Development of a Deep Learning Algorithm for Periapical Disease Detection in Dental Radiographs
Abstract
1. Introduction
2. Materials and Methods
2.1. Assessing the Reliability of OMF Surgeons’ Diagnoses of Periapical Radiolucenies in Panoramic Radiographs
2.2. Development of a Deep Learning Algorithm for the Automated Detection of Periapical Radiolucencies in Panoramic Radiographs
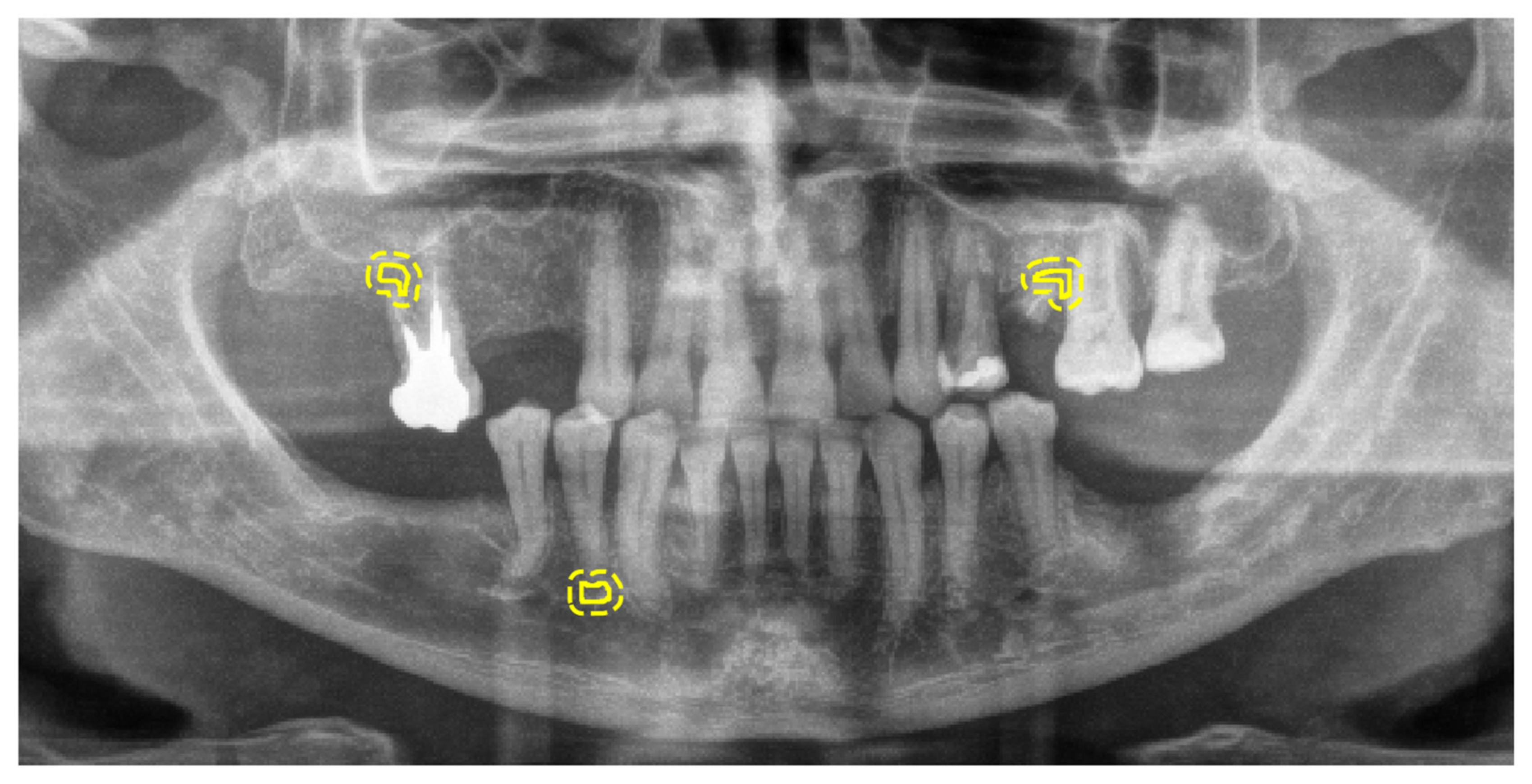
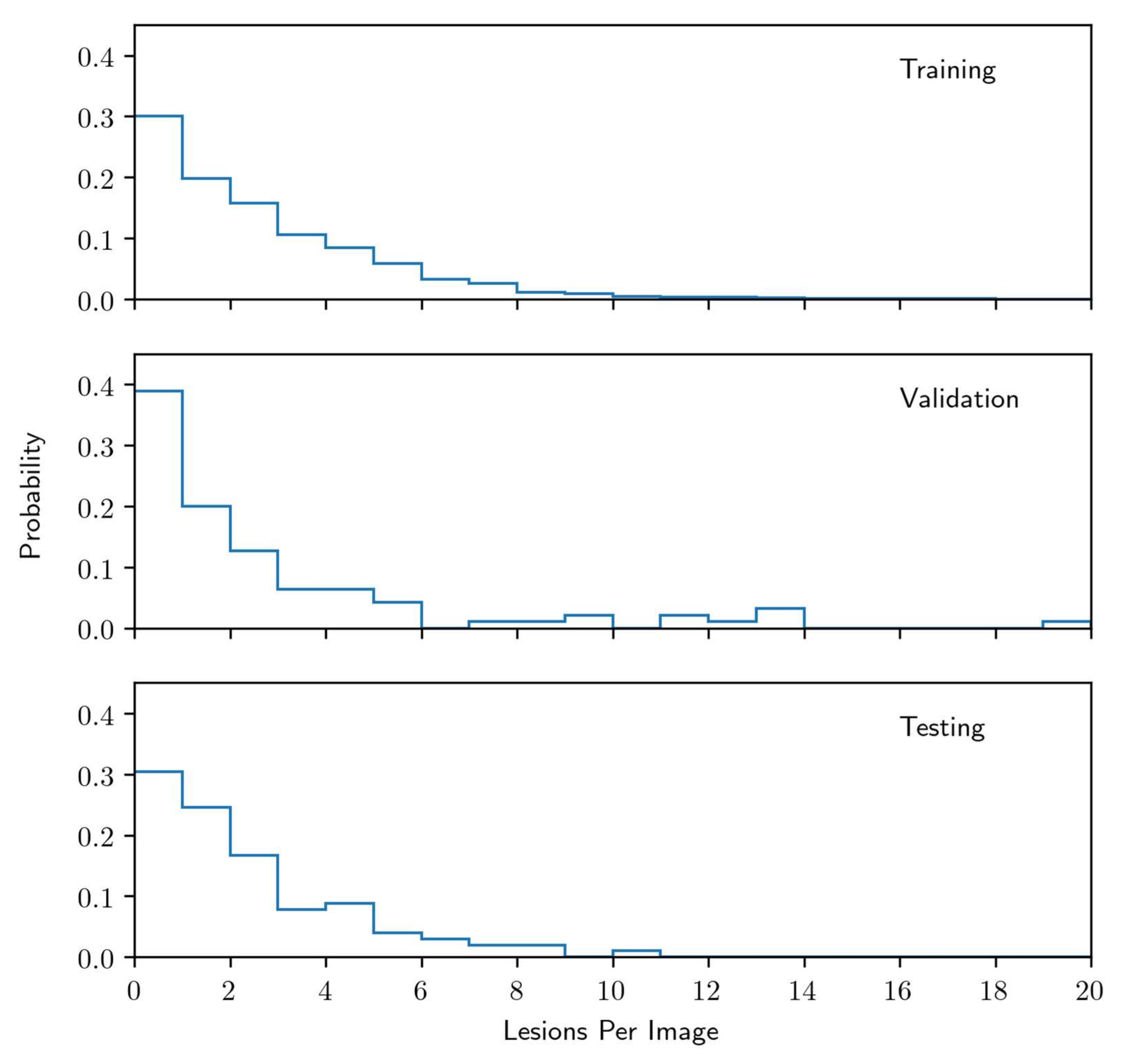
2.3. Radiographic Images and Labelling for Model Training
2.4. Reference Standard for Model Selection and Evaluation
2.5. Benchmarks for Model Comparison
2.6. Model
2.7. Evaluation Metrics
2.8. Evaluation of Correlations between Model and OMF Surgeons’ Performance
3. Results
3.1. The Reliability of OMF Surgeons’ Diagnoses of Periapical Radiolucenies in Panoramic Radiographs
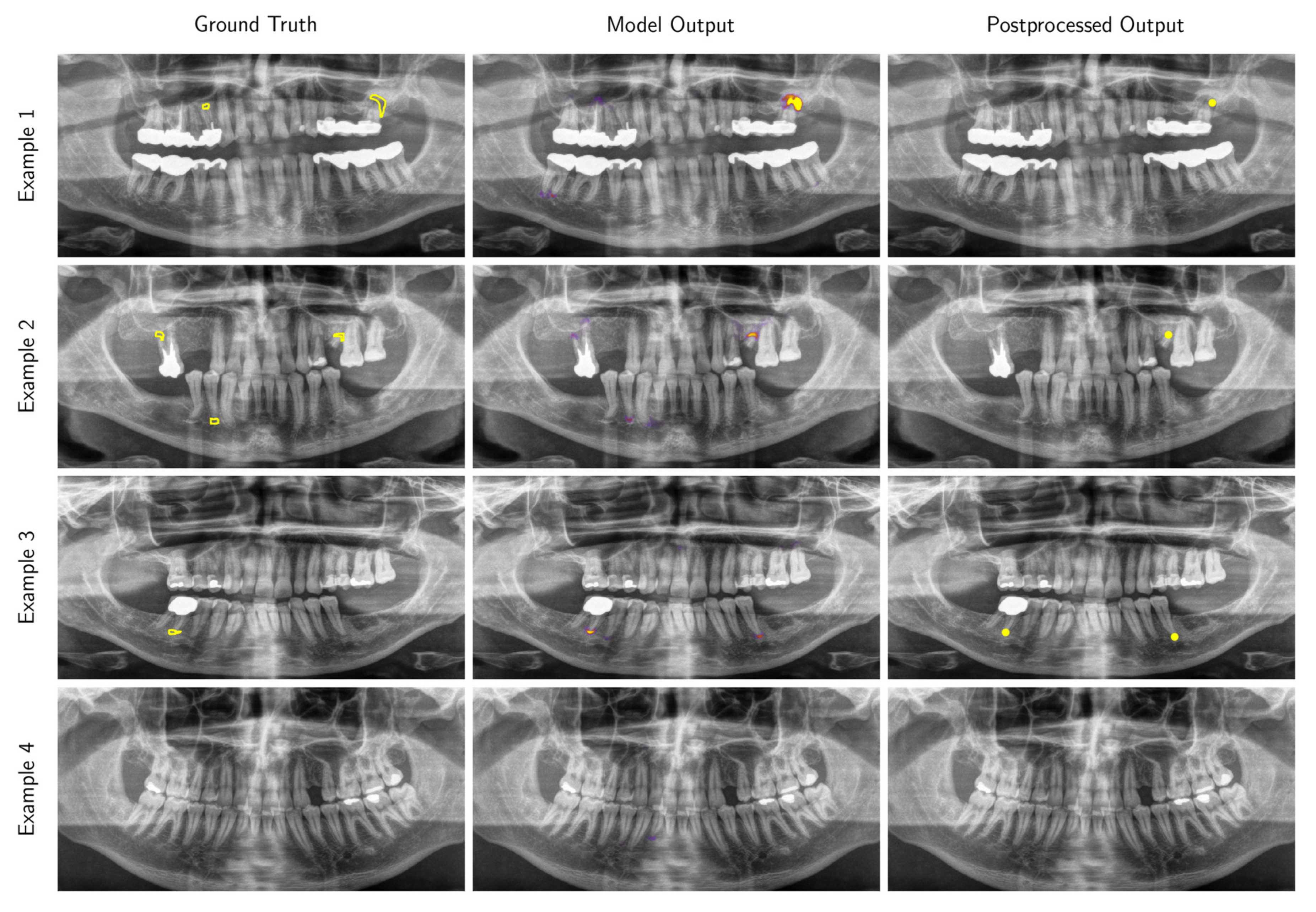
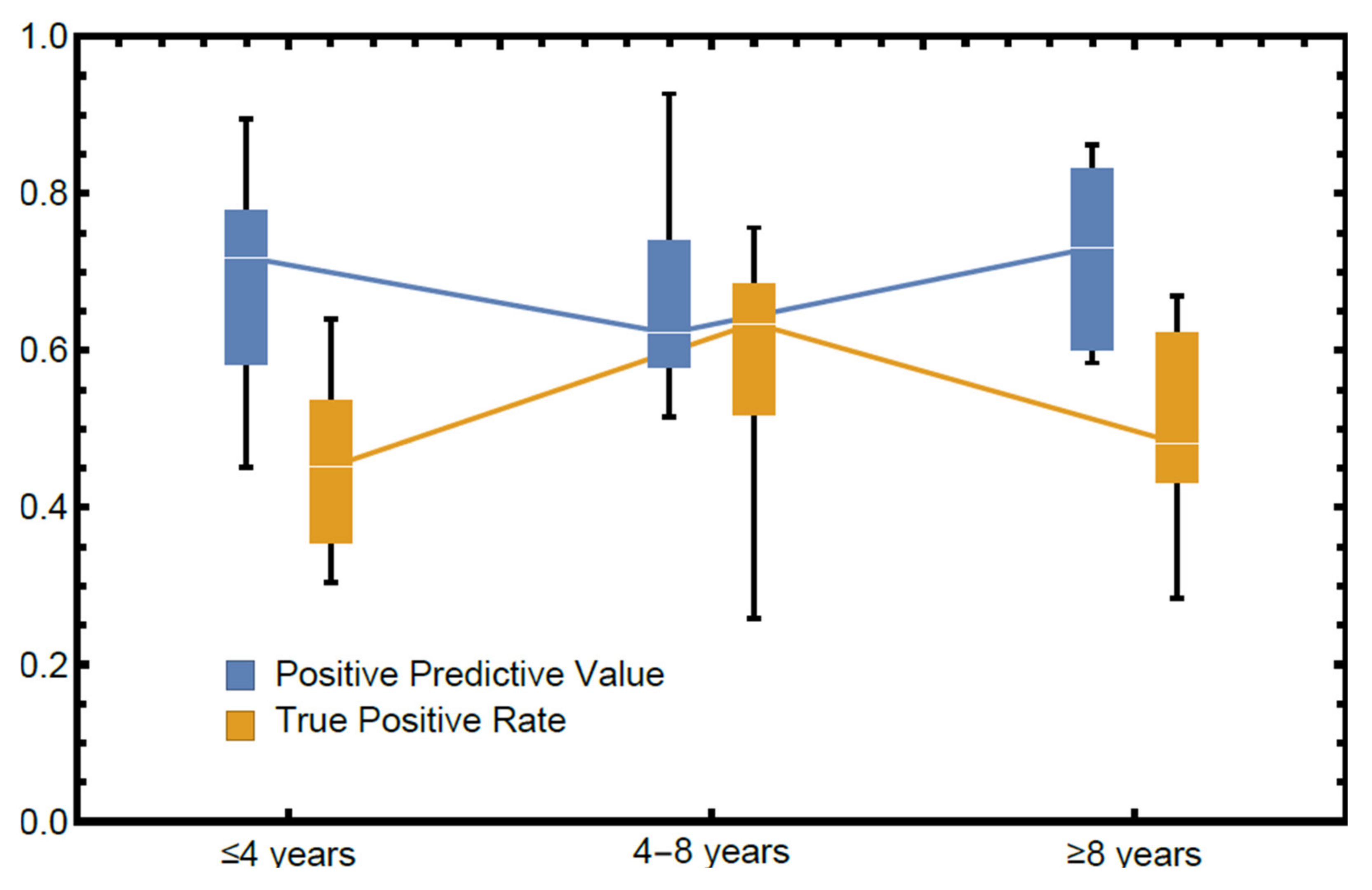
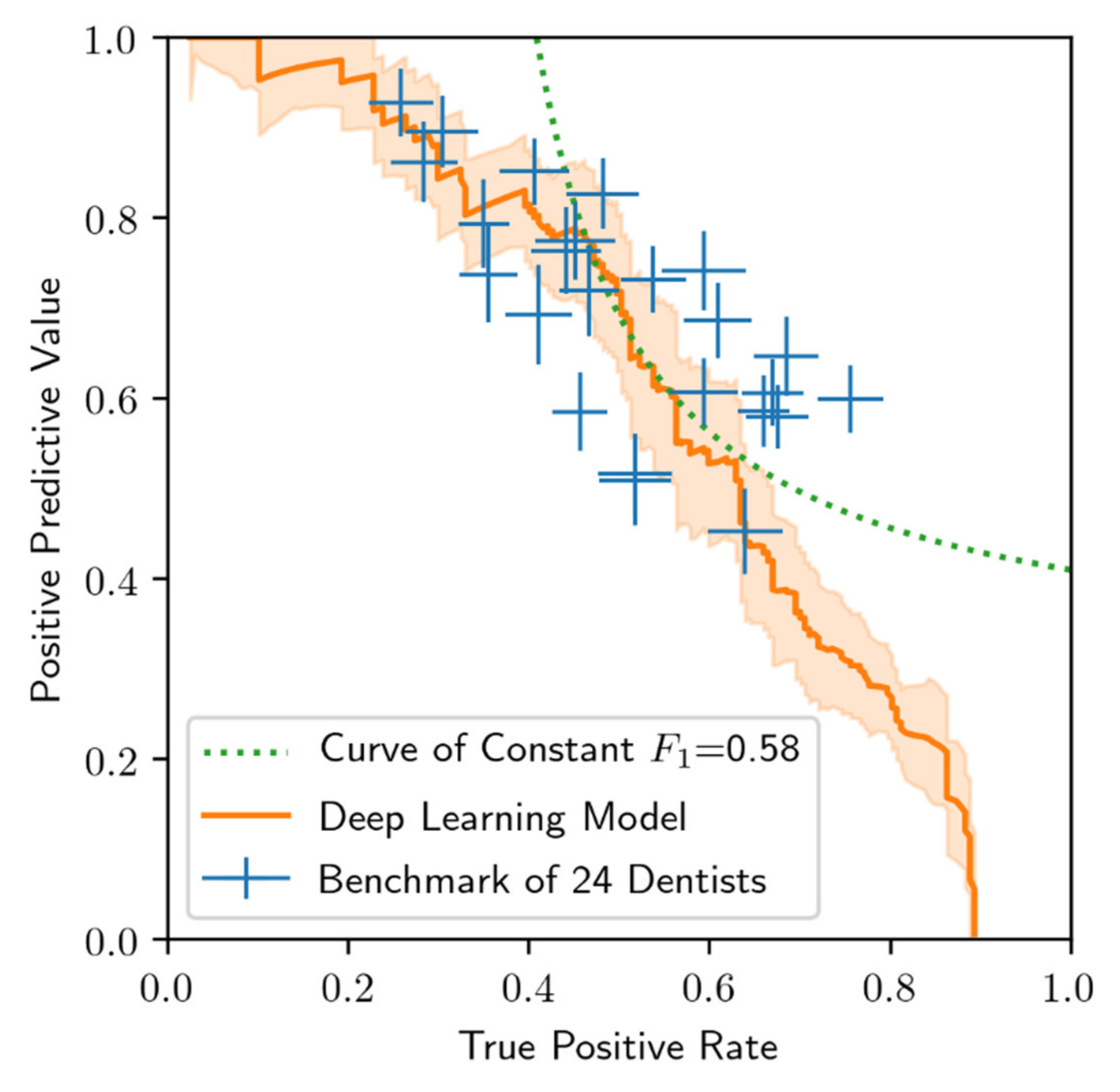
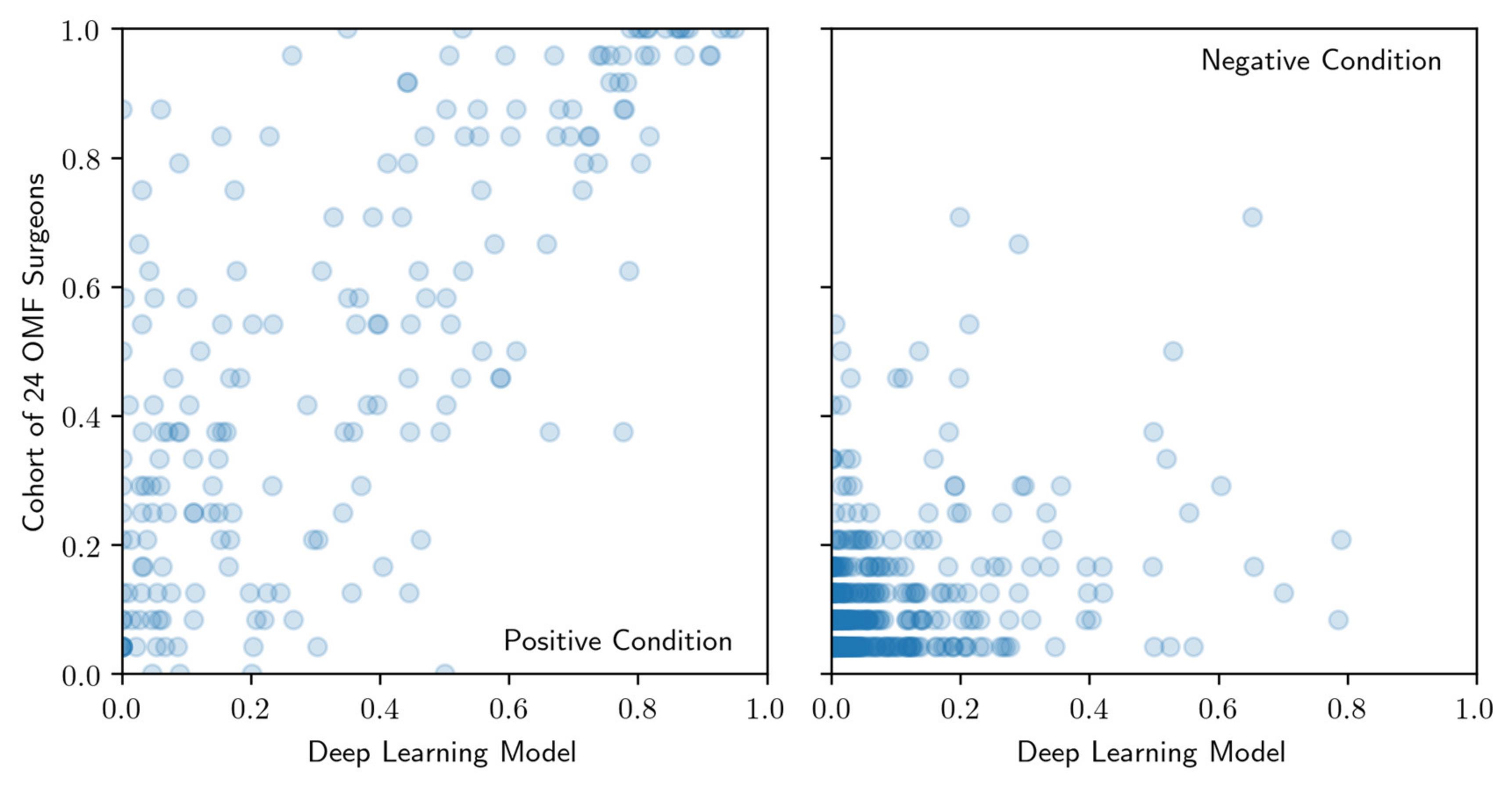
3.2. Performance of the Deep Learning Algorithm
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Material and Methods
Appendix A.1.1. Model
| Layer | Type | Kernel Size | Number of Kernels | Input Dimensions | Activation Function |
|---|---|---|---|---|---|
| 1 | Conv2D | 3 × 3 | 64 | 256 × 512 | ReLU |
| 2 | Conv2D | 3 × 3 | 64 | 256 × 512 | ReLU |
| 3 | MaxPool | 2 × 2 | - | 256 × 512 | - |
| 4 | Conv2D | 3 × 3 | 128 | 128 × 256 | ReLU |
| 5 | Conv2D | 3 × 3 | 128 | 128 × 256 | ReLU |
| 6 | MaxPool | 2 × 2 | - | 128 × 256 | - |
| 7 | Conv2D | 3 × 3 | 256 | 64 × 128 | ReLU |
| 8 | Conv2D | 3 × 3 | 256 | 64 × 128 | ReLU |
| 9 | MaxPool | 2 × 2 | - | 64 × 128 | - |
| 10 | Conv2D | 3 × 3 | 512 | 32 × 64 | ReLU |
| 11 | Conv2D | 3 × 3 | 512 | 32 × 64 | ReLU |
| 12 | MaxPool | 2 × 2 | - | 32 × 64 | - |
| 13 | Conv2D | 3 × 3 | 1024 | 16 × 32 | ReLU |
| 14 | Conv2D | 3 × 3 | 1024 | 16 × 32 | ReLU |
| 15 | Dropout | - | - | 16 × 32 | - |
| 16 | UpConv2D | 2 × 2 | 512 | 16 × 32 | - |
| - | Concat(11) | - | - | - | - |
| 17 | Conv2D | 3 × 3 | 512 | 32 × 64 | ReLU |
| 18 | Conv2D | 3 × 3 | 512 | 32 × 64 | ReLU |
| 19 | UpConv2D | 2 × 2 | 256 | 32 × 64 | - |
| - | Concat(8) | - | - | - | - |
| 20 | Conv2D | 3 × 3 | 256 | 64 × 128 | ReLU |
| 21 | Conv2D | 3 × 3 | 256 | 64 × 128 | ReLU |
| 22 | UpConv2D | 2 × 2 | 128 | 64 × 128 | - |
| - | Concat(5) | - | - | - | - |
| 23 | Conv2D | 3 × 3 | 128 | 128 × 256 | ReLU |
| 24 | Conv2D | 3 × 3 | 128 | 128 × 256 | ReLU |
| 25 | UpConv2D | 2 × 2 | 64 | 128 × 256 | - |
| - | Concat(3) | - | - | - | - |
| 26 | Conv2D | 3 × 3 | 64 | 256 × 512 | ReLU |
| 27 | Conv2D | 3 × 3 | 64 | 256 × 512 | ReLU |
| 28 | Conv2D | 1 × 1 | 1 | 256 × 512 | Sigmoid |
Appendix A.1.2. Inference and Post-Processing
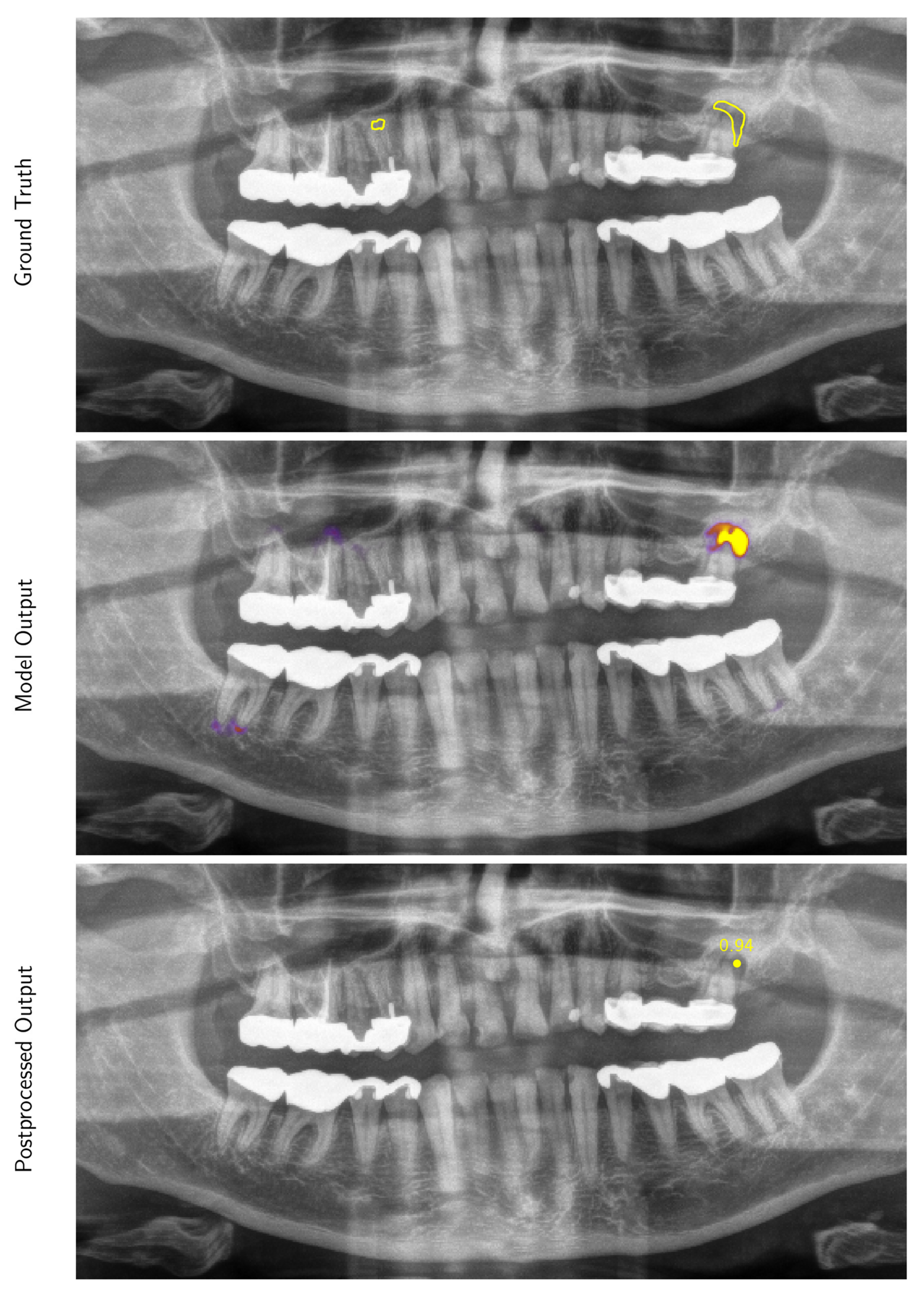
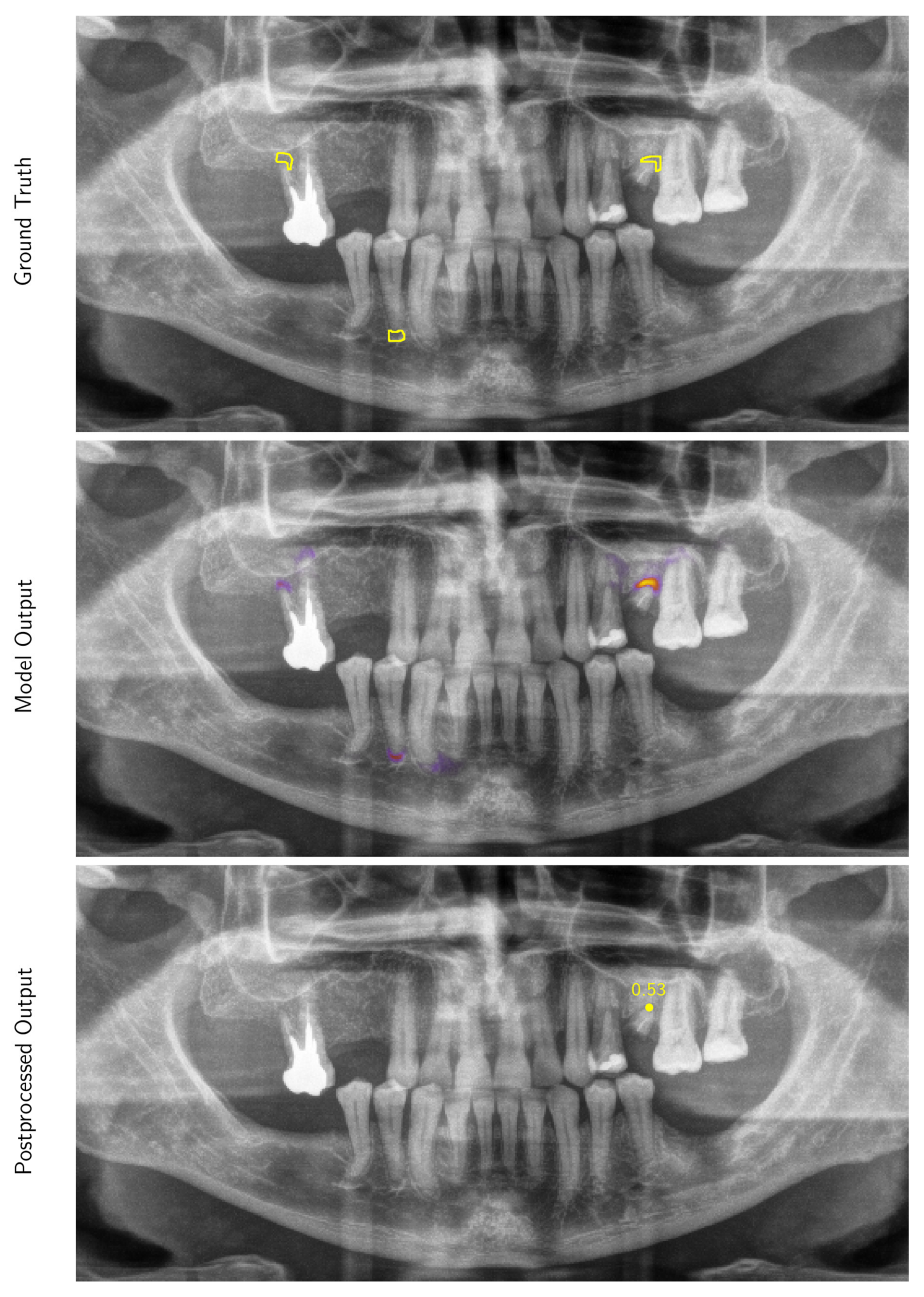
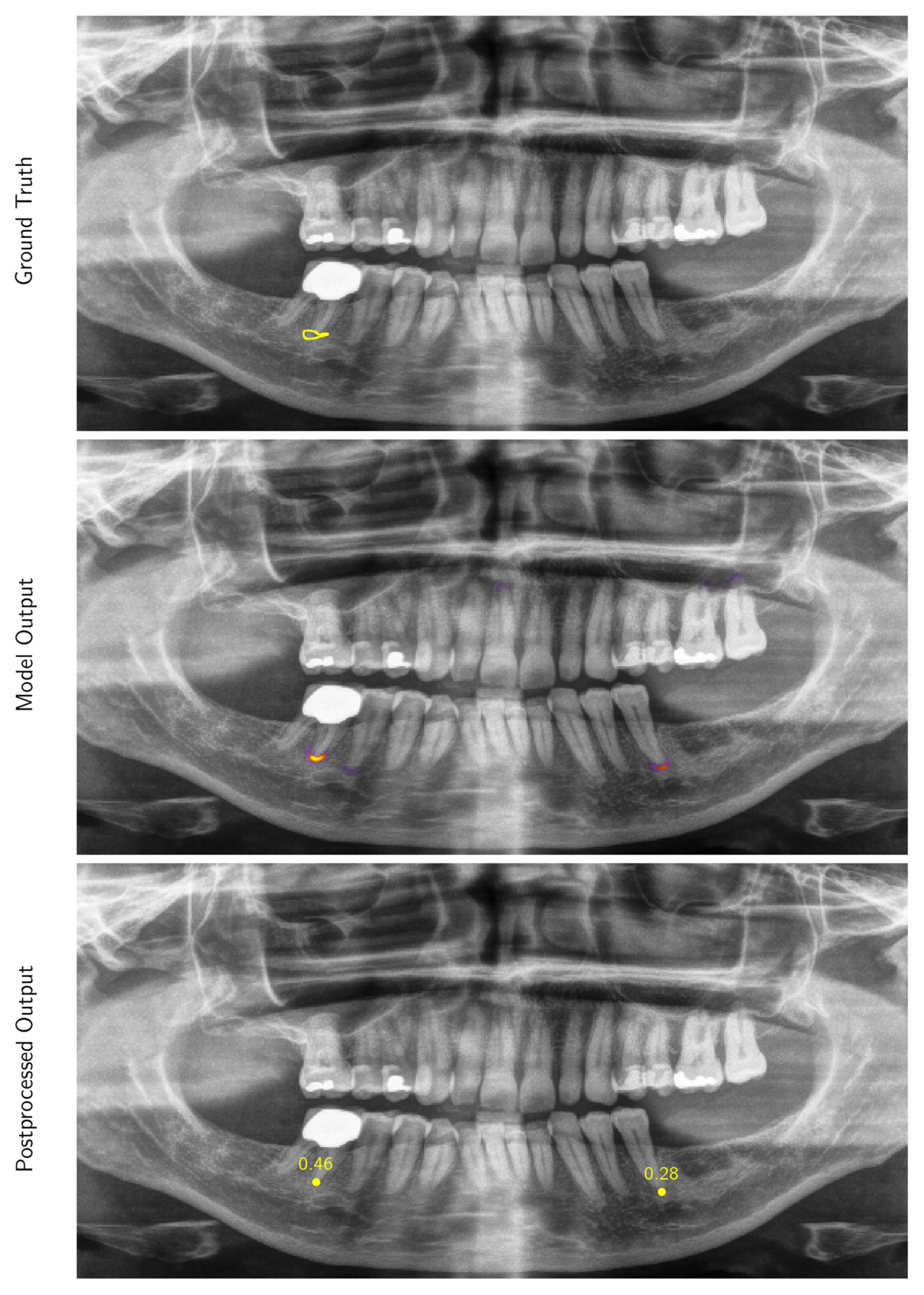
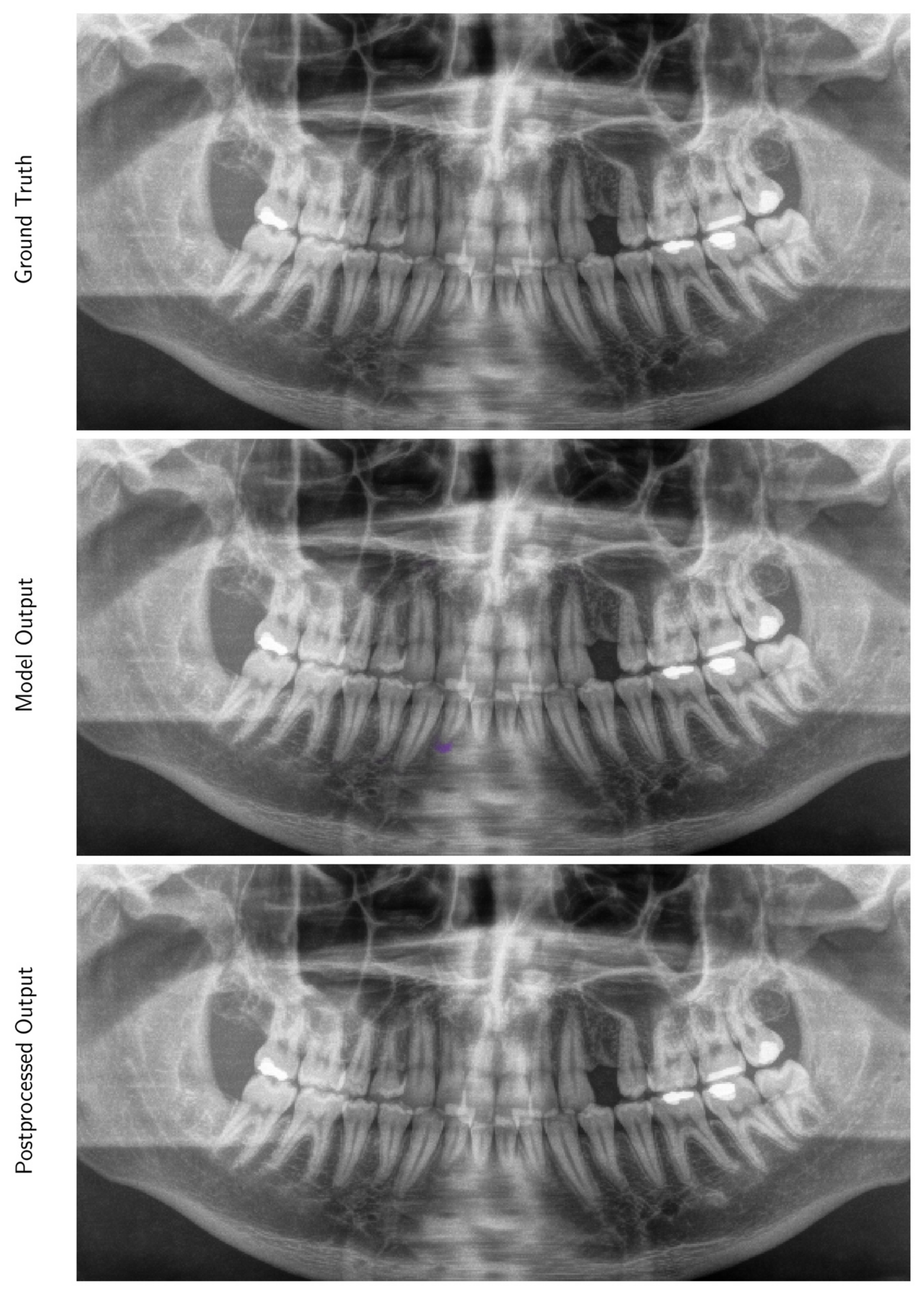
Appendix A.1.3. Evaluation Metrics
References
- Perschbacher, S. Interpretation of Panoramic Radiographs. Aust. Dent. J. 2012, 57, 40–45. [Google Scholar] [CrossRef]
- Molander, B. Panoramic Radiography in Dental Diagnostics. Swed. Dent. J. Suppl. 1996, 119, 1–26. [Google Scholar] [PubMed]
- Osman, F.; Scully, C.; Dowell, T.B.; Davies, R.M. Use of Panoramic Radiographs in General Dental Practice in England. Community Dent. Oral Epidemiol. 1986, 14, 8–9. [Google Scholar] [CrossRef] [PubMed]
- Rafferty, E.A.; Park, J.M.; Philpotts, L.E.; Poplack, S.P.; Sumkin, J.H.; Halpern, E.F.; Niklason, L.T. Assessing Radiologist Performance Using Combined Digital Mammography and Breast Tomosynthesis Compared with Digital Mammography Alone: Results of a Multicenter, Multireader Trial. Radiology 2013, 266, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Sabarudin, A.; Tiau, Y.J. Image Quality Assessment in Panoramic Dental Radiography: A Comparative Study between Conventional and Digital Systems. Quant. Imaging Med. Surg. 2013, 3, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Kantor, M.L.; Reiskin, A.B.; Lurie, A.G. A Clinical Comparison of X-Ray Films for Detection of Proximal Surface Caries. J. Am. Dent. Assoc. 1985, 111, 967–969. [Google Scholar] [CrossRef]
- Fitzgerald, R. Error in Radiology. Clin. Radiol. 2001, 56, 938–946. [Google Scholar] [CrossRef]
- Brady, A.; Laoide, R.Ó.; McCarthy, P.; McDermott, R. Discrepancy and Error in Radiology: Concepts, Causes and Consequences. Ulster Med. J. 2012, 81, 3–9. [Google Scholar]
- Valizadeh, S.; Goodini, M.; Ehsani, S.; Mohseni, H.; Azimi, F.; Bakhshandeh, H. Designing of a Computer Software for Detection of Approximal Caries in Posterior Teeth. Iran. J. Radiol. 2015, 12, e16242. [Google Scholar] [CrossRef]
- White, S.C.; Hollender, L.; Gratt, B.M. Comparison of Xeroradiographs and Film for Detection of Proximal Surface Caries. J. Am. Dent. Assoc. 1984, 108, 755–759. [Google Scholar] [CrossRef]
- Fiorellini, J.P.; Howell, T.H.; Cochran, D.; Malmquist, J.; Lilly, L.C.; Spagnoli, D.; Toljanic, J.; Jones, A.; Nevins, M. Randomized Study Evaluating Recombinant Human Bone Morphogenetic Protein-2 for Extraction Socket Augmentation. J. Periodontol. 2005, 76, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Yasaka, K.; Abe, O. Deep Learning and Artificial Intelligence in Radiology: Current Applications and Future Directions. PLoS Med. 2018, 15, e1002707. [Google Scholar] [CrossRef] [PubMed]
- Pesapane, F.; Codari, M.; Sardanelli, F. Artificial Intelligence in Medical Imaging: Threat or Opportunity? Radiologists Again at the Forefront of Innovation in Medicine. Eur. Radiol. Exp. 2018, 2, 35. [Google Scholar] [CrossRef] [PubMed]
- Nevin, L.; PLoS Medicine Editors. Advancing the Beneficial Use of Machine Learning in Health Care and Medicine: Toward a Community Understanding. PLoS Med. 2018, 15, e1002708. [Google Scholar] [CrossRef]
- Setio, A.A.A.; Traverso, A.; de Bel, T.; Berens, M.S.N.; van den Bogaard, C.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Comparison, and Combination of Algorithms for Automatic Detection of Pulmonary Nodules in Computed Tomography Images: The LUNA16 Challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef]
- Cruz-Roa, A.; Gilmore, H.; Basavanhally, A.; Feldman, M.; Ganesan, S.; Shih, N.N.C.; Tomaszewski, J.; González, F.A.; Madabhushi, A. Accurate and Reproducible Invasive Breast Cancer Detection in Whole-Slide Images: A Deep Learning Approach for Quantifying Tumor Extent. Sci. Rep. 2017, 7, 46450. [Google Scholar] [CrossRef]
- Komura, D.; Ishikawa, S. Machine Learning Methods for Histopathological Image Analysis. Comput. Struct. Biotechnol. J. 2018, 16, 34–42. [Google Scholar] [CrossRef]
- Hou, L.; Samaras, D.; Kurc, T.M.; Gao, Y.; Davis, J.E.; Saltz, J.H. Patch-Based Convolutional Neural Network for Whole Slide Tissue Image Classification. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. 2016, 2016, 2424–2433. [Google Scholar] [CrossRef]
- Xu, J.; Luo, X.; Wang, G.; Gilmore, H.; Madabhushi, A. A Deep Convolutional Neural Network for Segmenting and Classifying Epithelial and Stromal Regions in Histopathological Images. Neurocomputing 2016, 191, 214–223. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Askham, H.; Glorot, X.; O’Donoghue, B.; Visentin, D.; et al. Clinically Applicable Deep Learning for Diagnosis and Referral in Retinal Disease. Nat. Med. 2018, 24, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Nikolov, S.; Blackwell, S.; Mendes, R.; De Fauw, J.; Meyer, C.; Hughes, C.; Askham, H.; Romera-Paredes, B.; Karthikesalingam, A.; Chu, C.; et al. Deep Learning to Achieve Clinically Applicable Segmentation of Head and Neck Anatomy for Radiotherapy. arXiv 2018, arXiv:1809.04430. [Google Scholar]
- Wang, C.-W.; Huang, C.-T.; Lee, J.-H.; Li, C.-H.; Chang, S.-W.; Siao, M.-J.; Lai, T.-M.; Ibragimov, B.; Vrtovec, T.; Ronneberger, O.; et al. A Benchmark for Comparison of Dental Radiography Analysis Algorithms. Med. Image Anal. 2016, 31, 63–76. [Google Scholar] [CrossRef]
- Wenzel, A.; Hintze, H.; Kold, L.M.; Kold, S. Accuracy of Computer-Automated Caries Detection in Digital Radiographs Compared with Human Observers. Eur. J. Oral Sci. 2002, 110, 199–203. [Google Scholar] [CrossRef]
- Wenzel, A. Computer–Automated Caries Detection in Digital Bitewings: Consistency of a Program and Its Influence on Observer Agreement. Caries Res. 2001, 35, 12–20. [Google Scholar] [CrossRef]
- Murata, S.; Lee, C.; Tanikawa, C.; Date, S. Towards a Fully Automated Diagnostic System for Orthodontic Treatment in Dentistry. In Proceedings of the 2017 IEEE 13th International Conference on e-Science (e-Science), Auckland, New Zealand, 24–27 October 2017; pp. 1–8. [Google Scholar] [CrossRef]
- Behere, R.; Lele, S. Reliability of Logicon Caries Detector in the Detection and Depth Assessment of Dental Caries: An in-Vitro Study. Indian J. Dent. Res. 2011, 22, 362. [Google Scholar] [CrossRef]
- Cachovan, G.; Phark, J.-H.; Schön, G.; Pohlenz, P.; Platzer, U. Odontogenic Infections: An 8-Year Epidemiologic Analysis in a Dental Emergency Outpatient Care Unit. Acta Odontol. Scand. 2013, 71, 518–524. [Google Scholar] [CrossRef]
- Kirkevang, L.L.; Ørstavik, D.; Hörsted-Bindslev, P.; Wenzel, A. Periapical Status and Quality of Root Fillings and Coronal Restorations in a Danish Population. Int. Endod. J. 2000, 33, 509–515. [Google Scholar] [CrossRef]
- Lupi-Pegurier, L.; Bertrand, M.-F.; Muller-Bolla, M.; Rocca, J.P.; Bolla, M. Periapical Status, Prevalence and Quality of Endodontic Treatment in an Adult French Population. Int. Endod. J. 2002, 35, 690–697. [Google Scholar] [CrossRef]
- Chapman, M.N.; Nadgir, R.N.; Akman, A.S.; Saito, N.; Sekiya, K.; Kaneda, T.; Sakai, O. Periapical Lucency around the Tooth: Radiologic Evaluation and Differential Diagnosis. RadioGraphics 2013, 33, E15–E32. [Google Scholar] [CrossRef] [PubMed]
- Ekert, T.; Krois, J.; Meinhold, L.; Elhennawy, K.; Emara, R.; Golla, T.; Schwendicke, F. Deep Learning for the Radiographic Detection of Apical Lesions. J. Endod. 2019, 45, 917–922.e5. [Google Scholar] [CrossRef] [PubMed]
- Nardi, C.; Calistri, L.; Pradella, S.; Desideri, I.; Lorini, C.; Colagrande, S. Accuracy of Orthopantomography for Apical Periodontitis without Endodontic Treatment. J. Endod. 2017, 43, 1640–1646. [Google Scholar] [CrossRef] [PubMed]
- Nardi, C.; Calistri, L.; Grazzini, G.; Desideri, I.; Lorini, C.; Occhipinti, M.; Mungai, F.; Colagrande, S. Is Panoramic Radiography an Accurate Imaging Technique for the Detection of Endodontically Treated Asymptomatic Apical Periodontitis? J. Endod. 2018, 44, 1500–1508. [Google Scholar] [CrossRef]
- Choi, B.R.; Choi, D.H.; Huh, K.H.; Yi, W.J.; Heo, M.S.; Choi, S.C.; Bae, K.H.; Lee, S.S. Clinical Image Quality Evaluation for Panoramic Radiography in Korean Dental Clinics. Imaging Sci. Dent. 2012, 42, 183–190. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Langlotz, C.P.; Allen, B.; Erickson, B.J.; Kalpathy-Cramer, J.; Bigelow, K.; Cook, T.S.; Flanders, A.E.; Lungren, M.P.; Mendelson, D.S.; Rudie, J.D.; et al. A Roadmap for Foundational Research on Artificial Intelligence in Medical Imaging: From the 2018 NIH/RSNA/ACR/The Academy Workshop. Radiology 2019, 291, 781–791. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Reader study level-I and level-II Groups. Man against Machine: Diagnostic Performance of a Deep Learning Convolutional Neural Network for Dermoscopic Melanoma Recognition in Comparison to 58 Dermatologists. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Kanagasingam, S.; Hussaini, H.M.; Soo, I.; Baharin, S.; Ashar, A.; Patel, S. Accuracy of Single and Parallax Film and Digital Periapical Radiographs in Diagnosing Apical Periodontitis—A Cadaver Study. Int. Endod. J. 2017, 50, 427–436. [Google Scholar] [CrossRef]
- Leonardi Dutra, K.; Haas, L.; Porporatti, A.L.; Flores-Mir, C.; Nascimento Santos, J.; Mezzomo, L.A.; Corrêa, M.; De Luca Canto, G. Diagnostic Accuracy of Cone-Beam Computed Tomography and Conventional Radiography on Apical Periodontitis: A Systematic Review and Meta-Analysis. J. Endod. 2016, 42, 356–364. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. arXiv 2015, arXiv:1502.03167. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. arXiv 2016, arXiv:1606.04797. [Google Scholar]
- Kingma, D.P.; Adam, B.A. A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]

| Radiolucent Periapical Alterations | Characteristics [32] |
|---|---|
| Periapical inflammation/infection | Widened periodontal ligament |
| Periapical granuloma | Small lucent lesion with undefined borders (<200 mm3) |
| Periapical cysts | Round-shaped and well-defined lesions with sclerotic borders around the tooth root (>200 mm3) |
| Osteomyelitis | Lesion with irregular borders and irregular density, often spread over more than one root |
| Tumor | Lesion with irregular borders and irregular density, often spread over more than one root |
| Dentist | A | B | C | TPR | PPV |
|---|---|---|---|---|---|
| 1 | ≤4 | 23 | 8 | 0.36 | 0.74 |
| 2 | ≤4 | 22 | 11 | 0.35 | 0.79 |
| 3 | ≤4 | 43 | 2 | 0.59 | 0.79 |
| 4 | ≤4 | 41 | 8 | 0.52 | 0.51 |
| 5 | ≤4 | 54 | 10 | 0.30 | 0.90 |
| 6 | ≤4 | 69 | 19 | 0.45 | 0.77 |
| 7 | ≤4 | 30 | 2 | 0.64 | 0.45 |
| 8 | ≤4 | 59 | 10 | 0.41 | 0.69 |
| 9 | ≤4 | 27 | 8 | 0.47 | 0.72 |
| 10 | 4–8 | 32 | 2 | 0.68 | 0.58 |
| 11 | 4–8 | 51 | 4 | 0.69 | 0.65 |
| 12 | 4–8 | 119 | 0 | 0.59 | 0.74 |
| 13 | 4–8 | 22 | 0 | 0.52 | 0.42 |
| 14 | 4–8 | 25 | 8 | 0.26 | 0.93 |
| 15 | 4–8 | 27 | 8 | 0.76 | 0.60 |
| 16 | ≥8 | 58 | 8 | 0.46 | 0.58 |
| 17 | ≥8 | 17 | 7 | 0.48 | 0.83 |
| 18 | ≥8 | 34 | 10 | 0.54 | 0.73 |
| 19 | ≥8 | 45 | 14 | 0.44 | 0.76 |
| 20 | ≥8 | 43 | 12 | 0.67 | 0.61 |
| 21 | ≥8 | 25 | 9 | 0.28 | 0.86 |
| 22 | ≥8 | 63 | 5 | 0.61 | 0.69 |
| 23 | ≥8 | 21 | 1 | 0.41 | 0.85 |
| 24 | ≥8 | 58 | 9 | 0.66 | 0.59 |
| Mean | 7.6 | 42 | 6.9 | 0.51 | 0.69 |
| Median | 6.0 | 38 | 8.0 | 0.50 | 0.71 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Endres, M.G.; Hillen, F.; Salloumis, M.; Sedaghat, A.R.; Niehues, S.M.; Quatela, O.; Hanken, H.; Smeets, R.; Beck-Broichsitter, B.; Rendenbach, C.; et al. Development of a Deep Learning Algorithm for Periapical Disease Detection in Dental Radiographs. Diagnostics 2020, 10, 430. https://doi.org/10.3390/diagnostics10060430
Endres MG, Hillen F, Salloumis M, Sedaghat AR, Niehues SM, Quatela O, Hanken H, Smeets R, Beck-Broichsitter B, Rendenbach C, et al. Development of a Deep Learning Algorithm for Periapical Disease Detection in Dental Radiographs. Diagnostics. 2020; 10(6):430. https://doi.org/10.3390/diagnostics10060430
Chicago/Turabian StyleEndres, Michael G., Florian Hillen, Marios Salloumis, Ahmad R. Sedaghat, Stefan M. Niehues, Olivia Quatela, Henning Hanken, Ralf Smeets, Benedicta Beck-Broichsitter, Carsten Rendenbach, and et al. 2020. "Development of a Deep Learning Algorithm for Periapical Disease Detection in Dental Radiographs" Diagnostics 10, no. 6: 430. https://doi.org/10.3390/diagnostics10060430
APA StyleEndres, M. G., Hillen, F., Salloumis, M., Sedaghat, A. R., Niehues, S. M., Quatela, O., Hanken, H., Smeets, R., Beck-Broichsitter, B., Rendenbach, C., Lakhani, K., Heiland, M., & Gaudin, R. A. (2020). Development of a Deep Learning Algorithm for Periapical Disease Detection in Dental Radiographs. Diagnostics, 10(6), 430. https://doi.org/10.3390/diagnostics10060430







