Non-Small-Cell Lung Cancer-Sensitive Detection of the p.Thr790Met EGFR Alteration by Preamplification before PNA-Mediated PCR Clamping and Pyrosequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples and DNA Extraction
2.2. Next-Generation Sequencing
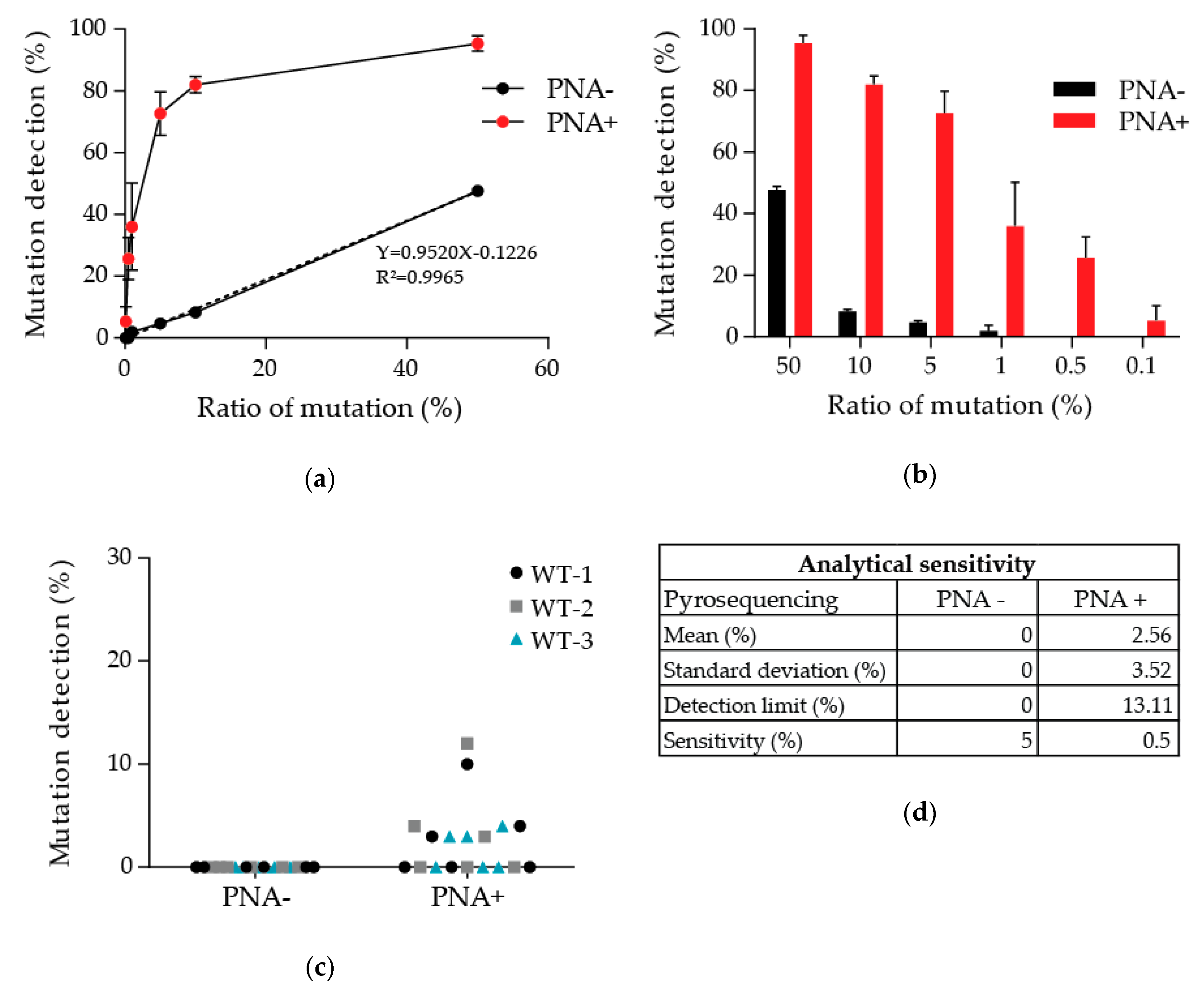
2.3. Analysis of the Sensitivity and Specificity of the Detection of the p.Thr790Met Alteration
2.4. PNA-Mediated PCR Clamping on the Wild-Type EGFR Sequence
2.5. Pyrosequencing
2.6. Statistics
3. Results
3.1. Determination of the Detection Limit of Our PNA-Mediated PCR Clamping
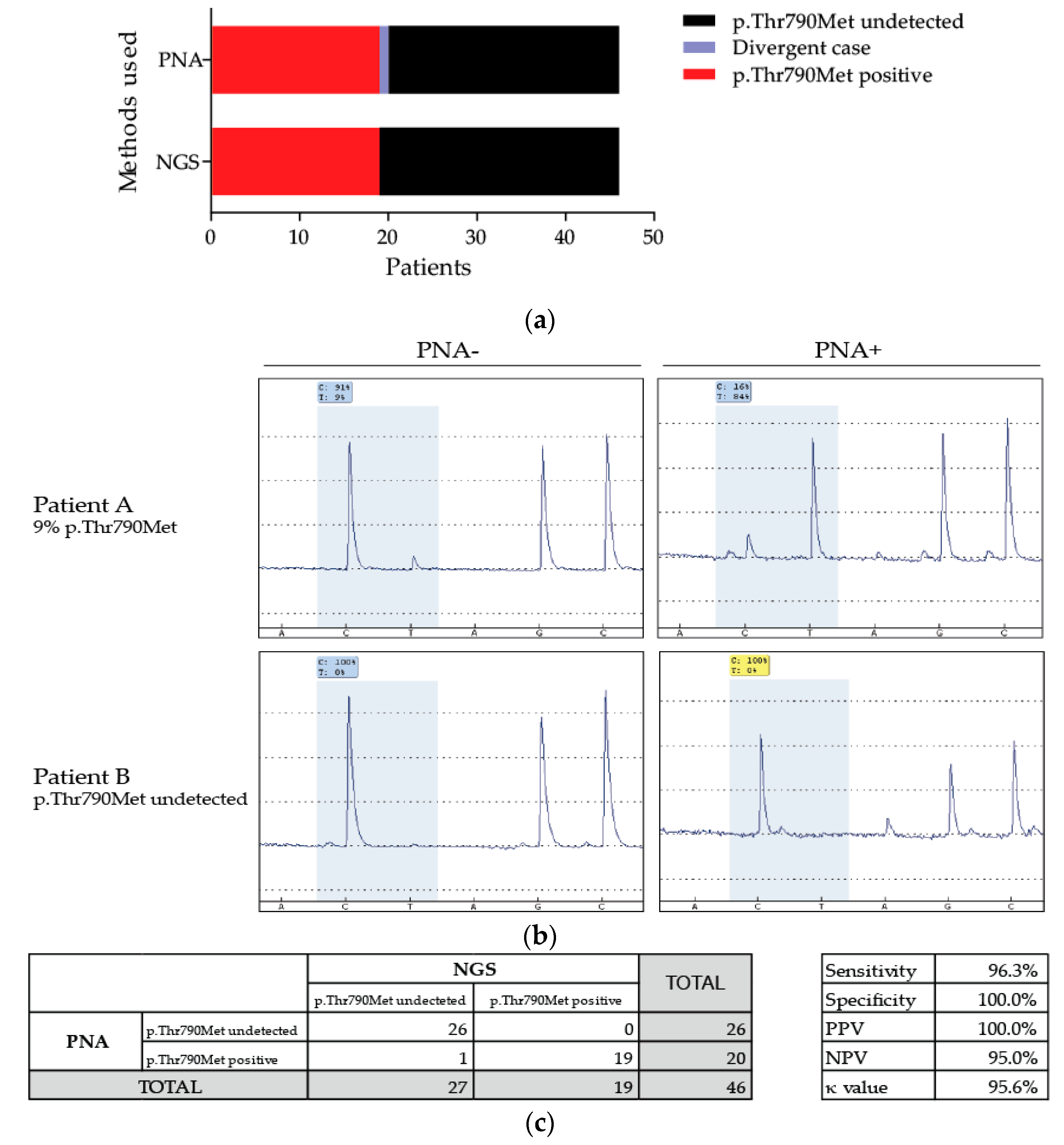
3.2. Detection of the p.Thr790Met Alteration of EGFR in Patients’ Somatic DNA
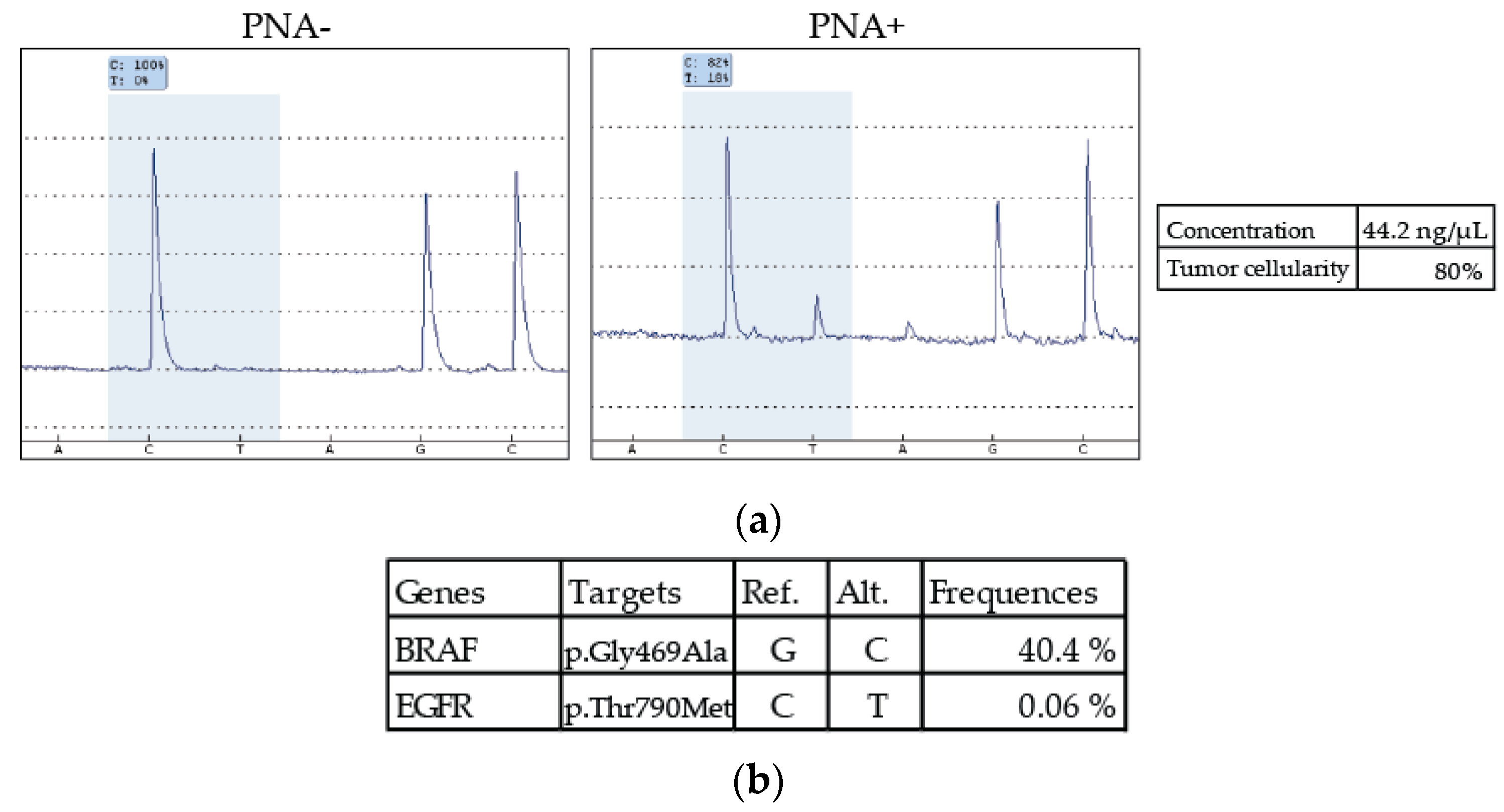
3.3. Patient with Discordant Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TKI | tyrosine kinase inhibitor |
| NSCLC | non-small-cell lung cancer |
| PNA | peptide nucleic acid |
| SCLC | small-cell lung cancer |
| ctDNA | circulating tumor DNA |
| NGS | next-generation sequencing |
| PBNC | peripheral blood nuclear cells |
| FFPE | formalin-fixed paraffin-embedded |
| ICO | Institut de Cancérologie de l’Ouest |
Appendix A

References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.; Torre, L.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Heymach, J.V.; Lippman, S.M. Molecular Origins of Cancer Lung Cancer. N. Engl. J. Med. 2008, 6, 53–55. [Google Scholar]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Rosell, R.; Moran, T.; Queralt, C.; Cardenal, F.; Camps, C.; Majem, M.; Lopez-Vivanco, G.; Isla, D.; Provencio, M.; Insa, A.; et al. Screening for Epidermal Growth Factor Receptor Mutations in Lung Cancer. N. Engl. J. Med. 2009, 361, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Barlesi, F.; Mazieres, J.; Merlio, J.P.; Debieuvre, D.; Mosser, J.; Lena, H.; Ouafik, L.; Besse, B.; Rouquette, I.; Westeel, V.; et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: Results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016, 387, 1415–1426. [Google Scholar] [CrossRef]
- Sharma, S.V.; Bell, D.; Settleman, J.; Haber, D.A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 2007, 7, 169–181. [Google Scholar] [CrossRef]
- Brewer, M.R.; Yun, C.-H.; Lai, D.; Lemmon, M.A.; Eck, M.J.; Pao, W. Mechanism for activation of mutated epidermal growth factor receptors in lung cancer. Proc. Natl. Acad. Sci. USA 2013, 110, E3595–E3604. [Google Scholar] [CrossRef]
- Shan, Y.; Eastwood, M.P.; Zhang, X.; Kim, E.T.; Arkhipov, A.; Dror, R.O.; Jumper, J.; Kuriyan, J.; Shaw, D.E. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell 2012, 149, 860–870. [Google Scholar] [CrossRef]
- Mok, T.S.; Wu, Y.-L.; Thongprasert, S.; Yang, C.-H.; Chu, D.-T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Ichinose, Y.; et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009, 361, 947–957. [Google Scholar] [CrossRef]
- Lee, C.K.; Davies, L.; Wu, Y.L.; Mitsudomi, T.; Inoue, A.; Rosell, R.; Zhou, C.; Nakagawa, K.; Thongprasert, S.; Fukuoka, M.; et al. Gefitinib or Erlotinib vs Chemotherapy for EGFR Mutation-Positive Lung Cancer: Individual Patient Data Meta-Analysis of Overall Survival. J. Natl. Cancer Inst. 2017, 109, 1–9. [Google Scholar] [CrossRef]
- Rosell, R.; Carcereny, E.; Gervais, R.; Vergnenegre, A.; Massuti, B.; Felip, E.; Palmero, R.; Garcia-Gomez, R.; Pallares, C.; Sanchez, J.M.; et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012, 13, 239–246. [Google Scholar] [CrossRef]
- Nguyen, K.H.; Kobayashi, S.; Costa, D.B. Acquired Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancers Dependent on the Epidermal Growth Factor Receptor Pathway. Clin. Lung Cancer 2009, 10, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Jotte, R.M.; Spigel, D.R. Advances in molecular-based personalized non-small-cell lung cancer therapy: Targeting epidermal growth factor receptor and mechanisms of resistance. Cancer Med. 2015, 4, 1621–1632. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2017, 15, 81–94. [Google Scholar] [CrossRef]
- Forde, P.M.; Ettinger, D.S. Managing Acquired Resistance in EGFR-Mutated Non-Small Cell Lung Cancer. Clin. Adv. Hematol. Oncol. 2015, 13, 528–532. [Google Scholar]
- Mok, T.S.; Wu, Y.-L.; Ahn, M.-J.; Garassino, M.C.; Kim, H.R.; Ramalingam, S.S.; Shepherd, F.A.; He, Y.; Akamatsu, H.; Theelen, W.S.M.E.; et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N. Engl. J. Med. 2017, 376, 629–640. [Google Scholar] [CrossRef]
- Blumenthal, G.M.; Keegan, P.; He, K.; Weinstock, C.; Khozin, S.; Pazdur, R.; Cheng, J.; Zhuang, L.; Charlab, R.; Zhao, H.; et al. Osimertinib for the Treatment of Metastatic EGFR T790M Mutation-Positive Non-Small Cell Lung Cancer. Clin. Cancer Res. 2016, 23, 2131–2135. [Google Scholar] [CrossRef]
- Maley, C.C.; Aktipis, A.; Graham, T.A.; Sottoriva, A.; Boddy, A.M.; Janiszewska, M.; Silva, A.S.; Gerlinger, M.; Yuan, Y.; Pienta, K.J.; et al. Classifying the evolutionary and ecological features of neoplasms. Nat. Rev. Cancer 2017, 17, 605–619. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating Tumor DNA as a Liquid Biopsy for Cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef]
- Amirouchene-Angelozzi, N.; Swanton, C.; Bardelli, A. Tumor evolution as a therapeutic target. Cancer Discov. 2017, 7, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Oxnard, G.R.; Thress, K.S.; Alden, R.S.; Lawrance, R.; Paweletz, C.P.; Cantarini, M.; Yang, J.C.H.; Barrett, J.C.; Jänne, P.A. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J. Clin. Oncol. 2016, 34, 3375–3382. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Shen, L.; Zheng, D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: A systematic review and meta-analysis. Sci. Rep. 2014, 4, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jovelet, C.; Ileana, E.; Le Deley, M.C.; Motté, N.; Rosellini, S.; Romero, A.; Lefebvre, C.; Pedrero, M.; Pata-Merci, N.; Droin, N.; et al. Circulating cell-free tumor DNA analysis of 50 genes by next-generation sequencing in the prospective MOSCATO trial. Clin. Cancer Res. 2016, 22, 2960–2968. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Chen, C.; Hulbert, A.; Brock, M.V.; Yu, F. Non-blood circulating tumor DNA detection in cancer. Oncotarget 2017, 8, 69162–69173. [Google Scholar] [CrossRef]
- Mouliere, F.; Rosenfeld, N. Circulating tumor-derived DNA is shorter than somatic DNA in plasma. Proc. Natl. Acad. Sci. USA 2015, 112, 3178–3179. [Google Scholar] [CrossRef]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef]
- Castiglia, M.; Bazan, V.; Passiglia, F.; Gulotta, L.; Fulfaro, F.; Badalamenti, G.; Di Maio, M.; Listì, A.; Russo, A.; Galvano, A.; et al. The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: A systematic review and meta-analysis. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Nagai, Y.; Miyazawa, H.; Huqun; Tanaka, T.; Udagawa, K.; Kato, M.; Fukuyama, S.; Yokote, A.; Kobayashi, K.; Kanazawa, M.; et al. Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res. 2005, 65, 7276–7282. [Google Scholar] [CrossRef]
- Chevalier, L.; Billaud, A.; Passot, C.; Renoult, A.; Bigot, F.; Verrièle, V.; Morel, A. Caractérisation moléculaire de l’EGFR dans les cancers bronchiques non à petites cellules: Étude prospective comparative des technologies NGS et automate Idylla. Ann. Pathol. 2020. [Google Scholar] [CrossRef]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, H.; Tanaka, T.; Nagai, Y.; Matsuoka, M.; Sutani, A.; Udagawa, K.; Zhang, J.; Hirama, T.; Murayama, Y.; Koyama, N.; et al. Peptide nucleic acid-locked nucleic acid polymerase chain reaction clamp-based detection test for gefitinib-refractory T790M epidermal growth factor receptor mutation. Cancer Sci. 2008, 29, 595–600. [Google Scholar] [CrossRef]
- Cohen, J. Weighted kappa: Nominal scale agreement with provision for scaled disagreement or partial credit. Psychol. Bull. 1968, 70, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Soria, J.-C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Thress, K.S.; Paweletz, C.P.; Felip, E.; Cho, B.C.; Stetson, D.; Dougherty, B.; Lai, Z.; Markovets, A.; Vivancos, A.; Kuang, Y.; et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non–small cell lung cancer harboring EGFR T790M. Nat. Med. 2015, 21, 560–562. [Google Scholar] [CrossRef]
- Lazzaeri, C.; Gregore, V.; Karachaliou, N.; Rosell, R.; Santarpia, M. Mechanisms of resistance to osimertinib. J. Thorac. Dis. 2020, 12, 2851–2858. [Google Scholar] [CrossRef]
- Costa, C.; Molina, M.A.; Drozdowskyj, A.; Gimeńez-Capitán, A.; Bertran-Alamillo, J.; Karachaliou, N.; Gervais, R.; Massuti, B.; Wei, J.; Moran, T.; et al. The impact of EGFR T790M mutations and BIM mRNA expression on outcome in patients with EGFR-Mutant NSCLC treated with erlotinib or chemotherapy in the randomized phase III EURTAC trial. Clin. Cancer Res. 2014, 20, 2001–2010. [Google Scholar] [CrossRef]
- Maheswaran, S.; Sequist, L.V.; Nagrath, S.; Ulkus, L.; Brannigan, B.; Collura, C.V.; Inserra, E.; Diederichs, S.; Iafrate, A.J.; Bell, D.W.; et al. Detection of Mutations in EGFR in Circulating Lung-Cancer Cells. N. Engl. J. Med. 2008, 359, 366–377. [Google Scholar] [CrossRef]
| Names | Sequences |
|---|---|
| PCR forward primer | 5′-GCA TCT GCC TCA CCT CCA A-3′ |
| PCR reverse primer | 5′-biotin-CGA TCT GCA CAC ACC AGT TG-3′ |
| PNA sequence | NH2-CTCATCACGCAGCTCA-COOH [32] |
| Sequencing primer (pyrosequencing) | CCG TGC AGC TCA TCA |
| Dispensation order (pyrosequencing) | ACTAGCAGC |
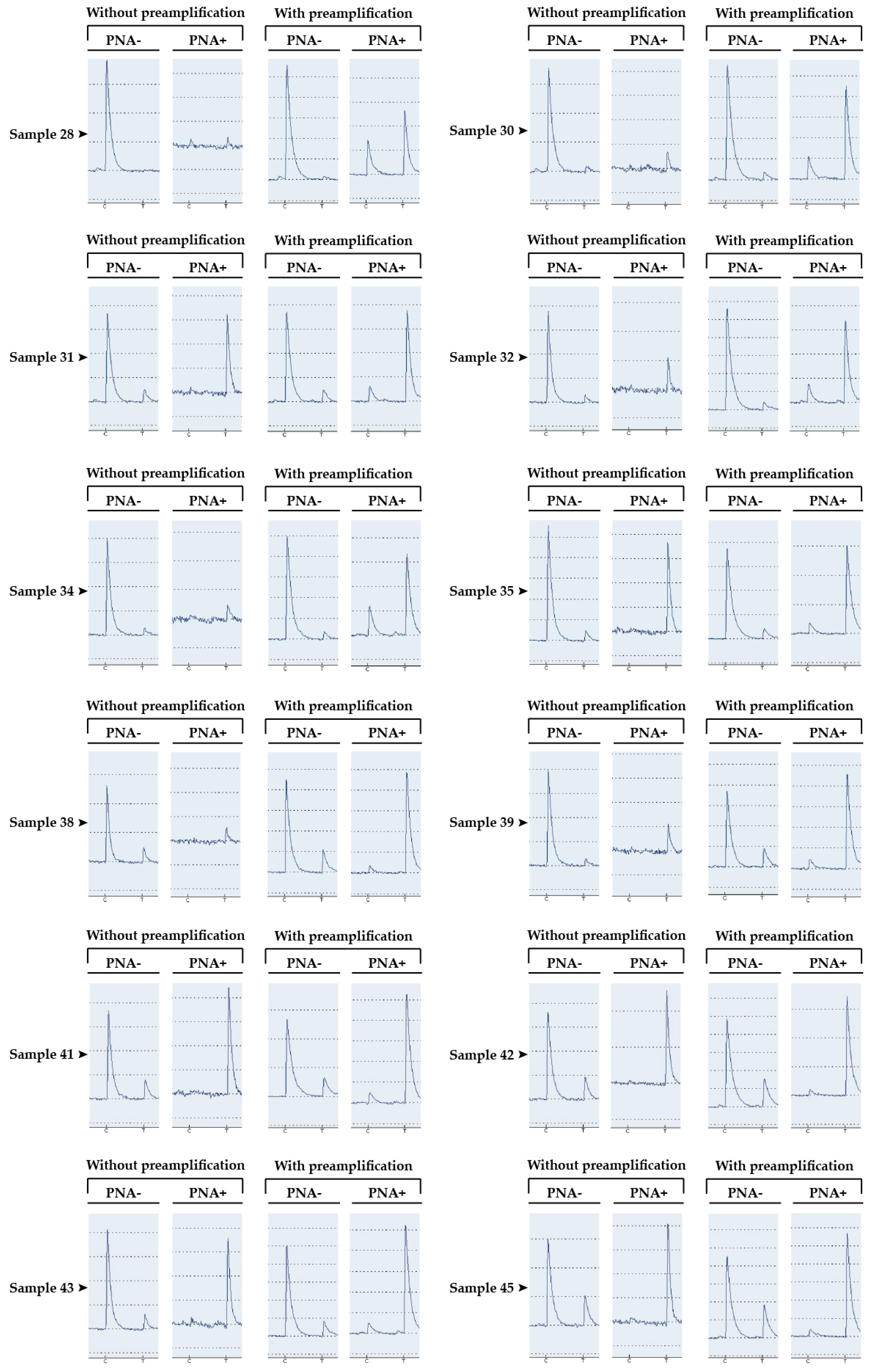
| Samples | Without Preamplification | With Preamplification | ||
|---|---|---|---|---|
| PNA− (%) | PNA+ (%) | PNA− (%) | PNA+ (%) | |
| Sample 28 | 0 | NA | 2 | 28 |
| Sample 30 | 5 | NA | 5 | 59 |
| Sample 31 | 12 | 100 | 10 | 68 |
| Sample 32 | 8 | NA | 7 | 72 |
| Sample 34 | 7 | NA | 9 | 84 |
| Sample 35 | 8 | 100 | 9 | 84 |
| Sample 38 | 16 | NA | 22 | 88 |
| Sample 39 | 22 | 100 | 20 | 90 |
| Sample 41 | 18 | 100 | 20 | 91 |
| Sample 42 | 21 | 100 | 23 | 91 |
| Sample 43 | 13 | 92 | 11 | 92 |
| Sample 45 | 26 | 100 | 28 | 93 |
| Samples | Ages | Sexes | [DNA] (ng/µL) | Cellularity | NGS Results | PNA-Mediated PCR | Concordance Results | |
|---|---|---|---|---|---|---|---|---|
| PNA− | PNA+ | |||||||
| Sample 1 | 50.8 | M | 10.7 | 30 | WT | 0 | 0 | OK-WT |
| Sample 2 | 53.7 | W | 14.2 | 40 | WT | 0 | 0 | OK-WT |
| Sample 3 | 62.1 | M | 25.9 | 20 | WT | 0 | 0 | OK-WT |
| Sample 4 | 55.5 | W | 10.2 | 20 | WT | 0 | 0 | OK-WT |
| Sample 5 | 62.7 | M | 13.3 | 30 | WT | 0 | 0 | OK-WT |
| Sample 6 | 73.4 | M | 39 | 10 | WT | 0 | 0 | OK-WT |
| Sample 7 | 64.2 | M | 6.4 | 20 | WT | 0 | 0 | OK-WT |
| Sample 8 | 67.2 | M | 6.6 | 40 | WT | 0 | 0 | OK-WT |
| Sample 9 | 51.4 | W | 11.6 | 40 | WT | 0 | 0 | OK-WT |
| Sample 10 | 81.6 | M | 11.4 | 20 | WT | 0 | 0 | OK-WT |
| Sample 11 | 56.6 | M | 52.8 | 70 | WT | 0 | 0 | OK-WT |
| Sample 12 | 83.6 | W | 1.4 | 10 | WT | 0 | 0 | OK-WT |
| Sample 13 | 56 | M | 15.6 | 30 | WT | 0 | 0 | OK-WT |
| Sample 14 | 52.7 | M | 7.2 | 30 | WT | 0 | 0 | OK-WT |
| Sample 15 | 67.6 | M | 10.7 | 30 | WT | 0 | 0 | OK-WT |
| Sample 16 | 59 | W | 18.2 | 80 | WT | 0 | 0 | OK-WT |
| Sample 17 | 69.7 | M | 29.7 | 60 | WT | 0 | 0 | OK-WT |
| Sample 18 | 56.2 | M | 5.7 | 60 | WT | 0 | 0 | OK-WT |
| Sample 19 | 62.4 | W | 2.9 | 5 | WT | 0 | 0 | OK-WT |
| Sample 20 | 63.2 | W | 9.5 | 70 | WT | 0 | 5 | L-WT |
| Sample 21 | 71.4 | M | 24.5 | 40 | WT | 0 | 5 | L-WT |
| Sample 22 | 82 | M | 9.5 | 10 | WT | 0 | 5 | L-WT |
| Sample 23 | 61.1 | M | 16.1 | 60 | WT | 0 | 6 | L-WT |
| Sample 24 | 83 | W | 5.5 | 20 | WT | 0 | 6 | L-WT |
| Sample 25 | NA | W | NA | NA | WT | 0 | 6 | L-WT |
| Sample 26 | 62.3 | M | 24.8 | 30 | WT | 0 | 8 | L-WT |
| Sample 27 | 51.4 | W | 44.2 | 40 | WT | 0 | 18 | Discordant |
| Sample 28 | 86.7 | W | 27.3 | 5 | T790M | 2 | 28 | OK-Mutated |
| Sample 29 | NA | M | 7.4 | NA | T790M (1.3%) | 2 | 35 | OK-Mutated |
| Sample 30 | 76.4 | W | 10 | 40 | T790M (3.5%) | 5 | 59 | OK-Mutated |
| Sample 31 | 74.6 | M | 12.4 | 4 | T790M | 10 | 68 | OK-Mutated |
| Sample 32 | 78.2 | W | 2.2 | 10 | T790M | 7 | 72 | OK-Mutated |
| Sample 33 | 73.7 | W | 2.1 | 50 | T790M (9%) | 11 | 75 | OK-Mutated |
| Sample 34 | 67.2 | M | 4.1 | 10 | T790M | 9 | 84 | OK-Mutated |
| Sample 35 | 75.6 | W | 23.7 | 40 | T790M | 9 | 84 | OK-Mutated |
| Sample 36 | 87.8 | W | 12.4 | 20 | T790M (12%) | 24 | 87 | OK-Mutated |
| Sample 37 | 66.4 | W | 6 | 80 | T790M | 26 | 87 | OK-Mutated |
| Sample 38 | 64.3 | W | 2.5 | 60 | T790M | 22 | 88 | OK-Mutated |
| Sample 39 | 64.8 | W | 9.4 | 40 | T790M | 20 | 90 | OK-Mutated |
| Sample 40 | 87.6 | W | 4.4 | 50 | T790M (15.4%) | 17 | 90 | OK-Mutated |
| Sample 41 | 65.3 | M | 30.2 | 10 | T790M | 20 | 91 | OK-Mutated |
| Sample 42 | NA | M | 6.4 | NA | T790M (18.1%) | 23 | 91 | OK-Mutated |
| Sample 43 | 78.3 | W | 30.9 | 70 | T790M | 11 | 92 | OK-Mutated |
| Sample 44 | 77 | W | 0.6 | 10 | T790M (14.7%) | 23 | 92 | OK-Mutated |
| Sample 45 | 74 | W | 6.9 | 40 | T790M | 28 | 93 | OK-Mutated |
| Sample 46 | 78.5 | M | 3.8 | 60 | T790M (21.3%) | 26 | 95 | OK-Mutated |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Billaud, A.; Verriele, V.; Dauvé, J.; Chevalier, L.-M.; Morel, A. Non-Small-Cell Lung Cancer-Sensitive Detection of the p.Thr790Met EGFR Alteration by Preamplification before PNA-Mediated PCR Clamping and Pyrosequencing. Diagnostics 2020, 10, 527. https://doi.org/10.3390/diagnostics10080527
Billaud A, Verriele V, Dauvé J, Chevalier L-M, Morel A. Non-Small-Cell Lung Cancer-Sensitive Detection of the p.Thr790Met EGFR Alteration by Preamplification before PNA-Mediated PCR Clamping and Pyrosequencing. Diagnostics. 2020; 10(8):527. https://doi.org/10.3390/diagnostics10080527
Chicago/Turabian StyleBillaud, Amandine, Veronique Verriele, Jonathan Dauvé, Louise-Marie Chevalier, and Alain Morel. 2020. "Non-Small-Cell Lung Cancer-Sensitive Detection of the p.Thr790Met EGFR Alteration by Preamplification before PNA-Mediated PCR Clamping and Pyrosequencing" Diagnostics 10, no. 8: 527. https://doi.org/10.3390/diagnostics10080527
APA StyleBillaud, A., Verriele, V., Dauvé, J., Chevalier, L.-M., & Morel, A. (2020). Non-Small-Cell Lung Cancer-Sensitive Detection of the p.Thr790Met EGFR Alteration by Preamplification before PNA-Mediated PCR Clamping and Pyrosequencing. Diagnostics, 10(8), 527. https://doi.org/10.3390/diagnostics10080527





