Direct-RT-qPCR Detection of SARS-CoV-2 without RNA Extraction as Part of a COVID-19 Testing Strategy: From Sample to Result in One Hour
Abstract
:1. Introduction
2. Materials and Methods
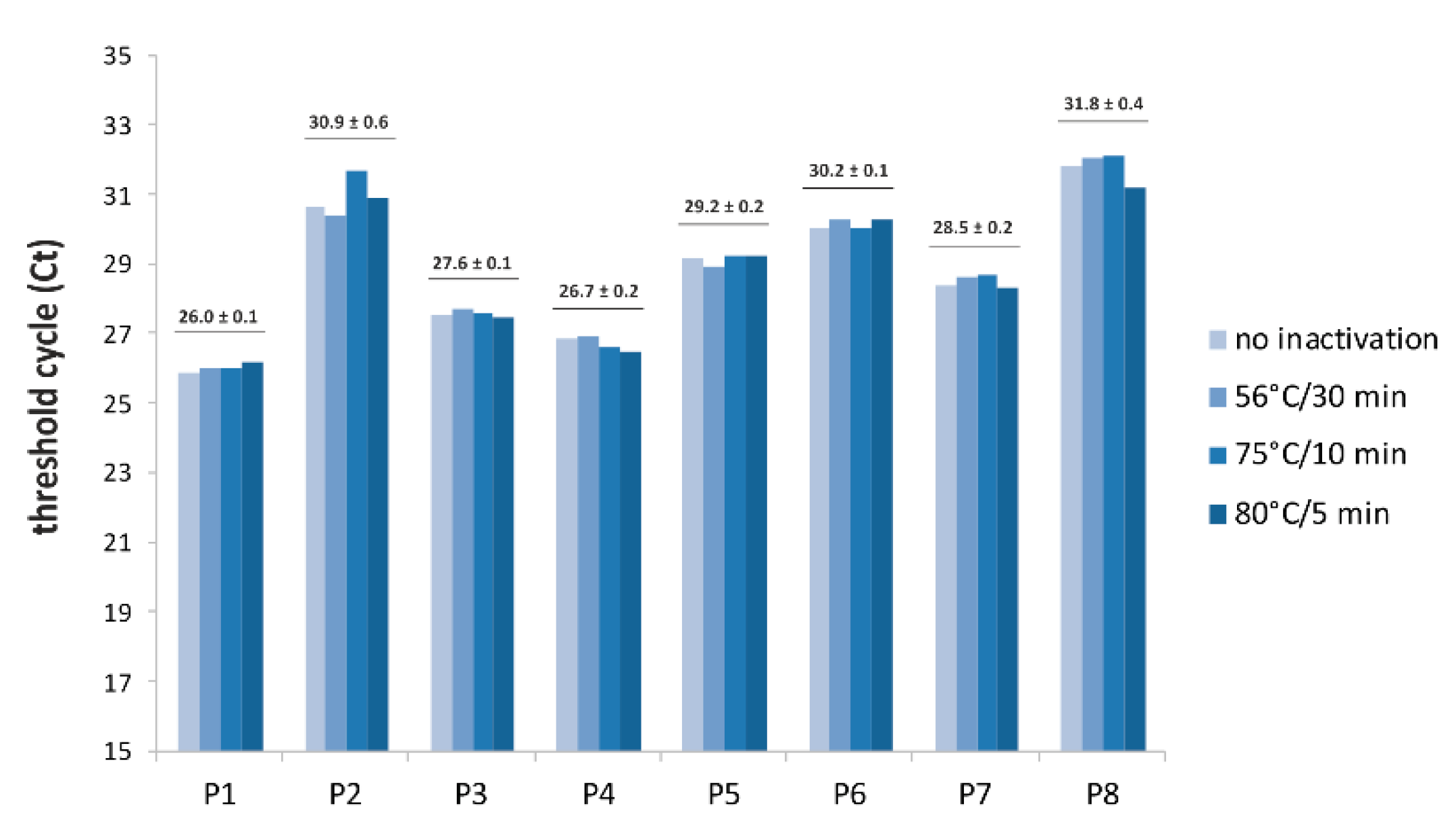
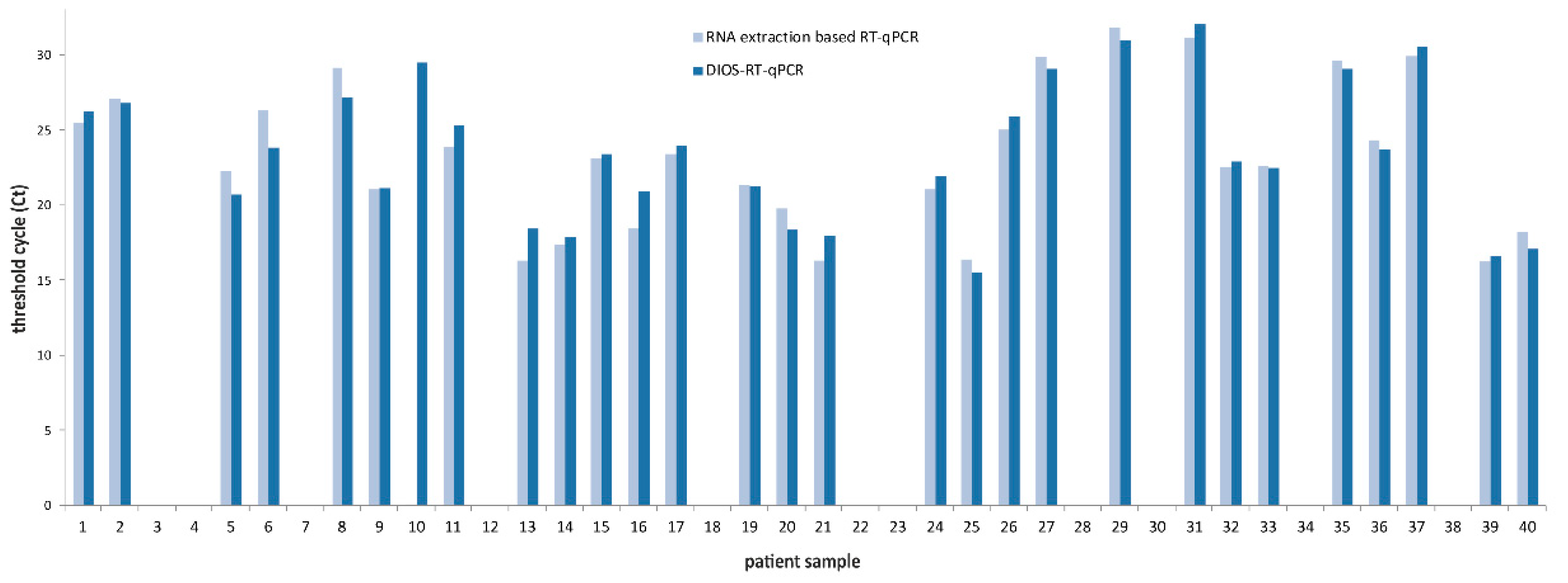
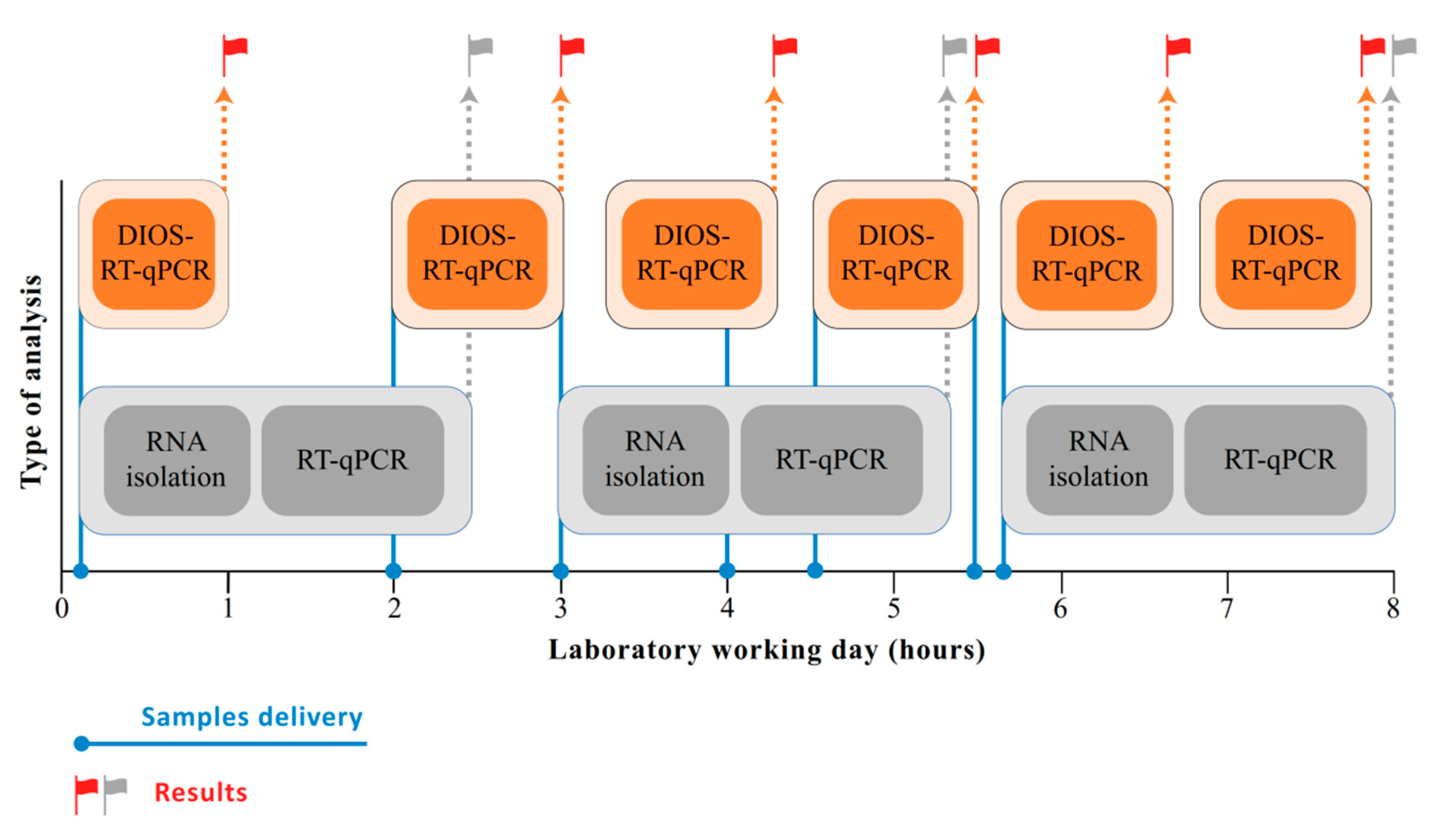
3. Results
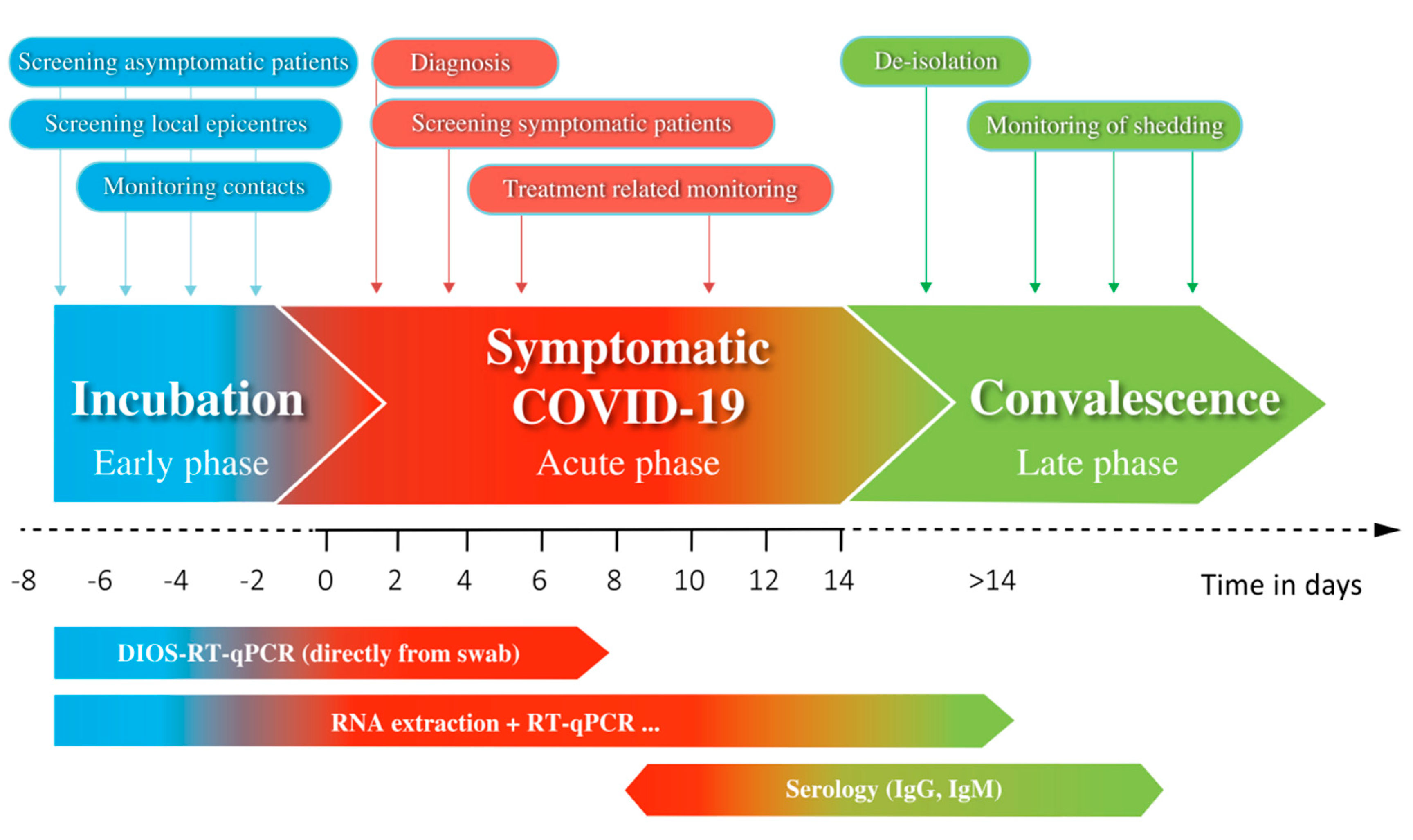
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Statement of Ethical Assurance
Ethics
References
- Johns Hopkins University. Available online: https://coronavirus.jhu.edu (accessed on 30 July 2020).
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Kubina, R.; Dziedzic, A. Molecular and Serological Tests for COVID-19 a Comparative Review of SARS-CoV-2 Coronavirus Laboratory and Point-of-Care Diagnostics. Diagnostics 2020, 10, 434. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Research Use only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (accessed on 28 June 2020).
- Hasan, M.R.; Mirza, F.; Al-Hail, H.; Sundararaju, S.; Xaba, T.; Iqbal, M.; Alhussain, H.; Yassine, H.M.; Perez-Lopez, A.; Tang, P. Detection of SARS-CoV-2 RNA by direct RT-qPCR on nasopharyngeal specimens without extraction of viral RNA. PLoS ONE 2020, 15, e0236564. [Google Scholar] [CrossRef]
- Li, L.; He, J.A.; Wang, W.; Xia, Y.; Song, L.; Chen, Z.H.; Zuo, H.Z.; Tan, X.P.; Ho, A.H.; Kong, S.K.; et al. Development of a Direct Reverse-Transcription Quantitative PCR (dirRT-qPCR) Assay for Clinical Zika Diagnosis. Int. J. Infect. Dis. 2019, 85, 167–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruce, E.A.; Huang, M.L.; Perchetti, G.A.; Tighe, S.; Laaguiby, P.; Hoffman, J.J.; Gerrard, D.L.; Nalla, A.K.; Wei, Y.; Greninger, A.L.; et al. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Fomsgaard, A.S.; Rosenstierne, M.W. An alternative workflow for molecular detection of SARS-CoV-2—Escape from the NA extraction kit-shortage. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Beltran-Pavez, C.; Marquez, C.L.; Munoz, G.; Valiente-Echeverria, F.; Gaggero, A.; Soto-Rifo, R.; Barriga, G.P. SARS-CoV-2 detection from nasopharyngeal swab samples without RNA extraction. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Grant, P.R.; Turner, M.A.; Shin, G.Y.; Nastouli, E.; Levett, L.J. Extraction-free COVID-19 (SARS-CoV-2) diagnosis by RT-PCR to increase capacity for national testing programmes during a pandemic. bioRxiv 2020. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Yan, L.; Wang, N.; Yang, S.; Wang, L.; Tang, Y.; Gao, G.; Wang, S.; Ma, C.; Xie, R.; et al. Quantitative Detection and Viral Load Analysis of SARS-CoV-2 in Infected Patients. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [Green Version]
- FDA, U.S. Policy for Coronavirus Disease-2019 Tests During the Public Health Emergency (Revised). Available online: https://www.fda.gov/media/135659/download (accessed on 28 June 2020).
- World Health Organisation. Laboratory Biosafety Guidance Related to Coronavirus Disease 2019 (COVID-19). Available online: https://apps.who.int/iris/bitstream/handle/10665/331138/WHO-WPE-GIH-2020.1-eng.pdf?sequence=1&isAllowed=y (accessed on 4 July 2020).
- Kampf, G.; Voss, A.; Scheithauer, S. Inactivation of coronaviruses by heat. J. Hosp. Infect. 2020, 105, 348–349. [Google Scholar] [CrossRef] [PubMed]
- Lista, M.J.; Page, R.; Sartkaya, H.; Matos, P.M.; Ortiz-Zapater, E.; Maguire, T.J.A.; Poulton, K.; O’Byrne, A.; Bouton, C.; Dickenson, R.E.; et al. Resilient SARS-CoV-2 diagnostics workflows including viral heat inactivation. medRxiv 2020. [Google Scholar] [CrossRef]
- Lu, R.; Wu, X.; Wan, Z.; Li, Y.; Jin, X.; Zhang, C. A Novel Reverse Transcription Loop-Mediated Isothermal Amplification Method for Rapid Detection of SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 2826. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Odiwuor, N.; Xiong, J.; Sun, L.; Nyaruaba, R.O.; Wei, H.; Tanner, N.A. Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP. medRxiv 2020. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Segel, M.; Bruneau, R.; Huang, M.W.; Kim, N.G.; Yu, X.; Li, J.; Walker, B.D.; et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv 2020. [Google Scholar] [CrossRef]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Boehm, C.K.; Petros, B.A.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.S.F.; Kemball, M.E.; et al. Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Version 1. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wikramaratna, P.; Paton, R.S.; Ghafari, M.; Lourenco, J. Estimating false-negative detection rate of SARS-CoV-2 by RT-PCR. medRxiv 2020. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Specimen Collection Guidelines. Available online: https://www.cdc.gov/urdo/downloads/SpecCollectionGuidelines.pdf (accessed on 28 June 2020).
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Geng, B.; Muenker, M.C.; Moore, A.J.; Vogels, C.B.F.; et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swap. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Yang, M.; Shen, C.; Wang, F.; Yuan, J.; Li, J.; Zhang, M.; Wang, Z.; Xing, L.; Wei, J.; et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Chin, A.W.H.; Chu, J.T.S.; Perera, M.R.A.; Hui, K.P.Y.; Yen, H.-L.; Chan, M.C.W.; Peiris, M.; Poon, L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 2020, 1, e10. [Google Scholar] [CrossRef]
- World Health Organisation. First Data on Stability and Resistance of SARS Coronavirus Complied by Members of WHO Laboratory Network. Available online: https://www.who.int/csr/sars/survival_2003_05_04/en/ (accessed on 17 July 2020).
| Name | Description | Oligonucleotide Sequence (5′ → 3′) | Final Concentration in Reaction |
|---|---|---|---|
| 2019-nCoV_N1 | Forward primer | GACCCCAAAATCAGCGAAAT | 333 nM |
| Reverse primer | TCTGGTTACTGCCAGTTGAATCTG | 333 nM | |
| Probe | FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ-1 | 83 nM | |
| 2019-nCoV_N2 | Forward primer | TTACAAACATTGGCCGCAAA | 200 nM |
| Reverse primer | GCGCGACATTCCGAAGAA | 200 nM | |
| Probe | FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ-1 | 50 nM | |
| RP | Forward primer | AGATTTGGACCTGCGAGCG | 133 nM |
| Reverse primer | GAGCGGCTGTCTCCACAAGT | 133 nM | |
| Probe | Cy5-TTCTGACCTGAAGGCTCTGCGCG-BHQ-2 | 33 nM |
| Mean Ct ± SD | Virus Copies/ Reaction | Virus Copies/mL in Swabs | ||
|---|---|---|---|---|
| qTOWER3 | LightCycler 480 | Rotor-Gene Q | ||
| 25.9 ± 0.1 | 28.2 ± 0.2 | 24.0 ± 0.1 | 125 | 8.9 × 103 |
| 26.8 ± 0.1 | 29.4 ± 0.2 | 25.2 ± 0.1 | 63 | 4.5 × 103 |
| 27.5 ± 0.1 | 30.7 ± 0.1 | 25.7 ± 0.1 | 31 | 2.2 × 103 |
| 28.3 ± 0.1 | 31.9 ± 0.2 | 26.9 ± 0.2 | 15.6 | 1.1 × 103 |
| 29.1 ± 0.1 | 32.8 ± 0.1 | 27.9 ± 0.2 | 7.8 | 554 |
| 30.0 ± 0.2 | 33.6 ± 0.2 | 28.8 ± 0.2 | 3.9 | 277 |
| 30.6 ± 0.3 | 34.2 ± 0.3 | 29.4 ± 0.3 | 2 | 141 |
| ND | ND | ND | 0 | 0 |
| QCMD EQA Specimens | DIOS-RT-qPCR Result | Study by Hasan et al. 2020 [5] | |||
|---|---|---|---|---|---|
| Sample ID | SARS-CoV-2 Result | SARS-CoV-2 Result | qTOWER3 (Ct) | QIAstat-Dx (Ct) | Direct Approach (Ct) |
| S 01 | Positive | Positive | 24.4 | 34.0 | 35.5 |
| S 02 | Negative | Negative | ND | ND | ND |
| S 03 | Positive | Positive | 27.1 | 35.4 | 37.1 |
| S 04 | Negative | Negative | ND | ND | ND |
| S 05 | Negative | Negative | ND | ND | ND |
| S 06 | Positive | Positive | 24.1 | 36.7 | 35.1 |
| S 07 | Positive | Positive | 20.8 | 31.5 | 31.7 |
| S 08 | Borderline | Positive | 30.2 | ND | ND |
| Target Subjects | Result Expected | Key Requirements for Methods Used | DIOS-RT-qPCR | |
|---|---|---|---|---|
| Screening | Local epicentres, contacts of positive cases, business, travelling, risk groups | Positive/negative for COVID-19 | Fast, cost-effective, high throughput, easy performance | Yes, suitable |
| Point-of-care testing | Patients before emergent hospitalisation, surgery, doctor/dentist visits | Positive/negative for COVID-19 | Ultra-fast, on-site | Yes, suitable |
| Diagnostics | Patients with suspected COVID-19 | Viral load, quantification of pathogens, positive/negative for COVID-19 and other respiratory pathogens (panels of targets) | Analysis of multiple targets and genes, ultra-sensitive, quantitative and qualitative evaluation | Not intended |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kriegova, E.; Fillerova, R.; Kvapil, P. Direct-RT-qPCR Detection of SARS-CoV-2 without RNA Extraction as Part of a COVID-19 Testing Strategy: From Sample to Result in One Hour. Diagnostics 2020, 10, 605. https://doi.org/10.3390/diagnostics10080605
Kriegova E, Fillerova R, Kvapil P. Direct-RT-qPCR Detection of SARS-CoV-2 without RNA Extraction as Part of a COVID-19 Testing Strategy: From Sample to Result in One Hour. Diagnostics. 2020; 10(8):605. https://doi.org/10.3390/diagnostics10080605
Chicago/Turabian StyleKriegova, Eva, Regina Fillerova, and Petr Kvapil. 2020. "Direct-RT-qPCR Detection of SARS-CoV-2 without RNA Extraction as Part of a COVID-19 Testing Strategy: From Sample to Result in One Hour" Diagnostics 10, no. 8: 605. https://doi.org/10.3390/diagnostics10080605
APA StyleKriegova, E., Fillerova, R., & Kvapil, P. (2020). Direct-RT-qPCR Detection of SARS-CoV-2 without RNA Extraction as Part of a COVID-19 Testing Strategy: From Sample to Result in One Hour. Diagnostics, 10(8), 605. https://doi.org/10.3390/diagnostics10080605





