Evaluation of Parameters for Estimating the Postmortem Interval of Skeletal Remains Using Bovine Femurs: A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Bone Sample Preparation
2.2. Environmental Conditions
2.3. DNA Extraction and Quantification
2.4. Elemental Analysis by ICP-OES
2.5. Bone Density Measurement by Micro X-Ray CT
2.6. Statistical Analysis
3. Results
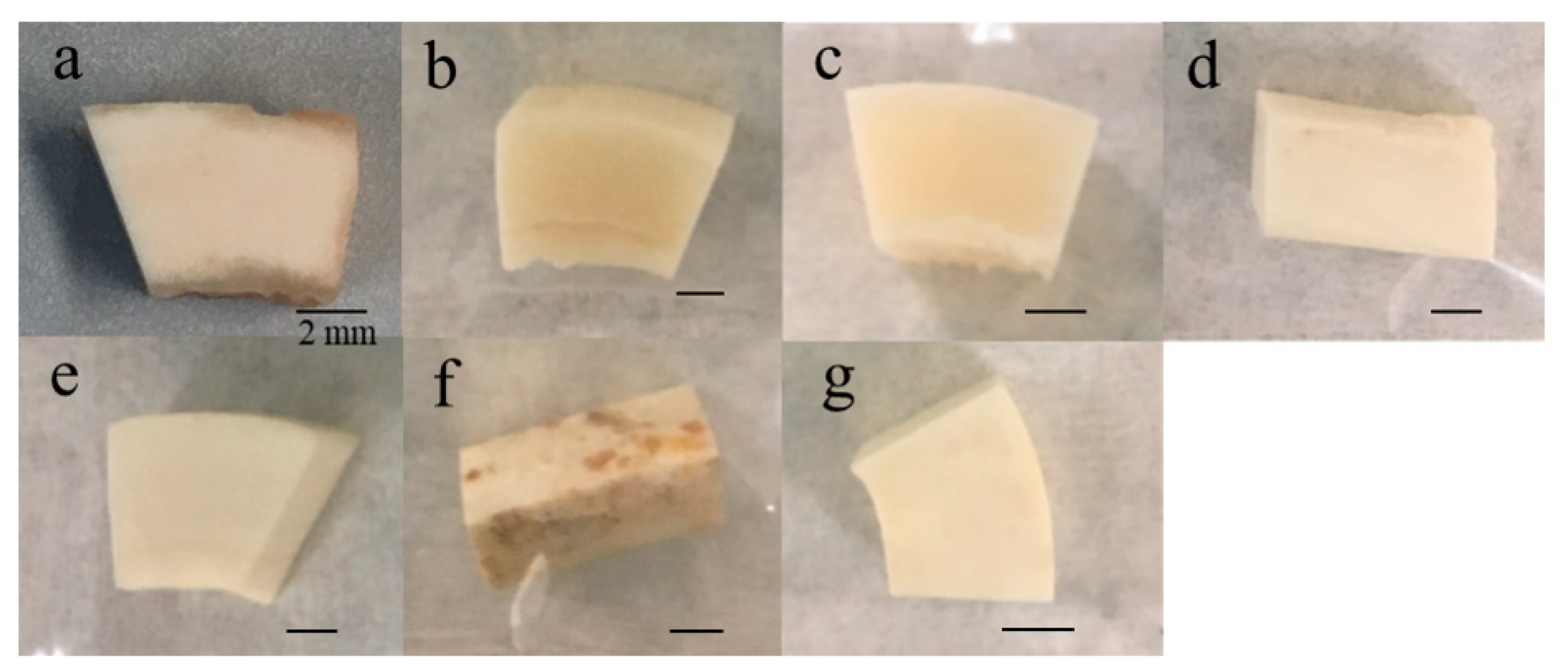
3.1. Visual Observation of Bone Samples in Five Environmental Conditions
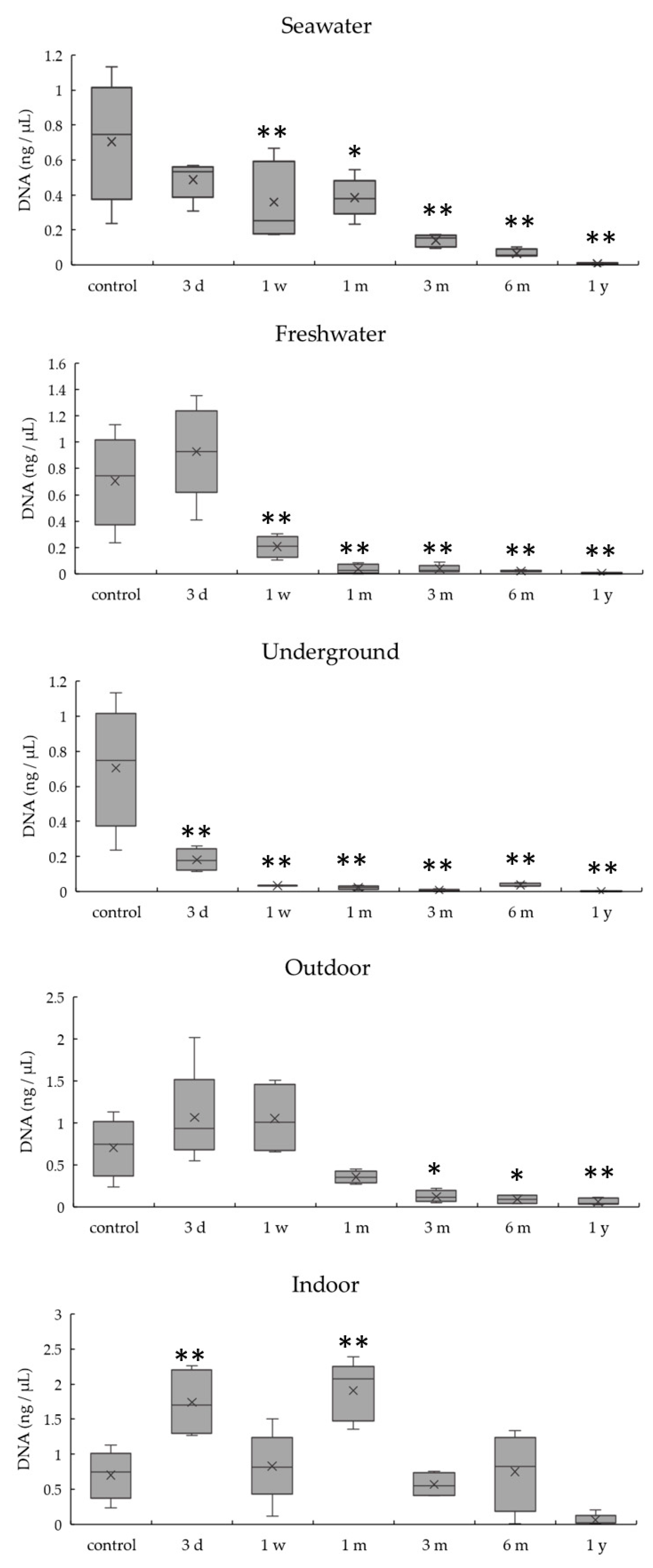
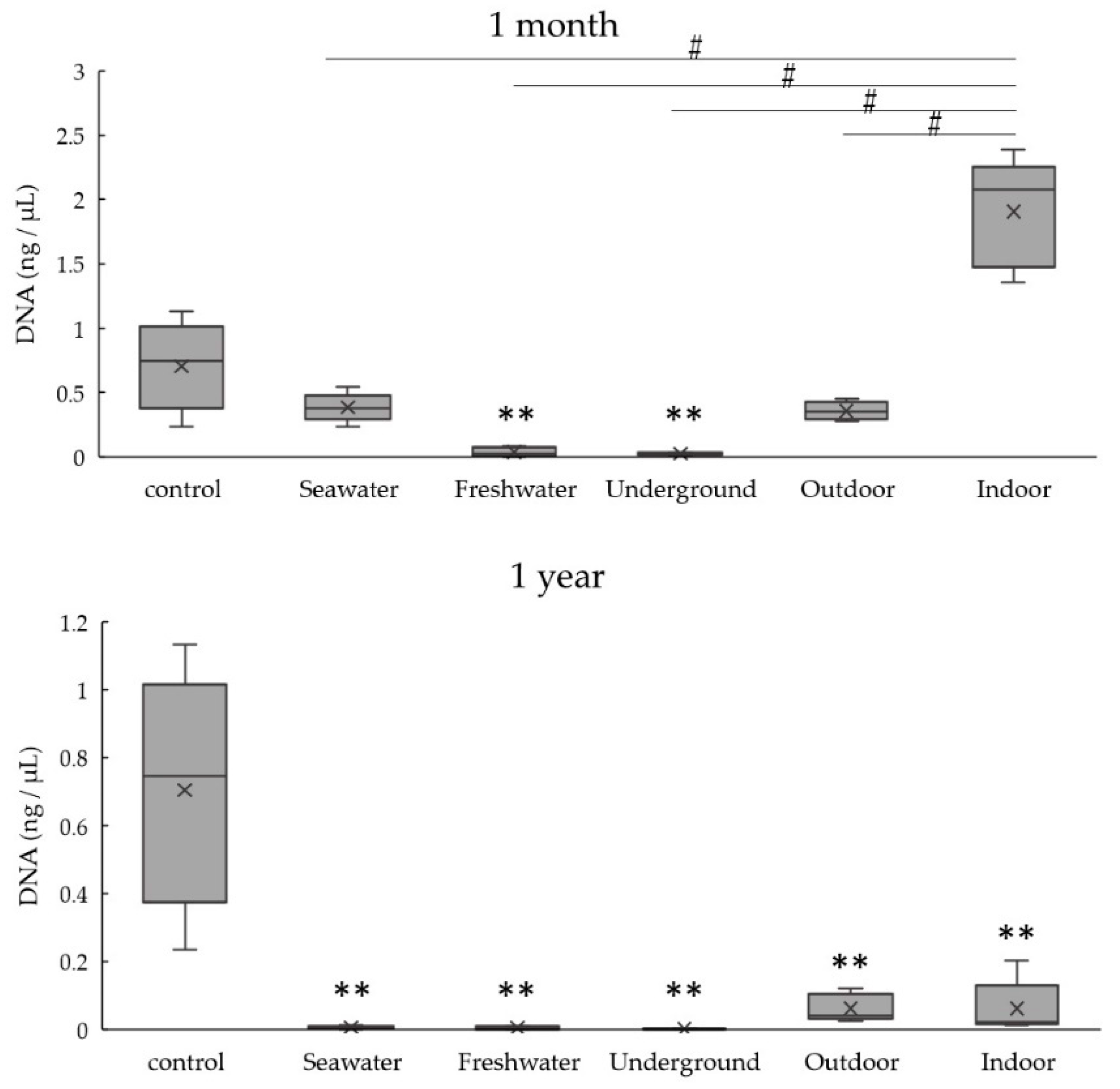
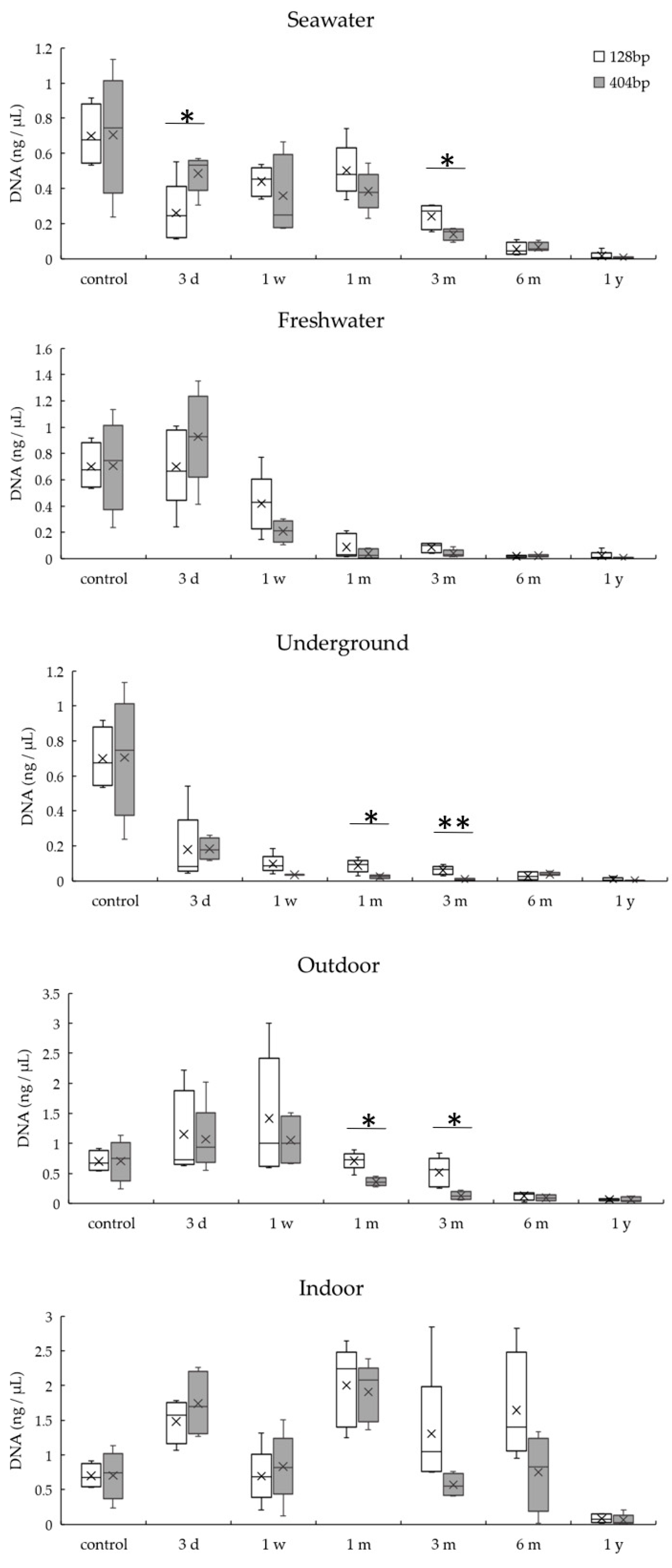
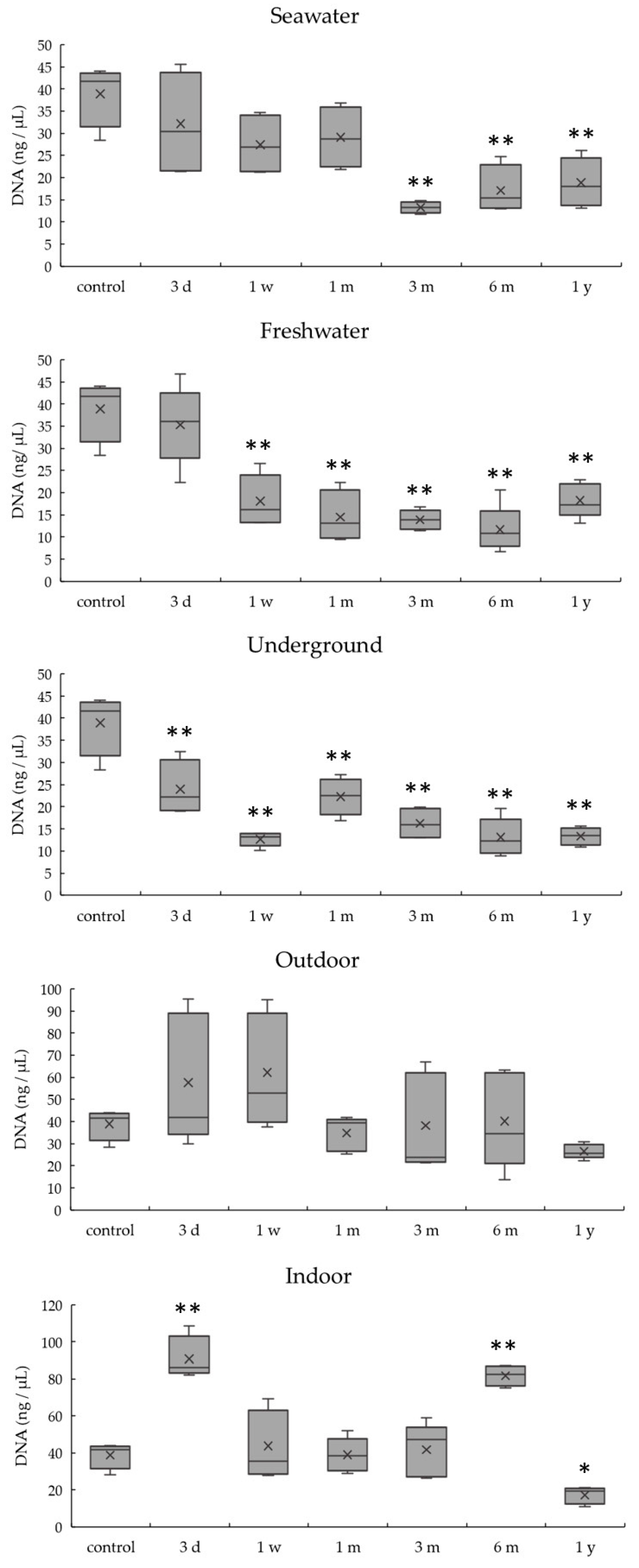
3.2. DNA Quantitation of Bones Exposed to Various Environmental Conditions
3.3. Elemental Analysis by ICP-OES
3.4. Bone Density
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Creagh, D.; Cameron, A. Estimating the Post-Mortem Interval of skeletonized remains: The use of Infrared spectroscopy and Raman spectro-microscopy. Radiat. Phys. Chem. 2017, 137, 225–229. [Google Scholar] [CrossRef]
- Woess, C.; Unterberger, S.H.; Roider, C.; Ritsch-Marte, M.; Pemberger, N.; Cemper-kiesslich, J.; Hatzer-Grubwieser, P.; Person, W.; Pallua, J.D. Assessing various Infrared (IR) microscopic imaging techniques for post-mortem interval evaluation of human skeletal remains. PLoS ONE 2017, 12, e0174552. [Google Scholar] [CrossRef] [PubMed]
- Walden, S.J.; Mulville, J.; Rowlands, J.P.; Evans, S.L. An analysis of systematic elemental changes in decomposing bone. J. Forensic Sci. 2018, 63, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Le Garff, E.; Mesli, V.; Marchand, E.; Behal, H.; Demondion, X.; Becart, A.; Hedouin, V. Is bone analysis with μ CT useful for short postmortem interval estimation? Int. J. Legal Med. 2018, 132, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Akbulut, N.; Çetin, S.; Bilecenoğlu, B.; Altan, A.; Akbulut, S.; Ocak, M.; Orhan, M.K. The micro-CT evaluation of enamel-cement thickness, abrasion, and mineral density in teeth in the postmortem interval (PMI): New parameters for the determination of PMI. Int. J. Legal Med. 2020, 134, 645–653. [Google Scholar] [CrossRef]
- Amadasi, A.; Cappella, A.; Cattaneo, C.; Cofrancesco, P.; Cucca, L.; Merli, D.; Milanese, C.; Pinto, A.; Profumo, A.; Scarpulla, V.; et al. Determination of the post mortem interval in skeletal remains by the comparative use of different physico-chemical methods: Are they reliable as an alternative to 14 C? HOMO 2017, 68, 213–221. [Google Scholar] [CrossRef]
- Cappella, A.; Gibelli, D.; Muccino, E.; Scarpulla, V.; Cerutti, E.; Caruso, V.; Sguazza, E.; Mazzarelli, D.; Cattaneo, C. The comparative performance of PMI estimation in skeletal remains by three methods (C-14, luminol test and OHI): Analysis of 20 cases. Int. J. Legal Med. 2018, 132, 1215–1224. [Google Scholar] [CrossRef]
- Sterzik, V.; Jung, T.; Jellinghaus, K.; Bohnert, M. Estimating the postmortem interval of human skeletal remains by analyzing their optical behavior. Int. J. Legal Med. 2016, 130, 1557–1566. [Google Scholar] [CrossRef]
- Longato, S.; Wöss, C.; Hatzer-Grubwieser, P.; Bauer, C.; Parson, W.; Unterberger, S.H.; Kuhn, V.; Pemberger, N.; Pallua, A.K.; Recheis, W.; et al. Post-mortem interval estimation of human skeletal remains by micro-computed tomography, mid-infrared microscopic imaging and energy dispersive X-ray mapping. Anal. Methods 2015, 7, 2917–2927. [Google Scholar] [CrossRef]
- Kaiser, C.; Bachmeier, B.; Conrad, C.; Nerlich, A.; Bratzke, H.; Eisenmenger, W.; Peschel, O. Molecular study of time dependent changes in DNA stability in soil buried skeletal residues. Forensic Sci. Int. 2008, 177, 32–36. [Google Scholar] [CrossRef]
- Mameli, A.; Piras, G.; Delogu, G. The Successful Recovery of Low Copy Number and Degraded DNA from Bones Exposed to Seawater Suitable for Generating a DNA STR Profile. J. Forensic Sci. 2014, 59, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Martínez, C.; Pérez-Cárceles, M.D.; Legaz, I.; Prieto-Bonete, G.; Luna, A. Quantification of nitrogenous bases, DNA and collagen type I for the estimation of the postmortem interval in bone remains. Forensic Sci. Int. 2017, 281, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.A.; Ferris, J.A.J. Analysis of postmortem DNA degradation by single-cell gel electrophoresis. Forensic Sci. Int. 2002, 126, 43–47. [Google Scholar] [CrossRef]
- Alaeddini, R.; Walsh, S.J.; Abbas, A. Forensic implications of genetic analyses from degraded DNA—A review. Forensic Sci. Int. Genet. 2010, 4, 148–157. [Google Scholar] [CrossRef]
- Alaeddini, R. Forensic implications of PCR inhibition—A review. Forensic Sci. Int. Genet. 2012, 6, 297–305. [Google Scholar] [CrossRef]
- Samsuwan, J.; Somboonchokepisal, T.; Akaraputtiporn, T.; Srimuang, T.; Phuengsukdaeng, P.; Suwannarat, A.; Mutirangura, A.; Kitkumthorn, N. A method for extracting DNA from hard tissues for use in forensic identification. Biomed. Rep. 2018, 9, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Gallelo, G.; Kuligowski, J.; Pastor, A.; Diez, A.; Bernabeu, J. Chemical element levels as a methodological tool in forensic science. J. Forensic Res. 2014, 6, 1. [Google Scholar]
- Miyaguchi, H.; Nakahara, H.; Sugita, R. Evaluation of the commercial kits for the extraction of extracellular DNA in soil. Jpn. J. Forensic Sci. Technol. 2019, 24, 63–72. [Google Scholar] [CrossRef]
- Imaizumi, K.; Akutsu, T.; Miyasaka, S.; Yoshino, M. Development of species identification tests targeting 16S ribosomal RNA coding region in mitochondrial DNA. Int. J. Legal Med. 2007, 121, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Watson, R.J.; Blackwell, B. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Can. J. Microbiol. 2000, 46, 633–642. [Google Scholar] [CrossRef]
- Braid, M.D.; Daniels, L.M.; Kitts, C.L. Removal of PCR inhibitors from soil DNA by chemical flocculation. J. Microbiol. Methods 2003, 52, 389–393. [Google Scholar] [CrossRef]
- Imaizumi, K.; Miyasaka, S.; Yoshino, M. Quantitative analysis of amplifiable DNA in tissues exposed to various environments using competitive PCR assays. Sci. Justice 2004, 44, 199–208. [Google Scholar] [CrossRef]
- Cartozzo, C.; Singh, B.; Boone, E.; Simmons, T. Evaluation of DNA Extraction Methods from Waterlogged Bones: A Pilot Study. J. Forensic Sci. 2018, 63, 1830–1835. [Google Scholar] [CrossRef] [PubMed]
- Campos, P.F.; Craig, O.E.; Turner-Walker, G.; Peacock, E.; Willerslev, E.; Gilbert, M.T.P. Annals of Anatomy DNA in ancient bone—Where is it located and how should we extract it? Ann. Anat. 2012, 194, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Korlević, P.; Gerber, T.; Marie-Theres, G.; Hajdinjak, M.; Nagel, S.; Aximu-Petri, A.; Meyer, M. Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. Biotechniques 2015, 93, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Sessa, F.; Varotto, E.; Salerno, M.; Vanin, S.; Bertozzi, G.; Galasi, F.M.; Maglietta, F.; Salerno, M.; Tuccia, F.; Pomara, C.; et al. First report of Heleomyzidae (Diptera) recovered from the inner cavity of an intact human femur. J. Forensic Legal Med. 2019, 66, 4–7. [Google Scholar] [CrossRef]
- Delannoy, Y.; Colard, T.; Le, E.; Mesli, V.; Aubernon, C.; Penel, G.; Hedouin, V.; Gosset, D. Effects of the environment on bone mass: A human taphonomic study. Leg. Med. 2016, 20, 61–67. [Google Scholar] [CrossRef]
- Aerssens, J.; Boonen, S.; Lowet, G.; Dequeker, J. Interspecies differences in bone composition, density, and quality: Potential implications for in vivo bone research. Endocrinology 1998, 139, 663–670. [Google Scholar] [CrossRef]


| PCR Product Size (bp) | Primer Sequences (5’–3’) | |
|---|---|---|
| Forward | Reverse | |
| 404 | CTTGTATGAATGGCCGCACG | TGATGGTGCAACCGCTATCA |
| 128 | AACCATTAAGGAATAACAACAA | AAATCACTCTATCGCTCATTG |
| Element | Estimated Concentration | Detection Limit | |||
|---|---|---|---|---|---|
| Control | Seawater | Freshwater | Underground | ||
| (μg/g) | (μg/g) | (μg/g) | (μg/g) | (μg/g) | |
| Li | ND * | ND | ND | ND | 10 |
| Be | ND | ND | ND | ND | 0.2 |
| B | ND | 60 | ND | ND | 20 |
| Na | 5000 | 7000 | 4000 | 6000 | 100 |
| Mg | 5000 | 5000 | 4000 | 5000 | 2 |
| Al | ND | ND | ND | ND | 10 |
| Si | ND | ND | ND | ND | 20 |
| P | 100,000 | 100,000 | 100,000 | 100,000 | 20 |
| K | ND | ND | ND | ND | 400 |
| Ca | 200,000 | 200,000 | 200,000 | 200,000 | 200 |
| Ti | ND | ND | ND | ND | 40 |
| V | ND | ND | ND | ND | 4 |
| Cr | 20 | ND | ND | 5 | 4 |
| Mn | 3 | ND | ND | ND | 1 |
| Fe | 300 | ND | ND | ND | 20 |
| Co | ND | ND | ND | ND | 10 |
| Ni | ND | ND | ND | ND | 10 |
| Cu | ND | ND | ND | ND | 10 |
| Zn | 60 | 70 | 90 | 60 | 10 |
| As | ND | ND | ND | ND | 40 |
| Se | ND | ND | ND | ND | 40 |
| Sr | 200 | 200 | 100 | 200 | 0.2 |
| Zr | ND | ND | ND | ND | 4 |
| Mo | ND | ND | ND | ND | 10 |
| Ag | ND | ND | ND | ND | 10 |
| Cd | ND | ND | ND | ND | 4 |
| Sn | ND | ND | ND | ND | 20 |
| Sb | ND | ND | ND | ND | 20 |
| Ba | 100 | 70 | 80 | 100 | 2 |
| Pb | ND | ND | ND | ND | 40 |
| Sample No. | Seawater | Freshwater | Underground | Outdoors | Indoors | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | 6 m * | 1 y † | 6 m | 1 y | 6 m | 1 y | 6 m | 1 y | 6 m | 1 y | |
| Sample 1 | 1.864 | 1.972 | 2.060 | 2.026 | 1.997 | 1.934 | 1.981 | 1.995 | 2.444 | 2.005 | 2.040 |
| Sample 2 | 1.717 | 1.929 | 2.020 | 1.965 | 2.079 | 1.961 | 2.042 | 1.984 | 1.985 | 2.018 | 2.054 |
| Sample 3 | 2.089 | 2.006 | 2.089 | 2.056 | 2.029 | 2.010 | 2.021 | 2.041 | 2.057 | 2.053 | 2.035 |
| Sample 4 | 2.162 | 2.027 | 2.048 | 2.027 | 2.072 | 2.002 | 2.003 | 2.003 | 2.004 | 2.045 | 2.026 |
| Sample 5 | 2.542 | 1.981 | 2.045 | 2.027 | 2.056 | 1.973 | 1.988 | 1.996 | 1.995 | 2.008 | 2.062 |
| Average | 2.075 | 1.983 | 2.053 | 2.020 | 2.047 | 1.976 | 2.007 | 2.004 | 2.097 | 2.026 | 2.043 |
| SD | 0.316 | 0.037 | 0.025 | 0.033 | 0.034 | 0.031 | 0.025 | 0.022 | 0.196 | 0.022 | 0.015 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagai, M.; Sakurada, K.; Imaizumi, K.; Ogawa, Y.; Uo, M.; Funakoshi, T.; Uemura, K. Evaluation of Parameters for Estimating the Postmortem Interval of Skeletal Remains Using Bovine Femurs: A Pilot Study. Diagnostics 2020, 10, 1066. https://doi.org/10.3390/diagnostics10121066
Nagai M, Sakurada K, Imaizumi K, Ogawa Y, Uo M, Funakoshi T, Uemura K. Evaluation of Parameters for Estimating the Postmortem Interval of Skeletal Remains Using Bovine Femurs: A Pilot Study. Diagnostics. 2020; 10(12):1066. https://doi.org/10.3390/diagnostics10121066
Chicago/Turabian StyleNagai, Midori, Koichi Sakurada, Kazuhiko Imaizumi, Yoshinori Ogawa, Motohiro Uo, Takeshi Funakoshi, and Koichi Uemura. 2020. "Evaluation of Parameters for Estimating the Postmortem Interval of Skeletal Remains Using Bovine Femurs: A Pilot Study" Diagnostics 10, no. 12: 1066. https://doi.org/10.3390/diagnostics10121066
APA StyleNagai, M., Sakurada, K., Imaizumi, K., Ogawa, Y., Uo, M., Funakoshi, T., & Uemura, K. (2020). Evaluation of Parameters for Estimating the Postmortem Interval of Skeletal Remains Using Bovine Femurs: A Pilot Study. Diagnostics, 10(12), 1066. https://doi.org/10.3390/diagnostics10121066




