 1701–1713. [Google Scholar] [CrossRef] [Green Version]
- Ortego, J.; Ceriani, J.E.; Patiño, C.; Plana, J.; Enjuanes, L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology 2007, 368, 296–308. [Google Scholar] [CrossRef] [Green Version]
- Younes, N.; Al-Sadeq, D.W.; Al-Jighefee, H.; Younes, S.; Al-Jamal, O.; Daas, H.I.; Yassine, H.M.; Nasrallah, G.K. Challenges in Laboratory Diagnosis of the Novel Coronavirus SARS-CoV-2. Viruses 2020, 12, 582. [Google Scholar] [CrossRef]
- Batéjat, C.; Grassin, Q.; Manuguerra, J.C.; Leclercq, I. Heat inactivation of the severe acute respiratory syndrome coronavirus 2. J. Biosaf. Biosecur. 2021, 3, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Darnell, M.E.; Taylor, D.R. Evaluation of inactivation methods for severe acute respiratory syndrome coronavirus in noncellular blood products. Transfusion 2006, 46, 1770–1777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittal, A.; Manjunath, K.; Ranjan, R.K.; Kaushik, S.; Kumar, S.; Verma, V. COVID-19 pandemic: Insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLoS Pathog. 2020, 16, e1008762. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Liu, G.; Ma, H. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020, 527, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Ye, G.; Shi, K. Structural basis of receptor recognition by SARS-CoV 2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Poland, G.A.; Ovsyannikova, I.G.; Kennedy, R.B. SARS-CoV-2 immunity: Review and applications to phase 3 vaccine candidates. Lancet 2020, 396, 1595–1606. [Google Scholar] [CrossRef]
- Bryan, A.; Pepper, G.; Wener, M.H. Performance characteristics of the Abbott architect SARS-CoV-2 IgG assay and seroprevalence testing in Idaho. J. Clin. Microbiol. 2020, 58, e00941-20. [Google Scholar] [CrossRef]
- Chan, C.W.; Parker, K.; Tesic, V. Analytical and clinical evaluation of the automated Elecsys anti-SARS-CoV-2 antibody assay on the Roche cobas e602 analyzer. Am. J. Clin. Pathol. 2020, 154, 620–626. [Google Scholar] [CrossRef]
- Muench, P.; Jochum, S.; Wenderoth, V. Development and validation of the Elecsys anti-SARS-CoV-2 immunoassay as a highly specific tool for determining past exposure to SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e01694-20. [Google Scholar] [CrossRef]
- Beavis, K.G.; Matushek, S.M.; Abeleda, A.P.F. Evaluation of the EUROIMMUN anti-SARS-CoV-2 ELISA assay for detection of IgA and IgG antibodies. J. Clin. Virol. 2020, 129, 104468. [Google Scholar] [CrossRef]
- Tan, Y.-J.; Lim, S.G.; Hong, W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral Res. 2005, 65, 69–78. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Leung, D.T.M.; Tam, F.C.H.; Ma, C.H. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 2004, 190, 379–386. [Google Scholar] [CrossRef] [Green Version]
- Burbelo, P.D.; Riedo, F.X.; Morishima, C. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J. Infect. Dis. 2020, 222, 206–213. [Google Scholar] [CrossRef]
- Atyeo, C.; Fischinger, S.; Zohar, T. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity 2020, 53, 524–532. [Google Scholar] [CrossRef]
- Weisberg, S.P.; Connors, T.J.; Zhu, Y. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 2020, 22, 25–31. [Google Scholar] [CrossRef]
- Yang, X.L.; Hu, B.; Wang, B.; Wang, M.N.; Zhang, Q.; Zhang, W.; Wu, L.J.; Ge, X.Y.; Zhang, Y.Z.; Daszak, P.; et al. Isolation and Characterization of a Novel Bat Coronavirus Closely Related to the Direct Progenitor of Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2015, 90, 3253–3256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, R.L.; Baric, R.S. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. J. Virol. 2010, 84, 3134–3146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bates, T.A.; Weinstein, J.B.; Farley, S.; Leier, H.C.; Messer, W.B.; Tafesse, F.G. Cross-reactivity of SARS-CoV structural protein antibodies against SARS-CoV-2. Cell Rep. 2021, 34, 108737. [Google Scholar] [CrossRef] [PubMed]
- Boson, B.; Legros, V.; Zhou, B.; Siret, E.; Mathieu, C.; Cosset, F.L.; Lavillette, D.; Denolly, S. The SARS-CoV-2 Envelope and Membrane Proteins Modulate Maturation and Retention of the Spike Protein, Allowing Assembly of Virus-Like Particles. J. Biol. Chem. 2021, 296, 100111. [Google Scholar] [CrossRef]
- Chow, S.C.; Ho, C.Y.; Tam, T.T.; Wu, C.; Cheung, T.; Chan, P.K.; Ng, M.H.; Hui, P.K.; Ng, H.K.; Au, D.M.; et al. Specific epitopes of the structural and hypothetical proteins elicit variable humoral responses in SARS patients. J. Clin. Pathol. 2006, 59, 468–476. [Google Scholar] [CrossRef] [Green Version]
- Liang, M.F.; Du, R.L.; Liu, J.Z.; Li, C.; Zhang, Q.F.; Han, L.L.; Yu, J.S.; Duan, S.M.; Wang, X.F.; Wu, K.X.; et al. SARS Patients-Derived Human Recombinant Antibodies to S and M Proteins Efficiently Neutralize SARS-Coronavirus Infectivity. Biomed. Environ. Sci. 2005, 18, 363–374. [Google Scholar]
- Wang, X.; Xu, W.; Tong, D.; Ni, J.; Gao, H.; Wang, Y.; Chu, Y.; Li, P.; Yang, X.; Xiong, S. A Chimeric Multi-Epitope DNA Vaccine Elicited Specific Antibody Response Against Severe Acute Respiratory Syndrome-Associated Coronavirus Which Attenuated the Virulence of SARS-CoV In Vitro. Immunol. Lett. 2008, 119, 71–77. [Google Scholar] [CrossRef]
- Westerbeck, J.W.; Machamer, C.E. A Coronavirus E Protein Is Present in Two Distinct Pools with Different Effects on Assembly and the Secretory Pathway. J. Virol. 2015, 89, 9313–9323. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Li, X.; Zhang, C.; Xu, S.; Shao, Y.; Zhuang, H.; Che, X.; Qiu, Y.; Yin, H.; Li, D.; et al. Establishment of a reference panel for the detection of anti-SARS-CoV antibodies. Biologicals 2007, 35, 203–210. [Google Scholar] [CrossRef]
- Vengesai, A.; Midzi, H.; Kasambala, M.; Mutandadzi, H.; Mduluza-Jokonya, T.L.; Rusakaniko, S.; Mutapi, F.; Naicker, T.; Mduluza, T. A systematic and meta-analysis review on the diagnostic accuracy of antibodies in the serological diagnosis of COVID-19. Syst. Rev. 2021, 10, 155. [Google Scholar] [CrossRef] [PubMed]
- Park, W.B.; Perera, R.A.; Choe, P.G.; Lau, E.H.; Choi, S.J.; Chun, J.Y.; Oh, H.S.; Song, K.H.; Bang, J.H.; Kim, E.S.; et al. Kinetics of Serologic Responses to MERS Coronavirus Infection in Humans, South Korea. Emerg. Infect. Dis. 2015, 21, 2186–2189. [Google Scholar]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.L.; Wang, H.J.; Deng, Y.; Song, T.; Lan, J.M.; Wu, G.Z.; Ke, C.W.; Tan, W.J. Serological Study of An Imported Case of Middle East Respiratory Syndrome and His Close Contacts in China, 2015. Biomed. Environ. Sci. 2016, 29, 219–223. [Google Scholar] [PubMed]
- Al-Abdely, H.M.; Midgley, C.M.; Alkhamis, A.M.; Abedi, G.R.; Lu, X.; Binder, A.M.; Alanazi, K.H.; Tamin, A.; Banjar, W.M.; Lester, S.; et al. Middle East Respiratory Syndrome Coronavirus Infection Dynamics and Antibody Responses among Clinically Diverse Patients, Saudi Arabia. Emerg. Infect. Dis. 2019, 25, 753–766. [Google Scholar] [CrossRef] [Green Version]
- Vogl, T.; Leviatan, S.; Segal, E. SARS-CoV-2 antibody testing for estimating COVID-19 prevalence in the population. Cell Rep. Med. 2021, 2, 100191. [Google Scholar] [CrossRef]
- Kharlamova, N.; Dunn, N.; Bedri, S.K.; Jerling, S.; Almgren, M.; Faustini, F.; Gunnarsson, I.; Rönnelid, J.; Pullerits, R.; Gjertsson, I.; et al. False Positive Results in SARS-CoV-2 Serological Tests for Samples from Patients With Chronic Inflammatory Diseases. Front. Immunol. 2021, 12, 666114. [Google Scholar] [CrossRef]
- Guevara-Hoyer, K.; Fuentes-Antrás, J.; De la Fuente-Muñoz, E.; Rodríguez de la Peña, A.; Viñuela, M.; Cabello-Clotet, N.; Estrada, V.; Culebras, E.; Delgado-Iribarren, A.; Martínez-Novillo, M.; et al. Serological Tests in the Detection of SARS-CoV-2 Antibodies. Diagnostics 2021, 11, 678. [Google Scholar] [CrossRef]
- Marklund, E.; Leach, S.; Axelsson, H.; Nyström, K.; Norder, H.; Bemark, M.; Angeletti, D.; Lundgren, A.; Nilsson, S.; Andersson, L.M.; et al. Serum-IgG responses to SARS-CoV-2 after mild and severe COVID-19 infection and analysis of IgG non-responders. PLoS ONE 2020, 15, e0241104. [Google Scholar] [CrossRef]
- Grzelak, L.; Temmam, S.; Planchais, C.; Demeret, C.; Tondeur, L.; Huon, C.; Guivel-Benhassine, F.; Staropoli, I.; Chazal, M.; Dufloo, J.; et al. A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations. Sci. Transl. Med. 2020, 12, eabc3103. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody Responses to SARS-CoV-2 in Patients with Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2027–2034. [Google Scholar] [CrossRef]
- To, K.K.; Tsang, O.T.; Leung, W.S.; Tam, A.R.; Wu, T.C.; Lung, D.C.; Yip, C.C.; Cai, J.P.; Chan, J.M.; Chik, T.S.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Farronato, M.; Dolci, C.; Boccalari, E.; Izadi, S.; Salvatierra Rios, L.H.; Festa, M.; Panetta, V.; De Vito, D.; Tartaglia, G.M. Serological Profile of Children and Young Adults with at Least One SARS-CoV-2 Positive Cohabitant: An Observational Study. Int. J. Environ. Res. Public Health 2021, 18, 1488. [Google Scholar] [CrossRef] [PubMed]
- Sinaei, R.; Pezeshki, S.; Parvaresh, S.; Sinaei, R. Why COVID-19 is less frequent and severe in children: A narrative review. World J. Pediatr. 2021, 17, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Castagnoli, R.; Votto, M.; Licari, A.; Brambilla, I.; Bruno, R.; Perlini, S.; Rovida, F.; Baldanti, F.; Marseglia, G.L. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection in Children and Adolescents. JAMA Pediatr. 2020, 174, 882. [Google Scholar] [CrossRef] [Green Version]
- Saleem, H.; Rahman, J.; Aslam, N.; Murtazaliev, S.; Khan, S. Coronavirus Disease 2019 (COVID-19) in Children: Vulnerable or Spared? A Systematic Review. Cureus 2020, 12, e8207. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Mo, X.; Hu, Y.; Qi, X.; Jiang, F.; Jiang, Z.; Tong, S. Epidemiology of COVID-19 Among Children in China. Paediatrics 2020, 145, e20200702. [Google Scholar] [CrossRef] [Green Version]
- Bialek, S.; Gierke, R.; Hughes, M.; McNamara, L.A.; Pilishvili, T.; Skoff, T. Coronavirus Disease 2019 in Children—United States, February 12–April 2, 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 422–426. [Google Scholar]
- Patel, N.A. Paediatric COVID-19: Systematic review of the literature. Am. J. Otolaryngol. 2020, 41, 102573. [Google Scholar] [CrossRef] [PubMed]
- Meena, J.; Yadav, J.; Saini, L.; Yadav, A.; Kumar, J. Clinical Features and Outcome of SARS-CoV-2 Infection in Children: A Systematic Review and Meta-analysis. Indian Pediatr. 2020, 57, 820–826. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Singhal, T. A Review of Coronavirus Disease-2019 (COVID-19). Indian J. Pediatr. 2020, 87, 281–286. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; Zhao, Z.; Zhang, T.; Guo, W.; Guo, W.; Zheng, J.; Zhang, J.; Dong, C.; Na, R.; Zheng, L.; et al. A systematic review and meta-analysis of children with coronavirus disease 2019 (COVID-19). J. Med. Virol. 2021, 93, 1057–1069. [Google Scholar] [CrossRef]
- Laws, R.L.; Chancey, R.J.; Rabold, E.M.; Chu, V.T.; Lewis, N.M.; Fajans, M.; Reses, H.E.; Duca, L.M.; Dawson, P.; Conners, E.E.; et al. Symptoms and Transmission of SARS-CoV-2 Among Children—Utah and Wisconsin, March–May 2020. Paediatrics 2021, 147, e2020027268. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lagousi, T.; Routsias, J.; Spoulou, V. Development of an Enzyme-Linked Immunosorbent Assay (ELISA) for Accurate and Prompt Coronavirus Disease 2019 (COVID-19) Diagnosis Using the Rational Selection of Serological Biomarkers. Diagnostics 2021, 11, 1970. https://doi.org/10.3390/diagnostics11111970
Lagousi T, Routsias J, Spoulou V. Development of an Enzyme-Linked Immunosorbent Assay (ELISA) for Accurate and Prompt Coronavirus Disease 2019 (COVID-19) Diagnosis Using the Rational Selection of Serological Biomarkers. Diagnostics. 2021; 11(11):1970. https://doi.org/10.3390/diagnostics11111970
Chicago/Turabian StyleLagousi, Theano, John Routsias, and Vana Spoulou. 2021. "Development of an Enzyme-Linked Immunosorbent Assay (ELISA) for Accurate and Prompt Coronavirus Disease 2019 (COVID-19) Diagnosis Using the Rational Selection of Serological Biomarkers" Diagnostics 11, no. 11: 1970. https://doi.org/10.3390/diagnostics11111970
APA StyleLagousi, T., Routsias, J., & Spoulou, V. (2021). Development of an Enzyme-Linked Immunosorbent Assay (ELISA) for Accurate and Prompt Coronavirus Disease 2019 (COVID-19) Diagnosis Using the Rational Selection of Serological Biomarkers. Diagnostics, 11(11), 1970. https://doi.org/10.3390/diagnostics11111970




