Intratumoral Genomic Heterogeneity May Hinder Precision Medicine Strategies in Patients with Serous Ovarian Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Analysis
2.2. Immunohistochemical HER2 Assessment
2.3. Reporting of Secondary Germline Findings
3. Results
3.1. Patient and Pathological Findings
3.2. Sequencing Results
3.3. Secondary Germline Findings
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Köbel, M.; Kalloger, S.E.; Lee, S.; Duggan, M.A.; Kelemen, L.E.; Prentice, L.; Kalli, K.R.; Fridley, B.L.; Visscher, D.W.; Keeney, G.L.; et al. Biomarker-based ovarian carcinoma typing: A histologic investigation in the ovarian tumor tissue analysis consortium. Cancer Epidem. Biomar. 2013, 22, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Prat, J. Ovarian carcinomas. five distinct diseases with different origins, genetic alterations, and clinicopathological features. Virchows Arch. 2012, 460, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Shih, I.M.; Kurman, R.J. Ovarian tumorigenesis: A proposed model based on morphological and molecular genetic analysis. Am. J. Pathol. 2004, 164, 1511–1518. [Google Scholar] [CrossRef]
- Kurman, R.J.; Shih, I.M. Pathogenesis of ovarian cancer: Lessons from morphology and molecular biology and their clinical implications. Int. J. Gynecol. Pathol. 2008, 27, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Kurman, R.J.; Shih, I.M. The origin and pathogenesis of epithelial ovarian cancer: A proposed unifying theory. Am. J. Surg. Pathol. 2010, 34, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.; Berchuck, A.; Birrer, M. The cancer genome atlas research network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Kanchi, K.L.; Johnson, K.J.; Lu, C.; McLellan, M.D.; Leiserson, M.D.M.; Wendl, M.C.; Zhang, Q.; Koboldt, D.C.; Xie, M.; Kandoth, C.; et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nat. Commun. 2014, 5, 3156. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Tanishima, S.; Fujii, K.; Mori, R.; Okamura, Y.; Yanagita, E.; Matsuoka, R.; Amano, T.; Kinoshita, I.; Komatsu, Y.; et al. Genomic testing for pancreatic cancer in clinical practice as real-world evidence. Pancreatology 2018, 18, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Von Roemeling, C.A.; Marlow, L.A.; Radisky, D.C.; Rohl, A.; Larsen, H.E.; Wei, J.; Sasinowska, H.; Zhu, H.; Drake, R.; Sasinowski, M.; et al. Functional genomics identifies novel genes essential for clear cell renal cell carcinoma tumor cell proliferation and migration. Oncotarget 2014, 5, 5320–5334. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. NEJM 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Yates, L.R. Intratumoral heterogeneity and subclonal diversification of early breast cancer. Breast 2017, 34, S36–S42. [Google Scholar] [CrossRef] [PubMed]
- Takaya, H.; Nakai, H.; Sakai, K.; Nishio, K.; Murakami, K.; Mandai, M.; Matsumura, N. Intratumor heterogeneity and homologous recombination deficiency of high-grade serous ovarian cancer are associated with prognosis and molecular subtype and change in treatment course. Gynecol. Oncol. 2020, 156, 415–422. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, R.L.; Kaufmann, A.; Woodhouse, L.; McCormick, A.; Cross, P.A.; Edmondson, R.J.; Curtin, N.J. Advanced ovarian cancer displays functional intratumor heterogeneity that correlates to ex vivo drug sensitivity. Int. J. Gynecol. Cancer 2016, 26, 1004–1011. [Google Scholar] [CrossRef] [PubMed]
- Masoodi, T.; Siraj, S.; Siraj, A.K.; Azam, S.; Qadri, Z.; Parvathareddy, S.K.; Tulbah, A.; Al-Dayel, F.; AlHusaini, H.; AlOmar, O.; et al. Genetic heterogeneity and evolutionary history of high-grade ovarian carcinoma and matched distant metastases. Br. J. Cancer 2020. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Messer, K.; Schwab, R.; Harismendy, O.; Pu, M.; Crain, B.; Yost, S.; Frazer, K.A.; Rana, B.; Hasteh, F.; et al. Mutational profiling can establish clonal or independent origin in synchronous bilateral breast and other tumors. PLoS ONE 2015, 10, e0142487. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Cerdá, J.L.; Hernández, M.; Sempere, A.; O’Connor, J.E.; Kimler, B.F.; Jiménez-Cruz, F. Intratumoral heterogeneity of DNA content in renal cell carcinoma and its prognostic significance. Cancer 1999, 86, 664–671. [Google Scholar] [CrossRef]
- Jiang, W.; Dulaimi, E.; Devarajan, K.; Parsons, T.; Wang, Q.; Liao, L.; Cho, E.A.; O’Neill, R.; Solomides, C.; Peiper, S.C.; et al. Immunohistochemistry successfully uncovers intratumoral heterogeneity and widespread co-losses of chromatin regulators in clear cell renal cell carcinoma. PLoS ONE 2016, 11, e0164554. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Horswell, S.; Larkin, J.; Rowan, A.J.; Salm, M.P.; Varela, I.; Fisher, R.; McGranahan, N.; Matthews, N.; Santos, C.R.; et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014, 46, 225–533. [Google Scholar] [CrossRef] [PubMed]
- Martinez, P.; Birkbak, N.J.; Gerlinger, M.; McGranahan, N.; Burrell, R.A.; Rowan, A.J.; Joshi, T.; Fisher, R.; Larkin, J.; Szallasi, Z.; et al. Parallel evolution of tumor subclones mimics diversity between tumors. J. Pathol. 2013, 230, 356–364. [Google Scholar] [CrossRef] [PubMed]
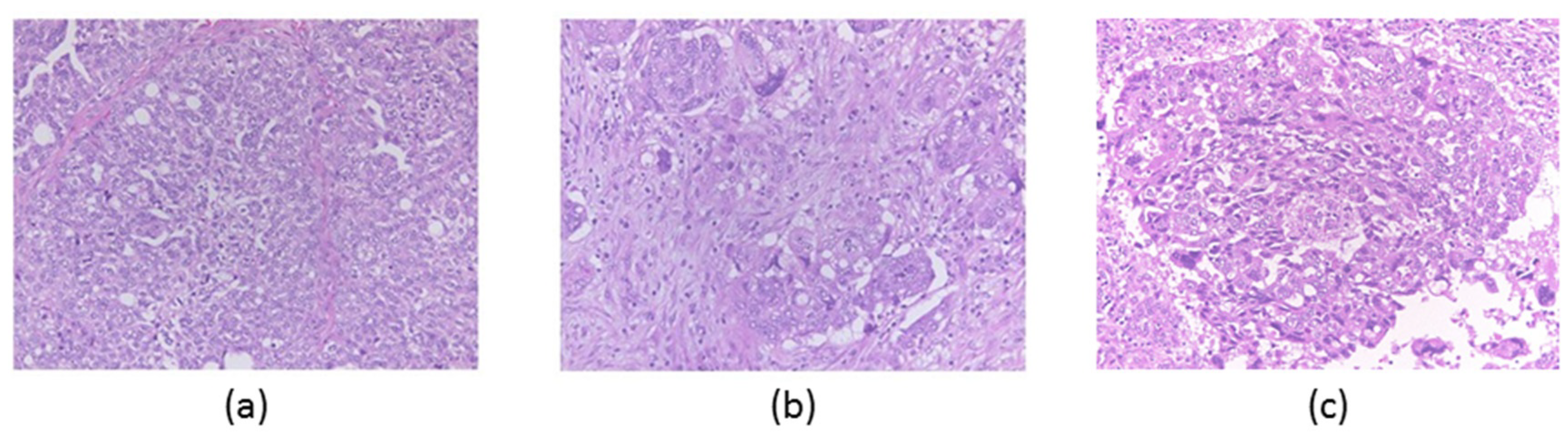

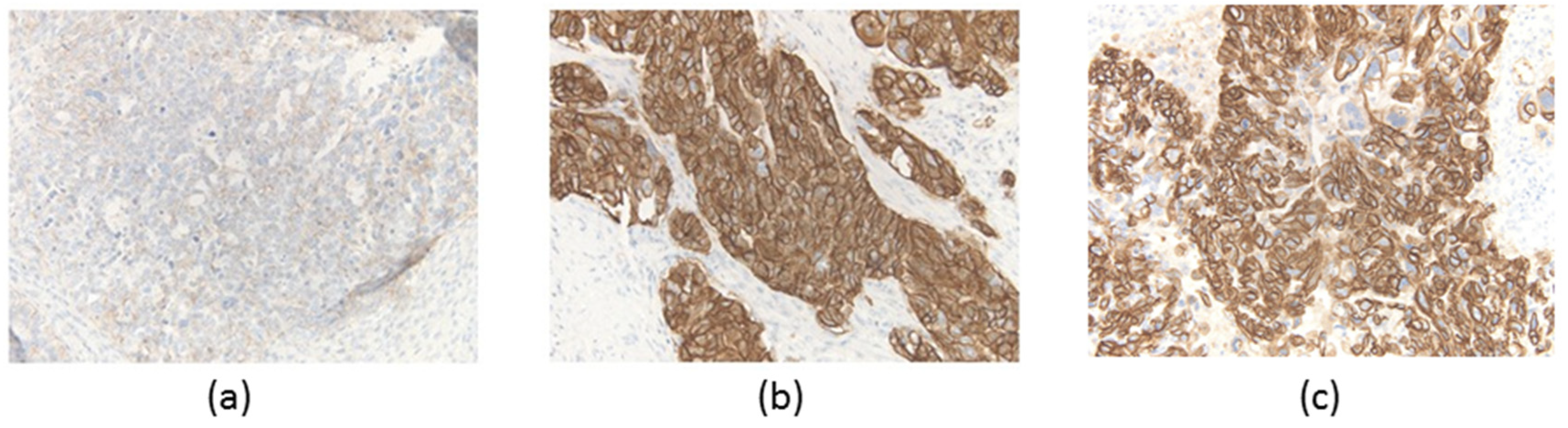
| Area | Actionable Gene Alterations | Tumor Mutation Burden (SNVs/Mbp) |
|---|---|---|
| a | TP53 V272M, ERBB2 amp (CN = 6), CDH1 loss, KDM6A loss, WT1 loss | 2.7 |
| b | TP53 V272M, ERBB2 amp (CN = 32), CDH1 loss, KDM6A loss, WT1 loss | 2.7 |
| c | TP53 V272M, ERBB2 amp (CN = 26), CDH1 loss, KDM6A loss, WT1 loss, KRAS amp (CN = 4) | 2.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakamura, K.; Aimono, E.; Tanishima, S.; Imai, M.; Nagatsuma, A.K.; Hayashi, H.; Yoshimura, Y.; Nakayama, K.; Kyo, S.; Nishihara, H. Intratumoral Genomic Heterogeneity May Hinder Precision Medicine Strategies in Patients with Serous Ovarian Carcinoma. Diagnostics 2020, 10, 200. https://doi.org/10.3390/diagnostics10040200
Nakamura K, Aimono E, Tanishima S, Imai M, Nagatsuma AK, Hayashi H, Yoshimura Y, Nakayama K, Kyo S, Nishihara H. Intratumoral Genomic Heterogeneity May Hinder Precision Medicine Strategies in Patients with Serous Ovarian Carcinoma. Diagnostics. 2020; 10(4):200. https://doi.org/10.3390/diagnostics10040200
Chicago/Turabian StyleNakamura, Kohei, Eriko Aimono, Shigeki Tanishima, Mitsuho Imai, Akiko Kawano Nagatsuma, Hideyuki Hayashi, Yuki Yoshimura, Kentaro Nakayama, Satoru Kyo, and Hiroshi Nishihara. 2020. "Intratumoral Genomic Heterogeneity May Hinder Precision Medicine Strategies in Patients with Serous Ovarian Carcinoma" Diagnostics 10, no. 4: 200. https://doi.org/10.3390/diagnostics10040200
APA StyleNakamura, K., Aimono, E., Tanishima, S., Imai, M., Nagatsuma, A. K., Hayashi, H., Yoshimura, Y., Nakayama, K., Kyo, S., & Nishihara, H. (2020). Intratumoral Genomic Heterogeneity May Hinder Precision Medicine Strategies in Patients with Serous Ovarian Carcinoma. Diagnostics, 10(4), 200. https://doi.org/10.3390/diagnostics10040200





