Assessment of DCE Utility for PCa Diagnosis Using PI-RADS v2.1: Effects on Diagnostic Accuracy and Reproducibility
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Population
2.2. Biopsy Protocol
2.3. Imaging Protocol
2.4. MR Image Analysis and Interpretation Using PI-RADS v2.1
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics and Clinical Findings
3.2. Inter-Reader and Inter-Method Agreement
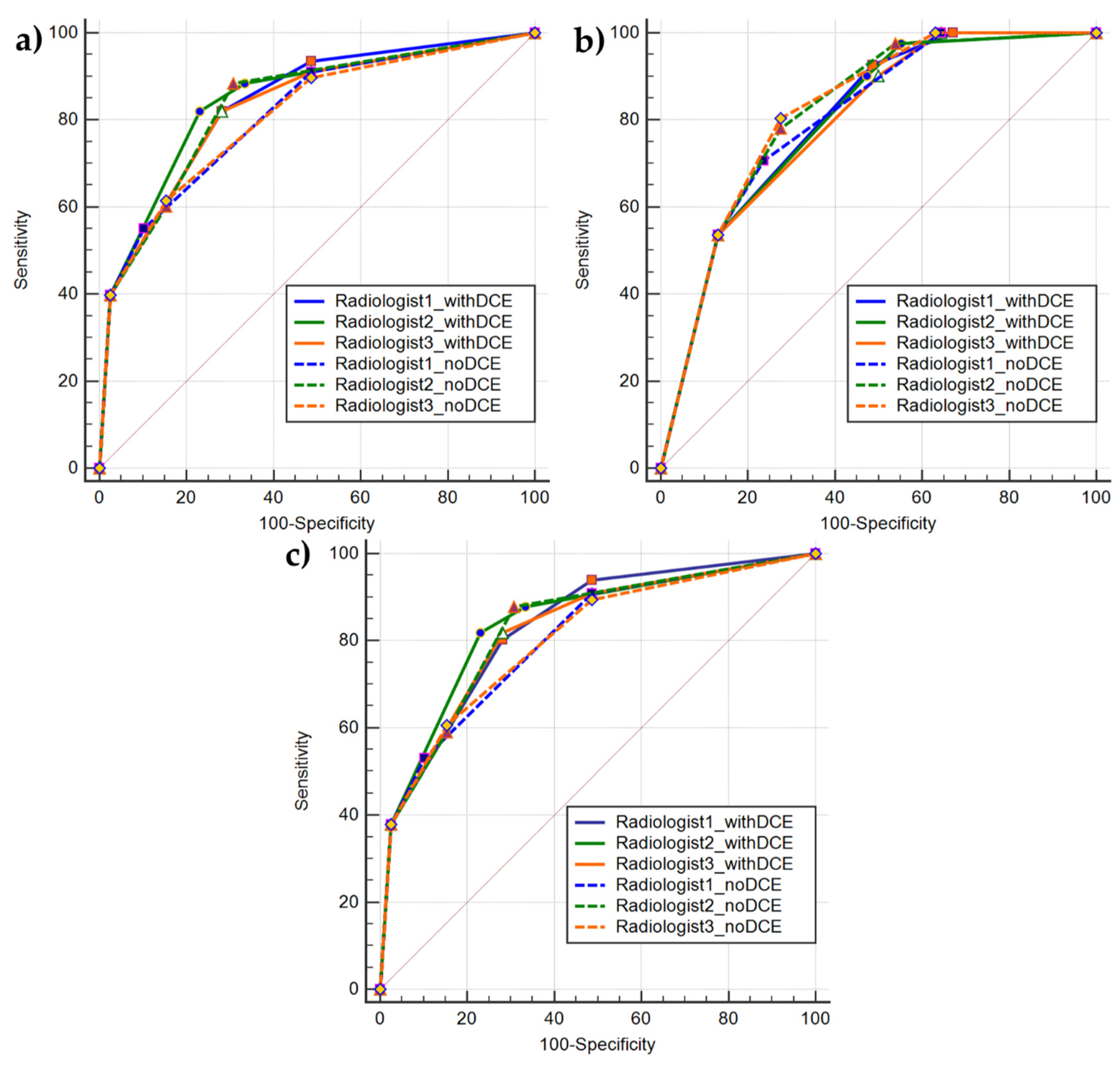
3.3. Diagnostic Accuracy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Johnson, L.M.; Turkbey, B.; Figg, W.D.; Choyke, P.L. Multiparametric MRI in prostate cancer management. Nat. Rev. Clin. Oncol. 2014, 11, 346–353. [Google Scholar] [CrossRef]
- Brancato, V.; Cavaliere, C.; Salvatore, M.; Monti, S. Non-Gaussian models of diffusion weighted imaging for detection and characterization of prostate cancer: A systematic review and meta-analysis. Sci. Rep. 2019, 9, 16837. [Google Scholar] [CrossRef] [PubMed]
- Hegde, J.V.; Mulkern, R.V.; Panych, L.P.; Fennessy, F.M.; Fedorov, A.; Maier, S.E.; Tempany, C.M. Multiparametric MRI of prostate cancer: An update on state-of-the-art techniques and their performance in detecting and localizing prostate cancer. J. Magn. Reson. Imaging 2013, 37, 1035–1054. [Google Scholar] [CrossRef] [PubMed]
- De Rooij, M.; Hamoen, E.H.; Fütterer, J.J.; Barentsz, J.O.; Rovers, M. Accuracy of Multiparametric MRI for Prostate Cancer Detection: A Meta-Analysis. Am. J. Roentgenol. 2014, 202, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Futterer, J.J.; Briganti, A.; De Visschere, P.; Emberton, M.; Giannarini, G.; Kirkham, A.; Taneja, S.S.; Thoeny, H.; Villeirs, G.; Villers, A. Can Clinically Significant Prostate Cancer Be Detected with Multiparametric Magnetic Resonance Imaging? A Systematic Review of the Literature. Eur. Urol. 2015, 68, 1045–1053. [Google Scholar] [CrossRef]
- Barentsz, J.O.; Richenberg, J.; Clements, R.; Choyke, P.; Verma, S.; Villeirs, G.; Rouvière, O.; Løgager, V.B.; Fütterer, J.J.; Radiology, E.S.O.U. ESUR prostate MR guidelines 2012. Eur. Radiol. 2012, 22, 746–757. [Google Scholar] [CrossRef]
- Barentsz, J.O.; Choyke, P.L.; Cornud, F.; Haider, M.A.; Macura, K.J.; Margolis, D.; Shtern, F.; Padhani, A.R.; Tempany, C.M.; Thoeny, H.C.; et al. Reply to Erik Rud and Eduard Baco’s Letter to the Editor re: PI-RADS Prostate Imaging—Reporting and Data System: 2015, Version 2. Eur. Urol. 2016, 70, e137–e138. [Google Scholar] [CrossRef]
- Feng, Z.-Y.; Wang, L.; Min, X.; Wang, S.-G.; Wang, G.-P.; Cai, J. Prostate Cancer Detection with Multiparametric Magnetic Resonance Imaging: Prostate Imaging Reporting and Data System Version 1 versus Version 2. Chin. Med. J. 2016, 129, 2451–2459. [Google Scholar] [CrossRef]
- Tewes, S.; Mokov, N.; Hartung, D.; Schick, V.; Peters, I.; Schedl, P.; Pertschy, S.; Wacker, F.; Voshage, G.; Hueper, K. Standardized Reporting of Prostate MRI: Comparison of the Prostate Imaging Reporting and Data System (PI-RADS) Version 1 and Version 2. PLoS ONE 2016, 11, e0162879. [Google Scholar] [CrossRef]
- Kasel-Seibert, M.; Lehmann, T.; Aschenbach, R.; Guettler, F.V.; Abubrig, M.; Grimm, M.-O.; Teichgraeber, U.; Franiel, T. Assessment of PI-RADS v2 for the Detection of Prostate Cancer. Eur. J. Radiol. 2016, 85, 726–731. [Google Scholar] [CrossRef]
- Becker, A.S.; Cornelius, A.; Reiner, C.S.; Stocker, D.; Ulbrich, E.J.; Barth, B.; Mortezavi, A.; Eberli, D.; Donati, O.F. Direct comparison of PI-RADS version 2 and version 1 regarding interreader agreement and diagnostic accuracy for the detection of clinically significant prostate cancer. Eur. J. Radiol. 2017, 94, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xin, C.; Zhang, L.; Dong, A.; Xu, H.; Wu, Y. Comparison of diagnostic performance between two prostate imaging reporting and data system versions: A systematic review. Eur. J. Radiol. 2019, 114, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Stanzione, A.; Imbriaco, M.; Cocozza, S.; Fusco, F.; Rusconi, G.; Nappi, C.; Mirone, V.; Mangiapia, F.; Brunetti, A.; Ragozzino, A.; et al. Biparametric 3T Magentic Resonance Imaging for prostatic cancer detection in a biopsy-naïve patient population: A further improvement of PI-RADS v2? Eur. J. Radiol. 2016, 85, 2269–2274. [Google Scholar] [CrossRef] [PubMed]
- Taghipour, M.; Ziaei, A.; Alessandrino, F.; Hassanzadeh, E.; Harisinghani, M.; Vangel, M.; Tempany, C.M.; Fennessy, F.M. Investigating the role of DCE-MRI, over T2 and DWI, in accurate PI-RADS v2 assessment of clinically significant peripheral zone prostate lesions as defined at radical prostatectomy. Abdom. Radiol. 2018, 44, 1520–1527. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.H.; Kim, C.K.; Lee, Y.J.; Jung, S.E. Prebiopsy Biparametric MRI for Clinically Significant Prostate Cancer Detection With PI-RADS Version 2: A Multicenter Study. Am. J. Roentgenol. 2019, 212, 839–846. [Google Scholar] [CrossRef]
- Junker, D.; Steinkohl, F.; Fritz, V.; Bektic, J.; Tokas, T.; Aigner, F.; Herrmann, T.R.W.; Rieger, M.; Nagele, U. Comparison of multiparametric and biparametric MRI of the prostate: Are gadolinium-based contrast agents needed for routine examinations? World J. Urol. 2018, 37, 691–699. [Google Scholar] [CrossRef]
- Gatti, M.; Faletti, R.; Calleris, G.; Giglio, J.; Berzovini, C.; Gentile, F.; Marra, G.; Misischi, F.; Molinaro, L.; Bergamasco, L.; et al. Prostate cancer detection with biparametric magnetic resonance imaging (bpMRI) by readers with different experience: Performance and comparison with multiparametric (mpMRI). Abdom. Radiol. 2019, 44, 1883–1893. [Google Scholar] [CrossRef]
- Di Campli, E.; Pizzi, A.D.; Seccia, B.; Cianci, R.; D’Annibale, M.; Colasante, A.; Cinalli, S.; Castellan, P.; Navarra, R.; Iantorno, R.; et al. Diagnostic accuracy of biparametric vs multiparametric MRI in clinically significant prostate cancer: Comparison between readers with different experience. Eur. J. Radiol. 2018, 101, 17–23. [Google Scholar] [CrossRef]
- Alabousi, M.; Salameh, J.-P.; Gusenbauer, K.; Samoilov, L.; Jafri, A.; Yu, H.; Alabousi, A. Biparametric vs multiparametric prostate magnetic resonance imaging for the detection of prostate cancer in treatment-naïve patients: A diagnostic test accuracy systematic review and meta-analysis. BJU Int. 2019, 124, 209–220. [Google Scholar] [CrossRef]
- Rosenkrantz, A.B.; Babb, J.S.; Taneja, S.S.; Ream, J.M. Proposed Adjustments to PI-RADS Version 2 Decision Rules: Impact on Prostate Cancer Detection. Radiology 2017, 283, 119–129. [Google Scholar] [CrossRef]
- Greer, M.D.; Shih, J.H.; Lay, N.; Barrett, T.; Bittencourt, L.K.; Borofsky, S.; Kabakus, I.M.; Law, Y.M.; Marko, J.; Shebel, H.; et al. Validation of the Dominant Sequence Paradigm and Role of Dynamic Contrast-enhanced Imaging in PI-RADS Version 2. Radiology 2017, 285, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Turkbey, B.; Rosenkrantz, A.B.; Haider, M.A.; Padhani, A.R.; Villeirs, G.; Macura, K.J.; Tempany, C.M.; Choyke, P.L.; Cornud, F.; Margolis, D.J.; et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur. Urol. 2019, 76, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, T.; Schimmöller, L. Perspective: A critical assessment of PI-RADS 2.1. Abdom. Radiol. 2020, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tamada, T.; Kido, A.; Takeuchi, M.; Yamamoto, A.; Miyaji, Y.; Kanomata, N.; Sone, T. Comparison of PI-RADS version 2 and PI-RADS version 2.1 for the detection of transition zone prostate cancer. Eur. J. Radiol. 2019, 121, 108704. [Google Scholar] [CrossRef]
- Byun, J.; Park, K.J.; Kim, M.-H.; Kim, J.K. Direct Comparison of PI-RADS Version 2 and 2.1 in Transition Zone Lesions for Detection of Prostate Cancer: Preliminary Experience. J. Magn. Reson. Imaging 2020. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data Data for Categorical of Observer Agreement. Measurement 2013, 33, 159–174. [Google Scholar]
- Rosenkrantz, A.B.; Ginocchio, L.; Cornfeld, D.; Froemming, A.T.; Gupta, R.T.; Turkbey, B.; Westphalen, A.C.; Babb, J.S.; Margolis, D.J. Interobserver Reproducibility of the PI-RADS Version 2 Lexicon: A Multicenter Study of Six Experienced Prostate Radiologists. Radiology 2016, 280, 793–804. [Google Scholar] [CrossRef]
- Sim, J.; Wright, C.C. The Kappa Statistic in Reliability Studies: Use, Interpretation, and Sample Size Requirements. Phys. Ther. 2005, 85, 257–268. [Google Scholar] [CrossRef]
- De Visschere, P.; Lumen, N.; Ost, P.; Decaestecker, K.; Pattyn, E.; Villeirs, G. Dynamic contrast-enhanced imaging has limited added value over T2-weighted imaging and diffusion-weighted imaging when using PI-RADSv2 for diagnosis of clinically significant prostate cancer in patients with elevated PSA. Clin. Radiol. 2017, 72, 23–32. [Google Scholar] [CrossRef]
- Kang, Z.; Min, X.; Weinreb, J.; Li, Q.; Feng, Z.; Wang, L. Abbreviated Biparametric Versus Standard Multiparametric MRI for Diagnosis of Prostate Cancer: A Systematic Review and Meta-Analysis. Am. J. Roentgenol. 2018, 212, 357–365. [Google Scholar] [CrossRef]
- Delgado-Rodríguez, M.; Llorca, J. Bias. J. Epidemiol. Community Health 2004, 58, 635–641. [Google Scholar]
- Steinkohl, F.; Pichler, R.; Junker, D. Short review of biparametric prostate MRI. Memo Mag. Eur. Med Oncol. 2018, 11, 309–312. [Google Scholar] [CrossRef] [PubMed]
| Sequence | Slices | TR (msec) | TE (msec) | ST (mm) | Avg. | BW | Matrix | FOV (mm) | FA |
|---|---|---|---|---|---|---|---|---|---|
| T2W Sagittal | 9 | 4740 | 102 | 3 | 2 | 200 | 320 × 310 | 200 × 200 | 137 |
| T2 Axial | 23 | 5610 | 102 | 3 | 2 | 200 | 320 × 272 | 200 × 200 | 123 |
| T2 Coronal | 16 | 4000 | 102 | 3 | 1 | 200 | 320 × 310 | 211 × 211 | 138 |
| T2 Lymph nodes | 36 | 7620 | 98 | 6.5 | 1 | 130 | 512 × 247 | 341 × 390 | 135 |
| DWI a | 19 | 3505 | 75 | 3 | 6 | 1698 | 128 × 128 | 250 × 250 | 90 |
| T1 vibe tra FA | 20 | 5.5 | 2.34 | 3.5 | 8 | 300 | 320 × 112 | 208 × 417 | 2 |
| T1 vibe tra FA | 20 | 5.5 | 2.34 | 3.5 | 8 | 300 | 320 × 112 | 208 × 417 | 15 |
| T1 vibe dyn b | 22 | 5.5 | 2.34 | 3.5 | 1 | 300 | 320 × 112 | 208 × 417 | 10 |
| Variable | Value (Patient Based) | Value (Lesion Based) |
|---|---|---|
| Clinical Variables | ||
| No. of patients/lesions [n] | 111 | 117 |
| Median age [y (range)] | 69 (50–81) | |
| Mean PSA density [ng/mL2] | 0.26 | |
| Prostate volume [cm3] | 57.5 | |
| Prostatic zone | ||
| PZ | 105 (89.7) | |
| TZ | 5 (4.3) | |
| CZ | 7 (6) | |
| Biopsy Results | ||
| PCa not detected [n (%)] | 39 (35.1) | 39 (33.3) |
| PCa detected [n (%)] | 72 (64.9) | 78 (66.7) |
| CS-PCa detected [n (%)] | 38 (34.2) | 41 (35) |
| Gleason scores for CS-PCa lesions [n (%)] | ||
| 3 + 4 | 6 (5.1) | |
| 4 + 3 | 15 (12.8) | |
| 4 + 4 | 15 (12.8) | |
| 4 + 5 | 3 (2.5) | |
| 5 + 3 | 1 (0.8) | |
| 5 + 4 | 1 (0.8) |
| PI-RADS v2.1 Type | Lesion Type | Agreement Rate, % [Number of Lesions in Agreement/Total No. of Lesions] | κ Value [95% CI] | ||||
|---|---|---|---|---|---|---|---|
| R1-R2 | R1-R3 | R2-R3 | R1-R2 | R1-R3 | R2-R3 | ||
| with DCE | All lesions (n = 117) | 85.5 (100/117) | 91.4 (107/117) | 90 (106/117) | 0.87 [0.82–0.93] | 0.92 [0.89–0.97] | 0.92 [0.87–0.97] |
| no DCE | All lesions (n = 117) | 82 (96/117) | 86.3 (101/117) | 92.3 (108/117) | 0.86 [0.8–0.92] | 0.89 [0.84–0.94] | 0.94 [0.9–0.98] |
| with DCE | PCa lesions (n = 78) | 87.1 (68/78) | 92.3 (72/78) | 94.9 (74/78) | 0.86 [0.79–0.94] | 0.92 [0.85–0.98] | 0.95 [0.9–1.00] |
| no DCE | PCa lesions (n = 78) | 87.2 (68/78) | 87.2 (68/78) | 97.4 (76/78) | 0.89 [0.82–0.95] | 0.88 [0.82–0.95] | 0.98 [0.95–1.00] |
| with DCE | PZ PCa lesions (n = 66) | 86.4 (57/66) | 92.4 (61/66) | 93.9 (62/66) | 0.86 [0.77–0.94] | 0.92 [0.85–0.99] | 0.94 [0.89–1.00] |
| no DCE | PZ PCa lesions (n = 66) | 87.9 (58/66) | 87.9 (58/66) | 97.4 (64/66) | 0.9 [0.83–0.97] | 0.89 [0.82–0.97] | 0.97 [0.94–1.00] |
| with DCE | CS-PCa lesions (n = 41) | 95.1 (39/41) | 97.6 (40/41) | 97.6 (40/41) | 0.93 [0.84–1.00] | 0.96 [0.89–1.00] | 0.96 [0.9–1.00] |
| no DCE | CS-PCa lesions (n = 41) | 85.4 (35/41) | 85.4 (35/41) | 92.1 (39/41) | 0.84 [0.73–0.95] | 0.83 [0.71–0.95] | 0.94 [0.87–1.00] |
| with DCE | non-PCa lesions (n = 39) | 82 (32/39) | 89.7 (35/39) | 82 (32/39) | 0.78 [0.62–0.94] | 0.89 [0.79–0.99] | 0.78 [0.62–0.94] |
| no DCE | non-PCa lesions (n = 39) | 71.8 (28/39) | 84.6 (33/39) | 82 (32/39) | 0.63 [0.44–0.83] | 0.8 [0.65–0.95] | 0.78 [0.61–0.94] |
| Lesion Type | Agreement Rate, % [Number of Lesions in Agreement/Total No. of Lesions] | κ Value [95% CI] | ||||
|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R1 | R2 | R3 | |
| All lesions (n = 117) | 74.3 (87/117) | 82 (96/117) | 81.2 (95/117) | 0.8 [0.73–0.87] | 0.86 [0.8–0.92] | 0.85 [0.79–0.91] |
| PCa lesions (n = 78) | 70.5 (55/78) | 78.2 (61/78) | 78.2 (61/78) | 0.73 [0.63–0.82] | 0.8 [0.72–0.89] | 0.8 [0.71–0.88] |
| PZ PCa lesions (n = 66) | 69.7 (46/66) | 77.3 (51/66) | 77.3 (51/66) | 0.72 [0.63–0.82] | 0.8 [0.7–0.89] | 0.79 [0.69–0.88] |
| CS-PCa lesions (n = 41) | 78 (32/41) | 87.8 (36/41) | 90.2 (37/41) | 0.73 [0.6–0.86] | 0.85 [0.73–0.97] | 0.87 [0.76–0.98] |
| non-PCa lesions (n = 39) | 82 (32/39) | 89.7 (35/39) | 87.2 (34/39) | 0.79 [0.67–0.92] | 0.87 [0.75–0.98] | 0.86 [0.74–0.97] |
| PI-RADS v2.1 Type | Pos Class | AUC [95% CI] | Youden Selected th | Sen at th | Specificity at th | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R1 | R2 | R3 | R1 | R2 | R3 | R1 | R2 | R3 | ||
| with DCE | PCa | 0.84 [0.86–0.9] | 0.85 [0.77–0.91] | 0.82 [0.75–0.89] | >3 | >3 | >3 | 0.80 | 0.82 | 0.82 | 0.72 | 0.77 | 0.72 |
| with DCE | PCa (PZ) | 0.83 [0.75–0.9] | 0.84 [0.76–0.91] | 0.82 [0.74–0.89] | >3 | >3 | >3 | 0.82 | 0.82 | 0.82 | 0.72 | 0.77 | 0.72 |
| with DCE | CS-PCa | 0.8 [0.72–0.87] | 0.8 [0.71–0.87] | 0.8 [0.71–0.86] | >3 | >3 | >4 | 0.92 | 0.9 | 0.54 | 0.51 | 0.53 | 0.87 |
| no DCE | PCa | 0.81 [0.73–0.88] | 0.84 [0.76–0.9] | 0.8 [0.73–0.88] | >3 | >2 | >3 | 0.55 | 0.88 | 0.61 | 0.9 | 0.69 | 0.85 |
| no DCE | PCa (PZ) | 0.81 [0.72–0.88] | 0.83 [0.75–0.9] | 0.8 [0.72–0.88] | >3 | >2 | >3 | 0.53 | 0.88 | 0.61 | 0.9 | 0.69 | 0.85 |
| no DCE | CS-PCa | 0.8 [0.72–0.87] | 0.82 [0.73–0.88] | 0.82 [0.74–0.89] | >3 | >3 | >3 | 0.71 | 0.78 | 0.8 | 0.76 | 0.72 | 0.72 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brancato, V.; Di Costanzo, G.; Basso, L.; Tramontano, L.; Puglia, M.; Ragozzino, A.; Cavaliere, C. Assessment of DCE Utility for PCa Diagnosis Using PI-RADS v2.1: Effects on Diagnostic Accuracy and Reproducibility. Diagnostics 2020, 10, 164. https://doi.org/10.3390/diagnostics10030164
Brancato V, Di Costanzo G, Basso L, Tramontano L, Puglia M, Ragozzino A, Cavaliere C. Assessment of DCE Utility for PCa Diagnosis Using PI-RADS v2.1: Effects on Diagnostic Accuracy and Reproducibility. Diagnostics. 2020; 10(3):164. https://doi.org/10.3390/diagnostics10030164
Chicago/Turabian StyleBrancato, Valentina, Giuseppe Di Costanzo, Luca Basso, Liberatore Tramontano, Marta Puglia, Alfonso Ragozzino, and Carlo Cavaliere. 2020. "Assessment of DCE Utility for PCa Diagnosis Using PI-RADS v2.1: Effects on Diagnostic Accuracy and Reproducibility" Diagnostics 10, no. 3: 164. https://doi.org/10.3390/diagnostics10030164
APA StyleBrancato, V., Di Costanzo, G., Basso, L., Tramontano, L., Puglia, M., Ragozzino, A., & Cavaliere, C. (2020). Assessment of DCE Utility for PCa Diagnosis Using PI-RADS v2.1: Effects on Diagnostic Accuracy and Reproducibility. Diagnostics, 10(3), 164. https://doi.org/10.3390/diagnostics10030164






