Head and Neck Squamous Cell Carcinoma: Epigenetic Landscape
Abstract
1. Introduction
2. The HNSCC Epigenetic Landscape and Its Clinical Implications
2.1. DNA Methylation
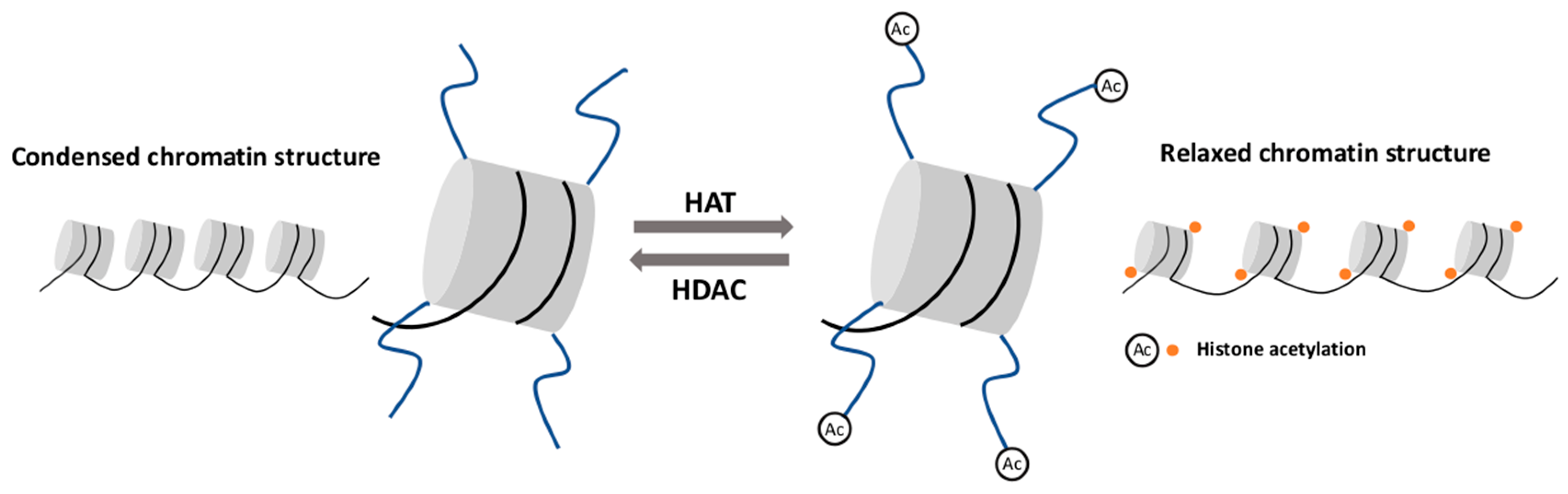
2.2. Histone Modifications
2.3. Non-Coding RNA Activity
2.4. RNA Methylation
- (1)
- main catalytic core enzyme which states methyltransferase like 3 (METTL3),
- (2)
- methyltransferase like 14 (METTL14) which structurally positions mRNA for methylation,
- (3)
- WT1-associated protein (WTAP) regulating the recruitment of methyltransferase complex to mRNA targets,
- (4)
- RNA-binding motif protein 15 (RBM15) which is responsible for moving the complex towards the appropriate m6A sites and the last “writer” protein,
- (5)
- Vir like m6A methyltransferase associated (VIRMA) with uncharacterized molecular function.
3. Conclusions
Funding
Conflicts of Interest
Abbreviations
| 3′UTR | 3′ untranslated region |
| 5-caC | 5-carboxylcytosine |
| 5-fC | 5-formylcytosine |
| 5-hMC | 5-hydroxymethylcytosine |
| 5-mC | 5-methylcytosine |
| AID/APOBEC | Activation- induced deaminase/apoplipoprotein B |
| ALKBH5 | AlkB homolog 5 |
| APC | Adenomatous polyposis coli |
| ATM | Ataxia-telangiectasia-mutated |
| BER | Base excision repair |
| BRCA1 | Breast cancer type I |
| CALML5 | Calmodulin like 5 |
| CCNA1 | Cyclin-A1 |
| CDKN2A | Cyclin Dependent Kinase Inhibitor 2A |
| CSC | Cancer steam cell |
| CT | Chemotherapy |
| ctDNA | Circulating DNA |
| CTLA4 | Cytotoxic T-Lymphocyte Associated Protein 4 |
| DAPK | Death-associated protein kinase |
| DNMT | DNA methyltrasnferase |
| EGFR | Epidermal growth factor receptor |
| EMT | Epithelial-mesenchymal transition |
| FAM135B | Family with sequence similarity 135 member B |
| FTO | FTO Alpha-ketoglutarate dependent dioxygenase |
| GLI1 | GLI1 Family Zinc Finger 1 |
| HAT | Histone acetyltransferase |
| HDAC | Histone deacetylase |
| HKMT | Histone lysine methyltransferase |
| HMT | Histone methyltransferase |
| HNSCC | Head and neck squamous cell carcinoma |
| HPV | Human papilloma virus |
| hTERT | Human telomerase reverse transcriptase |
| JMJD6 | Jumonji domain containing 6 |
| KDM | Lysine specific demethylase |
| LINE-1 | Long interspersed nuclear element 1 |
| LSCC | Laryngeal squamous cell carcinoma |
| LY6D | Lymphocyte antigen 6 family member D |
| m6A | N6-methyladenosine |
| MEIOC | Meiosis specific with coiled-coli domain |
| METTL | Methyltransferase like |
| MGMT | O-6-methylguanine-DNA methyltransferase |
| miRNA | microRNA |
| MLH1 | MutL homolog 1 |
| ncRNA | Non-coding RNA |
| NF-κB | Nuclear transcription factor-κB |
| NPC | Nasopharyngeal carcinoma |
| OSCC | Oral squamous cell carcinoma |
| PADI4 | Peptidyl arginine deiminase 4 |
| piRNA | PIWI-interacting RNA |
| Pl3K/Akt | Phosphatidylinositol 3-kinase/threonine protein kinase B |
| PRMT | Histone arginine methyltransferase |
| PROM1 | Prominin 1 |
| RASSF1 | Ras association domain family member 1 |
| RB | Retinoblastoma |
| RBM15 | RNA-binding motif protein 15 |
| RISC | RNA-induced silencing complex |
| RT | Radiotherapy |
| SAM | S-adenosylomethionine |
| SAT2 | Spermidine/spermine N1-acetyltransferase family member 2 |
| siRNA | Small interfering RNA |
| SMO | smoothened |
| snoRNA | Small nuclear RNA |
| STAT3 | Signal transducer and activator of transcription 3 |
| TDG | Thymine-DNA glycosylase |
| TET | Tet-methylcytosine dioxygenase |
| TGF-β | Transforming growth factor β |
| TIMP3 | TIMP metallopeptidase inhibitor 3 |
| TRBA | TAR RNA binding protein |
| TSCC | Tongue squamous cell carcinoma |
| TSG | Tumor suppressor gene |
| WTAP | WT1-associated protein |
| VIRMA | Vir like m6A methyltransferase associated |
| YTH | YT521-B homology domain |
| ZIC4 | Zic family member 4 |
| ZNF | Zinc-finger protein |
References
- Marur, S.; Forastiere, A.A. Head and neck squamous cell carcinoma: Update on epidemiology, diagnosis, and treatment. Mayo Clin. Proc. 2016, 91, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Sessions, D.G.; Spector, G.J.; Lenox, J.; Haughey, B.; Chao, C.; Marks, J. Analysis of treatment results for oral tongue cancer. Laryngoscope 2002, 112, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, S.; Talamini, R.; Barra, S.; Barón, A.E.; Negri, E.; Bidoli, E.; Serraino, D.; La Vecchia, C. Smoking and drinking in relation to cancers of the oral cavity, pharynx, larynx, and esophagus in northern Italy. Cancer Res. 1990, 50, 6502–6507. [Google Scholar] [PubMed]
- Argiris, A.; Karamouzis, M.V.; Raben, D.; Ferris, R.L. Head and neck cancer. Lancet 2008, 371, 1695–1709. [Google Scholar] [CrossRef]
- Gillison, M.L.; Koch, W.M.; Capone, R.B.; Spafford, M.; Westra, W.H.; Wu, L.; Zahurak, M.L.; Daniel, R.W.; Viglione, M.; Symer, D.E.; et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 2000, 92, 709–720. [Google Scholar] [CrossRef]
- Sisk, E.A.; Bradford, C.R.; Carey, T.E.; Paulino, A.; Robertson, E. Epstein-Barr virus detected in a head and neck squamous cell carcinoma cell line derived from an immunocompromised patient. Arch. Otolaryngol. Head Neck Surg. 2003, 129, 1115–1124. [Google Scholar] [CrossRef][Green Version]
- Moskovitz, J.; Moy, J.; Ferris, R.L. Immunotherapy for Head and Neck Squamous Cell Carcinoma. Curr. Oncol. Rep. 2018, 20, 22. [Google Scholar] [CrossRef]
- Alsahafi, E.; Begg, K.; Amelio, I.; Raulf, N.; Lucarelli, P.; Sauter, T.; Tavassoli, M. Clinical update on head and neck cancer: Molecular biology and ongoing challenges. Cell Death Dis. 2019, 10, 540. [Google Scholar] [CrossRef]
- Gleich, L.L.; Li, Y.Q.; Biddinger, P.W.; Gartside, P.S.; Stambrook, P.J.; Pavelic, Z.P.; Gluckman, J.L. The loss of heterozygosity in retinoblastoma and p53 suppressor genes as a prognostic indicator for head and neck cancer. Laryngoscope 1996, 106, 1378–1381. [Google Scholar] [CrossRef]
- Peltonen, J.K.; Helppi, H.M.; Pääkkö, P.; Turpeenniemi-Hujanen, T.; Vähäkangas, K.H. p53 in head and neck cancer: Functional consequences and environmental implications of TP53 mutations. Head Neck Oncol. 2010, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Balan, A.; Balaram, P. The expression of retinoblastoma tumor suppressor protein in oral cancers and precancers: A clinicopathological study. Dent. Res. J. 2015, 12, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.S.; Fredrik, P.; Ker, L.; Yu, F.G.; Wang, D.Y.; Goh, B.C.; Loh, K.S.; Lim, C.M. High-risk HPV genotypes and P16INK4a expression in a cohort of head and neck squamous cell carcinoma patients in Singapore. Oncotarget 2016, 7, 86730–86739. [Google Scholar] [CrossRef] [PubMed]
- Bossi, P.; Resteghini, C.; Paielli, N.; Licitra, L.; Pilotti, S.; Perrone, F. Prognostic and predictive value of EGFR in head and neck squamous cell carcinoma. Oncotarget 2016, 7, 74362–74379. [Google Scholar] [CrossRef] [PubMed]
- Fukusumi, T.; Califano, J.A. The NOTCH Pathway in Head and Neck Squamous Cell Carcinoma. J. Dent. Res. 2018, 97, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Marquard, F.E.; Jücker, M. PI3K/AKT/mTOR signaling as a molecular target in head and neck cancer. Biochem. Pharmacol. 2020, 172, 113729. [Google Scholar] [CrossRef] [PubMed]
- Geiger, J.L.; Grandis, J.R.; Bauman, J.E. The STAT3 pathway as a therapeutic target in head and neck cancer: Barriers and innovations. Oral Oncol. 2016, 56, 84–92. [Google Scholar] [CrossRef]
- Cho, Y.A.; Kim, E.K.; Heo, S.J.; Cho, B.C.; Kim, H.R.; Chung, J.M.; Yoon, S.O. Alteration status and prognostic value of MET in head and neck squamous cell carcinoma. J. Cancer 2016, 7, 2197–2206. [Google Scholar] [CrossRef]
- Allen, C.T.; Ricker, J.L.; Chen, Z.; Van Waes, C. Role of activated nuclear factor-kappaB in the pathogenesis and therapy of squamous cell carcinoma of the head and neck. Head Neck 2007, 29, 959–971. [Google Scholar] [CrossRef]
- Castilho, R.M.; Squarize, C.H.; Almeida, L.O. Epigenetic Modifications and Head and Neck Cancer: Implications for Tumor Progression and Resistance to Therapy. Int. J. Mol. Sci. 2017, 18, 1506. [Google Scholar] [CrossRef]
- Zhang, P.; He, Q.; Lei, Y.; Li, Y.; Wen, X.; Hong, M.; Zhang, J.; Ren, X.; Wang, Y.; Yang, X.; et al. m6A-mediated ZNF750 repression facilitates nasopharyngeal carcinoma progression. Cell Death Dis. 2018, 9, 1169. [Google Scholar] [CrossRef] [PubMed]
- Lorincz, M.C.; Dickerson, D.R.; Schmitt, M.; Groudine, M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004, 11, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- Antequera, F. Structure, function and evolution of CpG island promoters. Cell Mol. Life Sci. 2003, 60, 1647–1658. [Google Scholar] [CrossRef] [PubMed]
- Ha, P.K.; Califano, J.A. Promoter methylation and inactivation of tumour-suppressor genes in oral squamous-cell carcinoma. Lancet Oncol. 2006, 7, 77–82. [Google Scholar] [CrossRef]
- Zheng, Y.; Joyce, B.T.; Liu, L.; Zhang, Z.; Kibbe, W.A.; Zhang, W.; Hou, L. Prediction of genome-wide DNA methylation in repetitive elements. Nuc. Acids Res. 2017, 45, 8697–8711. [Google Scholar] [CrossRef]
- Jones, P. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Chodavarapu, R.K.; Feng, S.; Bernatavichute, Y.V.; Chen, P.Y.; Stroud, H.; Yu, Y.; Hetzel, J.A.; Kuo, F.; Kim, J.; Cokus, S.J.; et al. Relationship between nucleosome positioning and DNA methylation. Nature 2010, 466, 388–392. [Google Scholar] [CrossRef]
- Guo, J.U.; Su, Y.; Shin, J.H.; Shin, J.; Li, H.; Xie, B.; Zhong, C.; Hu, S.; Le, T.; Fan, G.; et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014, 17, 215–222. [Google Scholar] [CrossRef]
- Cheng, X.; Blumenthal, R.M. Mammalian DNA methyltransferases: A structural perspective. Structure 2008, 16, 341–350. [Google Scholar] [CrossRef]
- Feng, J.; Zhou, Y.; Campbell, S.L.; Le, T.; Li, E.; Sweatt, D.; Silva, A.J.; Fan, G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010, 13, 423–430. [Google Scholar] [CrossRef]
- Tuorto, F.; Liebers, R.; Musch, T.; Schaefer, M.; Hofmann, S.; Kellner, S.; Frye, M.; Helm, M.; Stoecklin, G.; Lyko, F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012, 19, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A.; Ehrenhofer-Murray, A.; Jurkowski, T.P.; Lyko, F.; Reuter, G.; Ankri, S.; Nellen, W.; Schaefer, M.; Helm, M. Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation. RNA Biol. 2017, 14, 1108–1123. [Google Scholar] [CrossRef]
- Kaiser, S.; Jurkowski, T.P.; Kellner, S.; Schneider, D.; Jeltsch, A.; Helm, M. The RNA methyltransferase Dnmt2 methylates DNA in the structural context of a tRNA. RNA Biol. 2017, 14, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Liu, Y.; Wu, F.; Liu, R.; Xie, Y.; Yang, Q.; Li, Y.; Liu, M.; Li, S.; Tang, H. miR-639 Expression Is Silenced by DNMT3A- Mediated Hypermethylation and Functions as a Tumor Suppressor in Liver Cancer Cells. Mol. Ther. 2019, 28, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gowher, H.; Jeltsch, A. Mammalian DNA methyltransferases: New discoveries and open questions. Biochem. Soc. Trans. 2018, 46, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Maiti, A. Mechanism of Active DNA Demethylation: Recent Progress in Epigenetics. J. Biomol. Res. Ther. 2012, 1, 1–2. [Google Scholar] [CrossRef]
- Rawłuszko-Wieczorek, A.A.; Siera, A.; Jagodziński, P.P. TET proteins in cancer: Current ‘state of the art’. Crit. Rev. Oncol. Hematol. 2015, 96, 425–436. [Google Scholar] [CrossRef]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef]
- Robertson, K. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef]
- Gaykalova, D.A.; Vatapalli, R.; Wei, Y.; Tsai, H.L.; Wang, H.; Zhang, C.; Hennessey, P.T.; Guo, T.; Tan, M.; Li, R.; et al. Outlier Analysis Defines Zinc Finger Gene Family DNA Methylation in Tumors and Saliva of Head and Neck Cancer Patients. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Sobecka, A.; Blaszczak, W.; Barczak, W.; Golusinski, P.; Rubis, B.; Masternak, M.M.; Suchorska, W.M.; Golusinski, W. hTERT promoter methylation status in peripheral blood leukocytes as a molecular marker of head and neck cancer progression. J. Appl. Genet. 2018, 59, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Ye, M.; Ni, S.; Li, Q.; Ye, D.; Li, J.; Shen, Z.; Deng, H. DNA methylation biomarkers for head and neck squamous cell carcinoma. Epigenetics 2018, 13, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Shen, Z.; Ye, D.; Li, Q.; Deng, H.; Liu, H.; Li, J. The Association and Clinical Significance of CDKN2A Promoter Methylation in Head and Neck Squamous Cell Carcinoma: A Meta-Analysis. Cell Physiol. Biochem. 2018, 50, 868–882. [Google Scholar] [CrossRef] [PubMed]
- Ai, L.; Vo, Q.N.; Zuo, C.; Li, L.; Ling, W.; Suen, J.Y.; Hanna, E.; Brown, K.D.; Fan, C.Y. Ataxia-telangiectasia-mutated (ATM) gene in head and neck squamous cell carcinoma: Promoter hypermethylation with clinical correlation in 100 cases. Cancer Epidemiol. Biomarkers Prev. 2004, 13, 150–156. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cai, F.; Xiao, X.; Niu, X.; Shi, H.; Zhong, Y. Aberrant Methylation of MGMT Promoter in HNSCC: A Meta-Analysis. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.; Santos, J.; Abreu, P.M.; Dettogni, R.S.; Santos, E.; Stur, E.; Agostini, L.P.; Anders, Q.S.; Alves, L.; Valle, I.; et al. Hypermethylation status of DAPK, MGMT and RUNX3 in HPV negative oral and oropharyngeal squamous cell carcinoma. Genet. Mol. Biol. 2020, 43, e20190334. [Google Scholar] [CrossRef] [PubMed]
- Koutsimpelas, D.; Pongsapich, W.; Heinrich, U.; Mann, S.; Mann, W.J.; Brieger, J. Promoter methylation of MGMT, MLH1 and RASSF1A tumor suppressor genes in head and neck squamous cell carcinoma: Pharmacological genome demethylation reduces proliferation of head and neck squamous carcinoma cells. Oncol. Rep. 2012, 27, 1135–1141. [Google Scholar] [CrossRef]
- Basu, B.; Chakraborty, J.; Chandra, A.; Katarkar, A.; Baldevbhai, J.; Dhar Chowdhury, D.; Ray, J.G.; Chaudhuri, K.; Chatterjee, R. Genome-wide DNA methylation profile identified a unique set of differentially methylated immune genes in oral squamous cell carcinoma patients in India. Clin. Epigenetics 2017, 9, 13. [Google Scholar] [CrossRef]
- Kim, Y.T.; Park, J.Y.; Jeon, Y.K.; Park, S.J.; Song, J.Y.; Kang, C.H.; Sung, S.W.; Kim, J.H. Aberrant promoter CpG island hypermethylation of the adenomatosis polyposis coli gene can serve as a good prognostic factor by affecting lymph node metastasis in squamous cell carcinoma of the esophagus. Dis. Esophagus 2009, 22, 143–150. [Google Scholar] [CrossRef]
- Rettori, M.M.; de Carvalho, A.C.; Longo, A.L.; de Oliveira, C.Z.; Kowalski, L.P.; Carvalho, A.L.; Vettore, A.L. TIMP3 and CCNA1 hypermethylation in HNSCC is associated with an increased incidence of second primary tumors. J. Trans. Med. 2013, 11, 316. [Google Scholar] [CrossRef]
- Paluszczak, J.; Wiśniewska, D.; Kostrzewska-Poczekaj, M.; Kiwerska, K.; Grénman, R.; Mielcarek-Kuchta, D.; Jarmuż-Szymczak, M. Prognostic significance of the methylation of Wnt pathway antagonists-CXXC4, DACT2, and the inhibitors of sonic hedgehog signaling-ZIC1, ZIC4, and HHIP in head and neck squamous cell carcinomas. Clin. Oral Investig. 2017, 21, 1777–1788. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Liu, H.; Zhang, X.; Hong, B.; Wu, Z.; Li, Q.; Zhou, C. Promoter hypermethylation of CD133/PROM1 is an independent poor prognosis factor for head and neck squamous cell carcinoma. Medicine 2020, 99, e19491. [Google Scholar] [CrossRef] [PubMed]
- Khongsti, S.; Lamare, F.A.; Shunyu, N.B.; Ghosh, S.; Maitra, A.; Ghosh, S. Whole genome DNA methylation profiling of oral cancer in ethnic population of Meghalaya, North East India reveals novel genes. Genomics 2018, 110, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.M.; Mydlarz, W.K.; Mithani, S.K.; Califano, J.A. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J. Cancer 2007, 121, 1724–1728. [Google Scholar] [CrossRef]
- Hsiung, D.T.; Marsit, C.J.; Houseman, E.A.; Eddy, K.; Furniss, C.S.; McClean, M.D.; Kelsey, K.T. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 2007, 16, 108–114. [Google Scholar] [CrossRef]
- Langevin, S.M.; Koestler, D.C.; Christensen, B.C.; Butler, R.A.; Wiencke, J.K.; Nelson, H.H.; Houseman, E.A.; Marsit, C.J.; Kelsey, K.T. Peripheral blood DNA methylation profiles are indicative of head and neck squamous cell carcinoma: An epigenome-wide association study. Epigenetics 2012, 7, 291–299. [Google Scholar] [CrossRef]
- Zheng, H.; Momeni, A.; Cedoz, P.L.; Vogel, H.; Gevaert, O. Whole slide images reflect DNA methylation patterns of human tumors. NPJ Genom. Med. 2020, 5, 11. [Google Scholar] [CrossRef]
- Colacino, J.A.; Dolinoy, D.C.; Duffy, S.A.; Sartor, M.A.; Chepeha, D.B.; Bradford, C.R.; McHugh, J.B.; Patel, D.A.; Virani, S.; Walline, H.M.; et al. Comprehensive analysis of DNA methylation in head and neck squamous cell carcinoma indicates differences by survival and clinicopathologic characteristics. PLoS ONE. 2013, 8, e54742. [Google Scholar] [CrossRef]
- Misawa, K.; Imai, A.; Matsui, H.; Kanai, A.; Misawa, Y.; Mochizuki, D.; Mima, M.; Yamada, S.; Kurokawa, T.; Nakagawa, T.; et al. Identification of novel methylation markers in HPV-associated oropharyngeal cancer: Genome-wide discovery, tissue verification and validation testing in ctDNA. Oncogene. 2020, 39, 4741–4755. [Google Scholar] [CrossRef]
- Khan, S.A.; Reddy, D.; Gupta, S. Global histone post-translational modifications and cancer: Biomarkers for diagnosis, prognosis and treatment. World J. Biol. Chem. 2015, 6, 333–345. [Google Scholar] [CrossRef]
- Le, J.M.; Squarize, C.H.; Castilho, R.M. Histone modifications: Targeting head and neck cancer stem cells. World J. Stem Cells. 2014, 6, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Verdone, L.; Caserta, M.; Di Mauro, E. Role of histone acetylation in the control of gene expression. Biochem. Cell Biol. 2005, 83, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, A.T.; Hansen, J.C. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 2000, 9, 37–61. [Google Scholar] [CrossRef] [PubMed]
- Marmorstein, R.; Zhou, M.M. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef]
- Hanigan, T.W.; Danes, J.M.; Taha, T.Y.; Frasor, J.; Petukhov, P.A. Histone deacetylase inhibitor-based chromatin precipitation for identification of targeted genomic loci. J. Biol. Methods. 2018, 5. [Google Scholar] [CrossRef]
- Bredell, M.G.; Ernst, J.; El-Kochairi, I.; Dahlem, Y.; Ikenberg, K.; Schumann, D.M. Current relevance of hypoxia in head and neck cancer. Oncotarget 2016, 7, 50781–50804. [Google Scholar] [CrossRef]
- Wang, J.Q.; Yan, F.Q.; Wang, L.H.; Yin, W.J.; Chang, T.Y.; Liu, J.P.; Wu, K.J. Identification of new hypoxia-regulated epithelial-mesenchymal transition marker genes labeled by H3K4 acetylation. Genes Chromosomes Cancer 2020, 59, 73–83. [Google Scholar] [CrossRef]
- Chen, F.; Qi, S.; Zhang, X.; Wu, J.; Yang, X.; Wang, R. lncRNA PLAC2 activated by H3K27 acetylation promotes cell proliferation and invasion via the activation of Wnt/β‑catenin pathway in oral squamous cell carcinoma. Int. J. Oncol. 2019, 54, 1183–1194. [Google Scholar] [CrossRef]
- Almeida, L.O.; Abrahao, A.C.; Rosselli-Murai, L.K.; Giudice, F.S.; Zagni, C.; Leopoldino, A.M.; Squarize, C.H.; Castilho, R.M. NFκB mediates cisplatin resistance through histone modifications in head and neck squamous cell carcinoma (HNSCC). FEBS Open Bio. 2013, 4, 96–104. [Google Scholar] [CrossRef]
- Giudice, F.S.; Pinto, D.S. Jr Nör, J.E.; Squarize, C.H.; Castilho, R.M. Inhibition of histone deacetylase impacts cancer stem cells and induces epithelial-mesenchyme transition of head and neck cancer. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- He, L.; Gao, L.; Shay, C.; Lang, L.; Lv, F.; Teng, Y. Histone deacetylase inhibitors suppress aggressiveness of head and neck squamous cell carcinoma via histone acetylation-independent blockade of the EGFR-Arf1 axis. J. Exp. Clin. Cancer Res. 2019, 38, 84. [Google Scholar] [CrossRef] [PubMed]
- Jambhekar, A.; Dhall, A.; Shi, Y. Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 2019, 20, 625–641. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.H.; Kubicek, S.; Mechtler, K.; O’Sullivan, R.J.; Derijck, A.A.; Perez-Burgos, L.; Kohlmaier, A.; Opravil, S.; Tachibana, M.; Shinkai, Y.; et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 2003, 12, 1577–1589. [Google Scholar] [CrossRef]
- Bannister, A.J.; Schneider, R.; Myers, F.A.; Thorne, A.W.; Crane-Robinson, C.; Kouzarides, T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 2005, 280, 17732–17736. [Google Scholar] [CrossRef]
- Song, Y.; Wu, F.; Wu, J. Targeting histone methylation for cancer therapy: Enzymes, inhibitors, biological activity and perspectives. J. Hematol. Oncol. 2016, 9, 49. [Google Scholar] [CrossRef]
- Zhang, J.; Jing, L.; Li, M.; He, L.; Guo, Z. Regulation of histone arginine methylation/demethylation by methylase and demethylase. Mol. Med. Rep. 2019, 19, 3963–3971. [Google Scholar] [CrossRef]
- Cai, L.; Ma, X.; Huang, Y.; Zou, Y.; Chen, X. Aberrant histone methylation and the effect of Suv39H1 siRNA on gastric carcinoma. Oncol. Rep. 2014, 31, 2593–2600. [Google Scholar] [CrossRef]
- Zhang, L.; Deng, L.; Chen, F.; Yao, Y.; Wu, B.; We, L.; Mo, Q.; Song, Y. Inhibition of histone H3K79 methylation selectively inhibits proliferation, self-renewal and metastatic potential of breast cancer. Oncotarget 2014, 5, 10665–10677. [Google Scholar] [CrossRef]
- Kang, K.A.; Piao, M.J.; Ryu, Y.S.; Kang, H.K.; Young, W. Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget 2016, 7, 40594–40620. [Google Scholar] [CrossRef]
- Mizuno, Y.; Maemura, K.; Tanaka, Y.; Hirata, A.; Futaki, S.; Hamamoto, H.; Taniguchi, K.; Hayashi, M.; Uchiyama, K.; Shibata, M.A.; et al. Expression of delta-like 3 is downregulated by aberrant DNA methylation and histone modification in hepatocellular carcinoma. Oncol. Rep. 2018, 39, 2209–2216. [Google Scholar] [CrossRef]
- Mancuso, M.; Matassa, D.S.; Conte, M.; Colella, G.; Rana, G.; Fucci, L.; Piscopo, M. H3K4 histone methylation in oral squamous cell carcinoma. Acta Biochem. Pol. 2009, 56, 405–410. [Google Scholar] [CrossRef]
- Liu, S.; Ye, D.; Guo, W.; Yu, W.; He, Y.; Hu, J.; Wang, Y.; Zhang, L.; Liao, Y.; Song, H.; et al. G9a is essential for EMT-mediated metastasis and maintenance of cancer stem cell-like characters in head and neck squamous cell carcinoma. Oncotarget 2015, 6, 6887–6901. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, C.D.; Kostiuk, M.A.; Harris, J.; O’Connell, D.A.; Seikaly, H.; Biron, V.L. Efficacy of EZH2 inhibitory drugs in human papillomavirus-positive and human papillomavirus-negative oropharyngeal squamous cell carcinomas. Clin. Epigenetics 2017, 9, 95. [Google Scholar] [CrossRef] [PubMed]
- Marcinkiewicz, K.M.; Gudas, L.J. Altered histone mark deposition and DNA methylation at homeobox genes in human oral squamous cell carcinoma. J. Cell Physiol. 2014, 229, 1405–1416. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long non-coding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Hammond, S.M. MicroRNAs as tumor suppressors. Nat. Genet. 2007, 39, 582–583. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef]
- Cannell, I.G.; Kong, Y.W.; Bushell, M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008, 36, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.; Victoria, B.; Lopez, Y.N.; Suchorska, W.; Barczak, W.; Sobecka, A.; Golusinski, W.; Masternak, M.M.; Golusinski, P. Tissue and serum microRNA profile of oral squamous cell carcinoma patients. Sci. Rep. 2018, 8, 675. [Google Scholar] [CrossRef] [PubMed]
- Lubov, J.; Maschietto, M.; Ibrahim, I.; Mlynarek, A.; Hier, M.; Kowalski, L.P.; Alaoui-Jamali, M.A.; da Silva, S.D. Meta-analysis of microRNAs expression in head and neck cancer: Uncovering association with outcome and mechanisms. Oncotarget 2017, 8, 55511–55524. [Google Scholar] [CrossRef] [PubMed]
- Sobecka, A.; Barczak, W.; Suchorska, W.M. RNA interference in head and neck oncology. Oncol. Lett. 2016, 12, 3035–3040. [Google Scholar] [CrossRef]
- Langevin, S.M.; Stone, R.A.; Bunker, C.H.; Lyons-Weiler, M.A.; LaFramboise, W.A.; Kelly, L.; Seethala, R.R.; Grandis, J.R.; Sobol, R.W.; Taioli, E. MicroRNA-137 promoter methylation is associated with poorer overall survival in patients with squamous cell carcinoma of the head and neck. Cancer 2011, 1, 1454–1462. [Google Scholar] [CrossRef]
- Metheetrairut, C.; Chotigavanich, C.; Amornpichetkul, K.; Keskool, P.; Ongard, S.; Metheetrairut, C. Expression levels of miR-34-family microRNAs are associated with TP53 mutation status in head and neck squamous cell carcinoma. Eur. Arch. Otorhinolaryngol. 2019, 276, 521–533. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, F.; Li, X.; Liu, Q.; Liu, W.; Song, P.; Qiu, Z.; Dong, Y.; Xiang, H. Prognostic role of miR-17-92 family in human cancers: Evaluation of multiple prognostic outcomes. Oncotarget 2017, 8, 69125–69138. [Google Scholar] [CrossRef]
- Gee, H.E.; Camps, C.; Buffa, F.M.; Patiar, S.; Winter, S.C.; Betts, G.; Homer, J.; Corbridge, R.; Cox, G.; West, C.M.L.; et al. hsa-mir-210 Is a Marker of Tumor Hypoxia and a Prognostic Factor in Head and Neck Cancer. Cancer 2010, 116, 2148–2158. [Google Scholar] [CrossRef]
- Koshizuka, K.; Kikkawa, N.; Hanazawa, T.; Yamada, Y.; Okato, A.; Arai, T.; Katada, K.; Okamoto, Y.; Seki, N. Inhibition of integrin β1-mediated oncogenic signalling by the antitumor microRNA-29 family in head and neck squamous cell carcinoma. Oncotarget 2017, 11, 3663–3676. [Google Scholar] [CrossRef]
- Arantes, L.M.; Laus, A.C.; Melendez, M.E.; de Carvalho, A.C.; Sorroche, B.P.; De Marchi, P.R.; Evangelista, A.F.; Scapulatempo-Neto, C.; de Souza Viana, L.; Carvalho, A.L. MiR-21 as prognostic biomarker in head and neck squamous cell carcinoma patients undergoing an organ preservation protocol. Oncotarget 2017, 7, 9911–9921. [Google Scholar] [CrossRef]
- Arunkumar, G.; Deva Magendhra Rao, A.K.; Manikandan, M.; Prasanna Srinivasa Rao, H.; Subbiah, S.; Ilangovan, R.; Murugan, A.K.; Munirajan, A.K. Dysregulation of miR-200 family microRNAs and epithelial-mesenchymal transition markers in oral squamous cell carcinoma. Oncol. Lett. 2018, 15, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Chang, J.T.; Ho, Y.F.; Shyu, A.B. MiR-26 down-regulates TNF-α/NF-κB signalling and IL-6 expression by silencing HMGA1 and MALT1. Nucleic. Acids Res. 2016, 44, 3772–3787. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Hanazawa, T.; Nohata, N.; Kikkawa, N.; Enokida, H.; Yoshino, H.; Yamasaki, T.; Hidaka, H.; Nakagawa, M.; Okamoto, Y.; et al. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma. Oncotarget 2012, 3, 1386–1400. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.F.; Wei, Y.Y.; Yang, C.C.; Liu, C.J.; Yeh, L.Y.; Chou, C.H.; Chang, K.W.; Lin, S.C. miR-125b suppresses oral oncogenicity by targeting the anti-oxidative gene PRXL2A. Redox Biol. 2019, 22, 101140. [Google Scholar] [CrossRef]
- González-Arriagada, W.A.; Olivero, P.; Rodríguez, B.; Lozano-Burgos, C.; de Oliveira, C.E.; Coletta, R.D. Clinicopathological significance of miR-26, miR-107, miR-125b, and miR-203 in head and neck carcinomas. Oral Dis. 2018, 24, 930–939. [Google Scholar] [CrossRef]
- Sannigrahi, M.K.; Sharma, R.; Singh, V.; Panda, N.K.; Rattan, V.; Khullar, M. Role of Host miRNA Hsa-miR-139-3p in HPV-16-Induced Carcinomas. Clin. Cancer Res. 2017, 23, 3884–3895. [Google Scholar] [CrossRef]
- Kolenda, T.; Guglas, K.; Teresiak, A.; Bliźniak, R.; Lamperska, K. Low let-7d and high miR-205 expression levels positively influence HNSCC patient outcome. J. Biomed. Sci. 2019, 26, 17. [Google Scholar] [CrossRef]
- Hess, A.K.; Müer, A.; Mairinger, F.D.; Weichert, W.; Stenzinger, A.; Hummel, M.; Budach, V.; Tinhofer, I. MiR-200b and miR-155 as predictive biomarkers for the efficacy of chemoradiation in locally advanced head and neck squamous cell carcinoma. Eur. J. Cancer 2017, 77, 3–12. [Google Scholar] [CrossRef]
- Vahabi, M.; Pulito, C.; Sacconi, A.; Donzelli, S.; D’Andrea, M.; Manciocco, V.; Pellini, R.; Paci, P.; Sanguineti, G.; Strigari, L.; et al. miR-96-5p targets PTEN expression affecting radio-chemosensitivity of HNSCC cells. J. Exp. Clin. Cancer Res. 2019, 29, 141. [Google Scholar] [CrossRef]
- Rather, M.I.; Nagashri, M.N.; Swamy, S.S.; Gopinath, K.S.; Kumar, A. Oncogenic microRNA-155 down-regulates tumor suppressor CDC73 and promotes oral squamous cell carcinoma cell proliferation: Implications for cancer therapeutics. J. Biol. Chem. 2013, 288, 608–618. [Google Scholar] [CrossRef]
- Wang, L.L.; Li, H.X.; Yang, Y.Y.; Su, Y.L.; Lian, J.S.; Li, T.; Xu, J.; Wang, X.N.; Jin, N.; Liu, X.F. MiR-31 is a potential biomarker for diagnosis of head and neck squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2018, 1, 4339–4345. [Google Scholar]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.C.; Ni, J.J.; Cui, W.Y.; Wang, B.Y.; Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366. [Google Scholar] [PubMed]
- Yang, B.; Shen, J.; Xu, L.; Chen, Y.; Che, X.; Qu, X.; Liu, Y.; Teng, Y.; Li, Z. Genome-Wide Identification of a Novel Eight-lncRNA Signature to Improve Prognostic Prediction in Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2019, 18, 898. [Google Scholar] [CrossRef]
- Li, Y.; Wan, Q.; Wang, W.; Mai, L.; Sha, L.; Mashrah, M.; Lin, Z.; Pan, C. LncRNA ADAMTS9-AS2 promotes tongue squamous cell carcinoma proliferation, migration and EMT via the miR-600/EZH2 axis. Biomed. Pharmacother. 2019, 112. [Google Scholar] [CrossRef]
- Jiang, Y.; Cao, W.; Wu, K.; Qin, X.; Wang, X.; Li, Y.; Binbin, Y.; Zhang, Z.; Wang, X.; Yan, H.; et al. LncRNA LINC00460 promotes EMT in head and neck squamous cell carcinoma by facilitating peroxiredoxin-1 into the nucleus. J. Exp. Clin. Cancer Res. 2019, 38, 365. [Google Scholar] [CrossRef]
- Wang, R.; Ma, Z.; Feng, L.; Yang, Y.; Tan, C.; Shi, Q.; Lian, M.; He, S.; Ma, H.; Fang, J. LncRNA MIR31HG targets HIF1A and P21 to facilitate head and neck cancer cell proliferation and tumorigenesis by promoting cell-cycle progression. Mol. Cancer 2018, 17, 162. [Google Scholar] [CrossRef]
- Ma, H.; Chang, H.; Yang, W.; Lu, Y.; Hu, J.; Jin, S. A novel IFNα-induced long non-coding RNA negatively regulates immunosuppression by interrupting H3K27 acetylation in head and neck squamous cell carcinoma. Mol. Cancer 2020, 19, 4. [Google Scholar] [CrossRef]
- Xiong, H.G.; Li, H.; Xiao, Y.; Yang, Q.C.; Yang, L.L.; Chen, L.; Bu, L.L.; Zhang, W.F.; Zhang, J.L.; Sun, Z.J. Long noncoding RNA MYOSLID promotes invasion and metastasis by modulating the partial epithelial-mesenchymal transition program in head and neck squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 278. [Google Scholar] [CrossRef]
- Zhang, C.; Cao, W.; Wang, J.; Liu, J.; Liu, J.; Wu, H.; Li, S.; Zhang, C. A prognostic long non-coding RNA-associated competing endogenous RNA network in head and neck squamous cell carcinoma. Peer J. 2020, 8, e9701. [Google Scholar] [CrossRef]
- Song, J.; Yi, C. Chemical Modifications to RNA: A New Layer of Gene Expression Regulation. ACS Chem. Biol. 2017, 12, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Mongan, N.P.; Emes, R.D.; Archer, N. Detection and analysis of RNA methylation. F1000Res 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.; Alemu, E.A.; Mertens, C.; Gantman, E.C.; Fak, J.J.; Mele, A.; Haripal, B.; Zucker-Scharff, I.; Moore, M.J.; Park, C.Y.; et al. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 2015, 1, 2037–2053. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkes, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015, 29, 1343–1355. [Google Scholar] [CrossRef]
- Chen, X.Y.; Zhang, J.; Zhu, J.S. The role of m6A RNA methylation in human cancer. Mol. Cancer 2019, 18, 103. [Google Scholar] [CrossRef]
- Lesbirel, S.; Wilson, S.A. The m6A‑methylase complex and mRNA export. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 319–328. [Google Scholar] [CrossRef]
- Liao, S.; Sun, H.; Xu, C. YTH Domain: A Family of N 6 -methyladenosine (m6A) Readers. Geno. Prot. Bioin. 2018, 16, 99–107. [Google Scholar] [CrossRef]
- Paris, J.; Morgan, M.; Campos, J.; Spencer, G.J.; Shmakova, A.; Ivanova, I.; Mapperley, C.; Lawson, H.; Wotherspoon, D.A.; Sepulveda, C.; et al. Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell 2019, 3, 137–148. [Google Scholar] [CrossRef]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.G.; et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017, 14, 2622–2634. [Google Scholar] [CrossRef]
- Liu, J.; Ren, D.; Du, Z.; Wang, H.; Zhang, H.; Jin, Y. m6A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem. Biophys. Res. Commun. 2018, 502, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wu, D.; Ning, J.; Liu, W.; Zhang, D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer 2019, 19, 326. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wei, L.; Law, C.T.; Tsang, F.H.; Shen, J.; Cheng, C.L.; Tsang, L.H.; Ho, D.W.; Chiu, D.K.; Lee, J.M.; et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent post-transcriptional silencing of SOCS2. Hepatology 2018, 67, 2254–2270. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Wang, J.Z.; Yang, X.; Yu, H.; Zhou, R.; Lu, H.C.; Yuan, W.B.; Lu, J.C.; Zhou, Z.J.; Lu, Q.; et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 2019, 22, 110. [Google Scholar] [CrossRef] [PubMed]
- Taketo, K.; Konno, M.; Asai, A.; Koseki, J.; Toratani, M.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H.; Ogawa, K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 2018, 52, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Cui, L. Development and validation of a m6A RNA methylation regulators-based signature for predicting the prognosis of head and neck squamous cell carcinoma. Am. J. Cancer Res. 2019, 9, 2156–2169. [Google Scholar] [PubMed]
- Zhao, W.; Cui, Y.; Liu, J.; Ma, X.; Qi, X.; Wang, Y.; Liu, Z.; Ma, S.; Liu, J.; Wu, J. METTL3 facilitates oral squamous cell carcinoma tumorigenesis by enhancing c-myc syability via YTHDF1-mediated m6A modification. Mol. Ther. Nucleic Acids. 2020, 20, 1–12. [Google Scholar] [CrossRef]
- Shriwas, O.; Priyadarshini, M.; Samal, S.K.; Rath, R.; Panda, S.; Das Majumdar, S.K.; Muduly, D.K.; Botlagunta, M.; Dash, R. DDX3 modulates cisplatin resistance in OSCC through ALKBH5-mediated m6A-demethylation of FOXM1 and NANOG. Apoptosis 2020, 25, 233–246. [Google Scholar] [CrossRef]
- Pilžys, T.; Marcinkowski, M.; Kukwa, W.; Garbicz, D.; Dylewska, M.; Ferenc, K.; Mieczkowski, A.; Kukwa, A.; Migacz, E.; Wołosz, D.; et al. ALKBH overexpression in head and neck cancer: Potential target for novel anticancer therapy. Sci. Rep. 2019, 9, 13249. [Google Scholar] [CrossRef]
- Ban, Y.; Tan, P.; Cai, J.; Li, J.; Hu, M.; Zhou, Y.; Mei, Y.; Tan, Y.; Li, X.; Zeng, Z.; et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol. Oncol. 2020, 14, 1282–1296. [Google Scholar] [CrossRef]


| Gene | Tissue | Type of Study | Diagnostic Significance | References |
|---|---|---|---|---|
| ZNF14, ZNF160, ZNF420 | Tumor and saliva | Meta-analysis confirmed in patient samples | HNSCC detection and surveillance | [40] |
| hTERT | Blood leukocytes | Patient study | HNSCC detection | [41] |
| FAM135B | Tumor | Meta-analysis | Overall survival of HNSCC patients | [42] |
| CDKN2A | Tumor and saliva | Meta-analysis | HNSCC progression and metastases | [43] |
| ATM | Tumor | Patients study | HNSCC detection in early age and early tumor stage | [44] |
| MGMT | Tumor | Meta-analysis | Risk of HNSCC | [45] |
| DAPK | Tumor | Patients study | HNSCC HPV(–) detection in early stage | [46] |
| RASSF1A, MLH1, MGMT | Tumor | Patients and in vitro study | HNSCC and high proliferative potential of tumor cells detection | [47] |
| CTLA4 | Tumor | Patients study | HNSCC detection and surveillance | [48] |
| APC | Tumor | Patients study | Lower number of metastatic lymph nodes | [49] |
| CCNA1, TIMP3 | Tumor | Patients study | Risk of second primary carcinomas | [50] |
| ZIC4 | Tumor | Patients study | Risk of lymph node involvement | [51] |
| PROM1 | Tumor | Meta-analysis | HNSCC detection in early stage and invasion potential | [52] |
| Process | microRNA | Diagnostic Significance | References |
|---|---|---|---|
| (Up- or Downregulated) | |||
| Apoptosis | miR137 | downregulated | [95] |
| miR34 | upregulated | [96] | |
| miR17-92 | upregulated | [97] | |
| Gene instability | miR210 | upregulated | [98] |
| miR29 | downregulated | [99] | |
| Immune evasion | miR21 | upregulated | [100] |
| miR210 | downregulated | [101] | |
| Inflammation | miR26 | downregulated | [102] |
| miR218 | downregulated | [103] | |
| Metabolism | miR26 | downregulated | [102] |
| miR125b | downregulated | [104] | |
| Metastases | miR26 | upregulated | [105] |
| miR125b | upregulated | [105] | |
| miR139 | downregulated | [106] | |
| let-7d | upregulated | [107] | |
| miR200b | upregulated | [108] | |
| miR218 | downregulated | [109] | |
| miR96 | upregulated | [109] | |
| miR29 | downregulated | [99] | |
| miR200 | downregulated | [101] | |
| Proliferation | miR21 | upregulated | [100] |
| miR29 | downregulated | [99] | |
| miR139 | downregulated | [106] | |
| miR155 | upregulated | [110] | |
| Resistance to the radiotherapy and chemotherapy | miR210 | downregulated | [101] |
| miR31 | upregulated | [111] | |
| miR125b | downregulated | [104,109] | |
| miR96 | upregulated | [110] | |
| let-7d | downregulated | [107] | |
| miR205 | upregulated | [107] | |
| miR96 | upregulated | [109] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romanowska, K.; Sobecka, A.; Rawłuszko-Wieczorek, A.A.; Suchorska, W.M.; Golusiński, W. Head and Neck Squamous Cell Carcinoma: Epigenetic Landscape. Diagnostics 2021, 11, 34. https://doi.org/10.3390/diagnostics11010034
Romanowska K, Sobecka A, Rawłuszko-Wieczorek AA, Suchorska WM, Golusiński W. Head and Neck Squamous Cell Carcinoma: Epigenetic Landscape. Diagnostics. 2021; 11(1):34. https://doi.org/10.3390/diagnostics11010034
Chicago/Turabian StyleRomanowska, Kamila, Agnieszka Sobecka, Agnieszka A. Rawłuszko-Wieczorek, Wiktoria M. Suchorska, and Wojciech Golusiński. 2021. "Head and Neck Squamous Cell Carcinoma: Epigenetic Landscape" Diagnostics 11, no. 1: 34. https://doi.org/10.3390/diagnostics11010034
APA StyleRomanowska, K., Sobecka, A., Rawłuszko-Wieczorek, A. A., Suchorska, W. M., & Golusiński, W. (2021). Head and Neck Squamous Cell Carcinoma: Epigenetic Landscape. Diagnostics, 11(1), 34. https://doi.org/10.3390/diagnostics11010034







