An Aggregated-Based Deep Learning Method for Leukemic B-lymphoblast Classification
Abstract
:1. Introduction
1.1. Background
1.2. Motivations and Contributions
- The proposed aggregated model, based on deep learning architectures, achieves a better performance in terms of accuracy, precision, and recall for the ALL leukemia images. To the best of our knowledge, this is the first study toward the implementation of an aggregated model based on well-tuned deep learning algorithms for classification of ALL leukemia images applied to the ISBI 2019 challenge dataset.
- To improve the prediction performance, we extensively investigated the effect of fine-tuning of the different architectures with different settings of optimizers and image normalization, in order to generate a more discriminative representation. With these changes, our proposed model achieved higher accuracy in ALL classification compared to other state-of-the-art deep learning-based architectures and machine learning models individually.
- The performance of models trained by features extracted automatically using deep CNN models was compared to the performance of models trained by hand-crafted features, e.g., local binary pattern (LBP) descriptors. The obtained hand-crafted features were separately fed into the five leading machine learning classifiers using a k-fold cross-validation strategy. Classification results of the proposed ensemble model were compared with widely-used machine learning classifiers to measure the overall performance of the proposed method.
1.3. Related Studies
2. Materials and Methods
2.1. Data Pre-Processing
2.2. Data Augmentation
2.3. Feature Extraction and Architectures
2.3.1. DCNN Models
2.3.2. Transfer Learning
2.3.3. Local Binary Patterns
2.4. Metrics for Performance Evaluation
3. Results
3.1. Experimental Dataset
3.2. Experimental Networks Parameter Settings
3.3. Study 1: Hand-Crafted Feature Extraction Based on Local Binary Patterns
3.3.1. Results on Local Binary Patterns
3.4. Study 2: Deep Feature Extraction Based on Transfer Learning
3.4.1. Results of Deep Feature Extraction Based on Transfer Learning
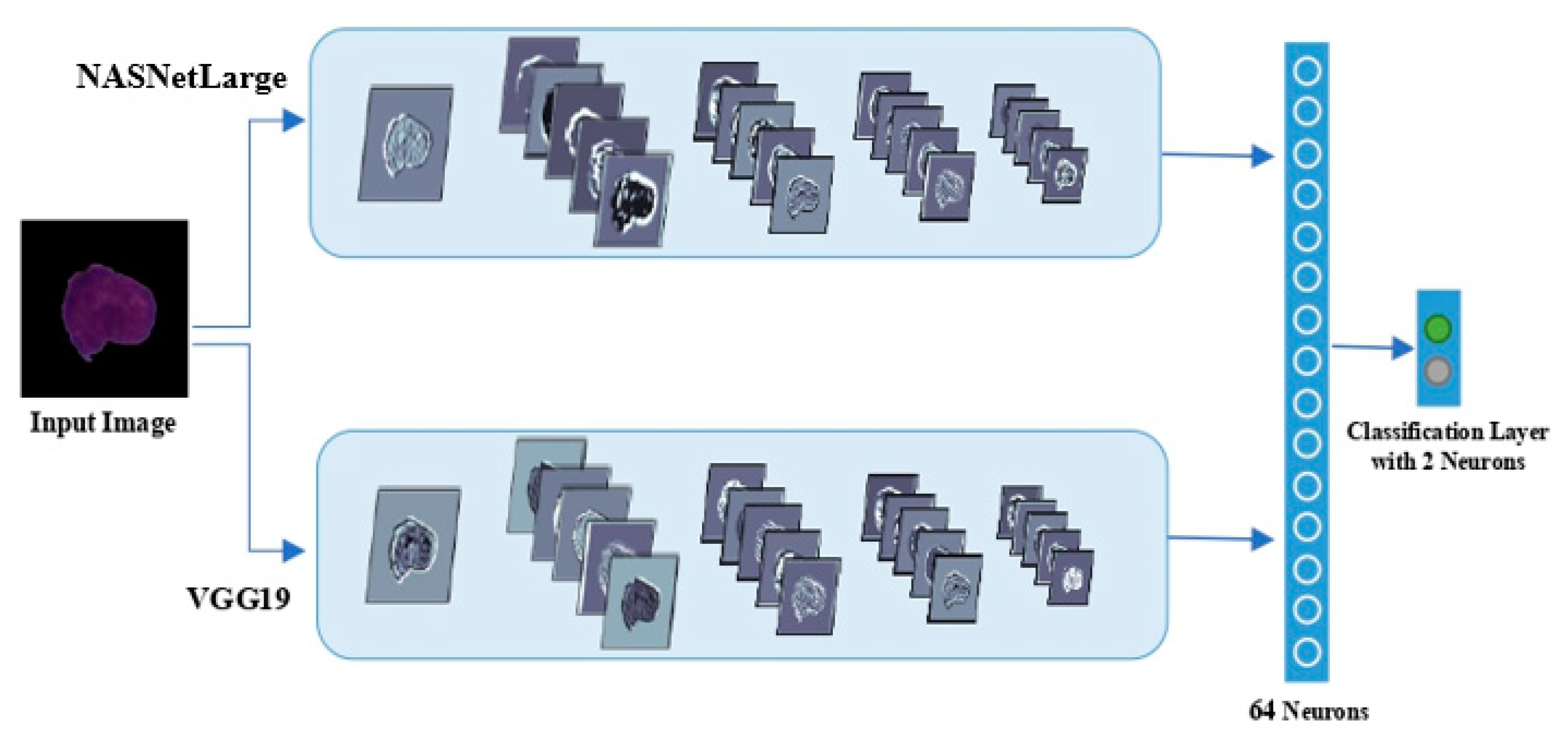
3.5. Study 3: Deep Feature Extraction Based on Deep Learning Multi-Model Ensemble
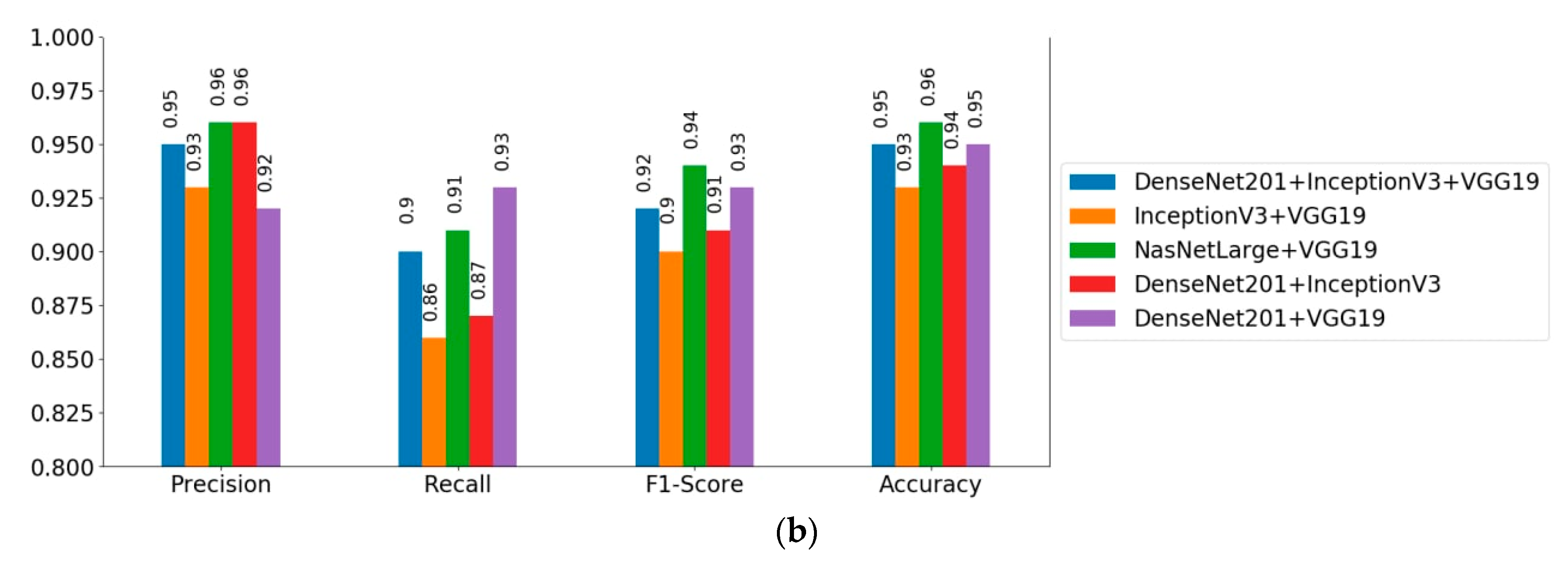
3.5.1. Results of the Deep Learning Multi-Model Ensemble
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability Statement
References
- Baytan:, B.; Evim, M.S.; Güler, S.; Güneş, A.M.; Okan, M. Acute Central Nervous System Complications in Pediatric Acute Lymphoblastic Leukemia. Pediatr. Neurol. 2015. [Google Scholar] [CrossRef]
- Tatar, A.S.; Nagy-Simon, T.; Tomuleasa, C.; Boca, S.; Astilean, S. Nanomedicine approaches in acute lymphoblastic leukemia. J. Control. Release 2016. [Google Scholar] [CrossRef]
- Key Statistics for Acute Lymphocytic Leukemia. Available online: https://www.cancer.org/cancer/acute-lymphocytic-leukemia/about/key-statistics (accessed on 10 July 2020).
- Schmiegelow, K.; van der Sluis, I. Pharmacokinetics, Pharmacodynamics and Pharmacogenetics of Antileukemic Drugs. In Childhood Acute Lymphoblastic Leukemia; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Aldoss, I.; Kamal, M.O.; Forman, S.J.; Pullarkat, V. Adults with Philadelphia Chromosome–Like Acute Lymphoblastic Leukemia: Considerations for Allogeneic Hematopoietic Cell Transplantation in First Complete Remission. Biol. Blood Marrow Transplant. 2019. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.M.; Kermani, S.; Talebi, A.; Oghli, M.G. Recognition of acute lymphoblastic leukemia cells in microscopic images using k-means clustering and support vector machine classifier. J. Med. Signals Sens. 2015. [Google Scholar] [CrossRef]
- Stein, A.P.; Norris, R.E.; Shah, J.R. Pediatric acute lymphoblastic leukemia presenting with periorbital edema. Otolaryngol. Case Rep. 2018. [Google Scholar] [CrossRef]
- Jaime-Pérez, J.C.; García-Arellano, G.; Herrera-Garza, J.L.; Marfil-Rivera, L.J.; Gómez-Almaguer, D. Revisiting the complete blood count and clinical findings at diagnosis of childhood acute lymphoblastic leukemia: 10-year experience at a single cente. Hematol. Transfus. Cell Ther. 2019. [Google Scholar] [CrossRef]
- Narayanan, S.; Shami, P.J. Treatment of acute lymphoblastic leukemia in adults. Crit. Rev. Oncol. /Hematol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Singhal, V.; Singh, P. Local Binary Pattern for automatic detection of Acute Lymphoblastic Leukemia. In Proceedings of the 2014 20th National Conference on Communications, NCC, Kanpur, India, 8 May 2014. [Google Scholar] [CrossRef]
- Gupta, A.; Duggal, R.; Gehlot, S.; Gupta, R.; Mangal, A.; Kumar, L.; Thakkar, N.; Satpathy, D. GCTI-SN: Geometry-inspired chemical and tissue invariant stain normalization of microscopic medical images. Med. Image Anal. 2020. [Google Scholar] [CrossRef]
- Gupta, R.; Mallick, P.; Duggal, R.; Gupta, A.; Sharma, O. Stain Color Normalization and Segmentation of Plasma Cells in Microscopic Images as a Prelude to Development of Computer Assisted Automated Disease Diagnostic Tool in Multiple Myeloma. Clin. Lymphoma Myeloma Leuk. 2017. [Google Scholar] [CrossRef]
- Kassani, P.H.; Kim, E. Pseudoinverse Matrix Decomposition Based Incremental Extreme Learning Machine with Growth of Hidden Nodes. Int. J. Fuzzy Log. Intell. Syst. 2016. [Google Scholar] [CrossRef]
- Mohamed, H.; Omar, R.; Saeed, N.; Essam, A.; Ayman, N.; Mohiy, T.; Abdelraouf, A. Automated detection of white blood cells cancer diseases. In Proceedings of the IWDRL 2018: 2018 1st International Workshop on Deep and Representation Learning, Cario, Egypt, 29 March 2018. [Google Scholar] [CrossRef]
- Kassani, S.H.; Kassani, P.H.; Najafi, S.E. Introducing a hybrid model of DEA and data mining in evaluating efficiency. Case study: Bank Branches. arXiv 2018, arXiv:1810.05524. [Google Scholar]
- Yu, W.; Chang, J.; Yang, C.; Zhang, L.; Shen, H.; Xia, Y.; Sha, J. Automatic classification of leukocytes using deep neural network. In Proceedings of the Proceedings of International Conference on ASIC, Guiyang, China, 25–28 October 2017. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015-Conference Track Proceedings, San Diego, CA, USA, 9 May 2015. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef] [Green Version]
- Honnalgere, A.; Nayak, G. Classification of normal versus malignant cells in B-ALL white blood cancer microscopic images. In Lecture Notes in Bioengineering; Springer: Singapore, 2019. [Google Scholar]
- Marzahl, C.; Aubreville, M.; Voigt, J.; Maier, A. Classification of leukemic B-Lymphoblast cells from blood smear microscopic images with an attention-based deep learning method and advanced augmentation techniques. In Lecture Notes in Bioengineering; Springer: Singapore, 2019. [Google Scholar]
- Shah, S.; Nawaz, W.; Jalil, B.; Khan, H.A. Classification of normal and leukemic blast cells in B-ALL cancer using a combination of convolutional and recurrent neural networks. In Lecture Notes in Bioengineering; Springer: Singapore, 2019. [Google Scholar]
- Pan, Y.; Liu, M.; Xia, Y.; Shen, D. Neighborhood-correction algorithm for classification of normal and malignant cells. In Lecture Notes in Bioengineering; Springer: Singapore, 2019. [Google Scholar]
- Mohapatra, S.; Patra, D.; Satpathy, S. An ensemble classifier system for early diagnosis of acute lymphoblastic leukemia in blood microscopic images. Neural Comput. Appl. 2014. [Google Scholar] [CrossRef]
- Kawahara, J.; Bentaieb, A.; Hamarneh, G. Deep features to classify skin lesions. Proc. Int. Symp. Biomed. Imaging 2016. [Google Scholar] [CrossRef]
- Yu, Z.; Jiang, X.; Wang, T.; Lei, B. Aggregating deep convolutional features for melanoma recognition in dermoscopy images. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Singapore, 2017. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems; Curran Associates Inc.: New York, NY, USA, 2012. [Google Scholar]
- Hosseinzadeh Kassani, S.; Hosseinzadeh Kassani, P. A comparative study of deep learning architectures on melanoma detection. Tissue Cell 2019. [Google Scholar] [CrossRef] [PubMed]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning Transferable Architectures for Scalable Image Recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [CrossRef] [Green Version]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef] [Green Version]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. Available online: http://arxiv.org/abs/1704.04861 (accessed on 17 April 2017).
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. ShuffleNet: An Extremely Efficient Convolutional Neural Network for Mobile Devices. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA,, 18–23 June 2018. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Lu, Z.; Zhang, Y.D. Pathological brain detection based on AlexNet and transfer learning. J. Comput. Sci. 2019. [Google Scholar] [CrossRef]
- Mardanisamani, S.; Maleki, F.; Kassani, S.H.; Rajapaksa, S.; Duddu, H.; Wang, M.; Shirtliffe, S.; Ryu, S.; Josuttes, A.; Zhang, T.; et al. Crop lodging prediction from UAV-acquired images of wheat and canola using a DCNN augmented with handcrafted texture features. In IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops; IEEE: Long Beach, CA, USA, 2019. [Google Scholar] [CrossRef] [Green Version]
- Ojala, T.; Pietikäinen, M.; Mäenpää, T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002. [Google Scholar] [CrossRef]
- Hassaballah, M.; Alshazly, H.A.; Ali, A.A. Ear recognition using local binary patterns: A comparative experimental study. Expert Syst. Appl. 2019. [Google Scholar] [CrossRef]
- Li, L.; Feng, X.; Xia, Z.; Jiang, X.; Hadid, A. Face spoofing detection with local binary pattern network. J. Vis. Commun. Image Represent. 2018. [Google Scholar] [CrossRef]
- Li, X.; Pang, T.; Xiong, B.; Liu, W.; Liang, P.; Wang, T. Convolutional neural networks based transfer learning for diabetic retinopathy fundus image classification. In Proceedings of the 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics, CISP-BMEI 2017, Shanghai, China, 14–16 October 2017. [Google Scholar] [CrossRef]
- Xiao, Y.; Wu, J.; Lin, Z.; Zhao, X. A deep learning-based multi-model ensemble method for cancer prediction. Comput. Methods Programs Biomed. 2018. [Google Scholar] [CrossRef]
- Yu, Y.; Lin, H.; Meng, J.; Wei, X.; Guo, H.; Zhao, Z. Deep transfer learning for modality classification of medical images. Information 2017, 8, 91. [Google Scholar] [CrossRef] [Green Version]
- Duggal, R.; Gupta, A.; Gupta, R.; Wadhwa, M.; Ahuja, C. Overlapping cell nuclei segmentation in microscopic images using deep belief networks. In ACM International Conference Proceeding Series; Association for Computing Machinery: New York, NY, USA, 2016. [Google Scholar] [CrossRef]
- Duggal, R.; Gupta, A.; Gupta, R.; Mallick, P. SD-Layer: Stain deconvolutional layer for CNNs in medical microscopic imaging. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Andrearczyk, V.; Whelan, P.F. Using filter banks in Convolutional Neural Networks for texture classification. Pattern Recognit. Lett. 2016. [Google Scholar] [CrossRef]
- “python”. Available online: https://www.python.org/ (accessed on 20 May 2020).
- “Keras”. Available online: https://keras.io/ (accessed on 20 May 2020).
- “tensorflow”. Available online: https://www.tensorflow.org/ (accessed on 20 May 2020).
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Russakovsky, O. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015. [Google Scholar] [CrossRef] [Green Version]
- Cover, T.M.; Hart, P.E. Nearest Neighbor Pattern Classification. IEEE Trans. Inf. Theory 1967. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001. [Google Scholar] [CrossRef]
- Chen, C.; Guestrin, T. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 3–17 August 2016. [Google Scholar]
- Friedman, N.; Geiger, D.; Goldszmidt, M. Bayesian Network Classifiers. Mach. Learn. 1997. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Yin, Z.; Wang, Y.; Zhang, W.; Wang, Y.; Zhang, J. Assessing cognitive mental workload via EEG signals and an ensemble deep learning classifier based on denoising autoencoders. Comput. Biol. Med. 2019. [Google Scholar] [CrossRef]
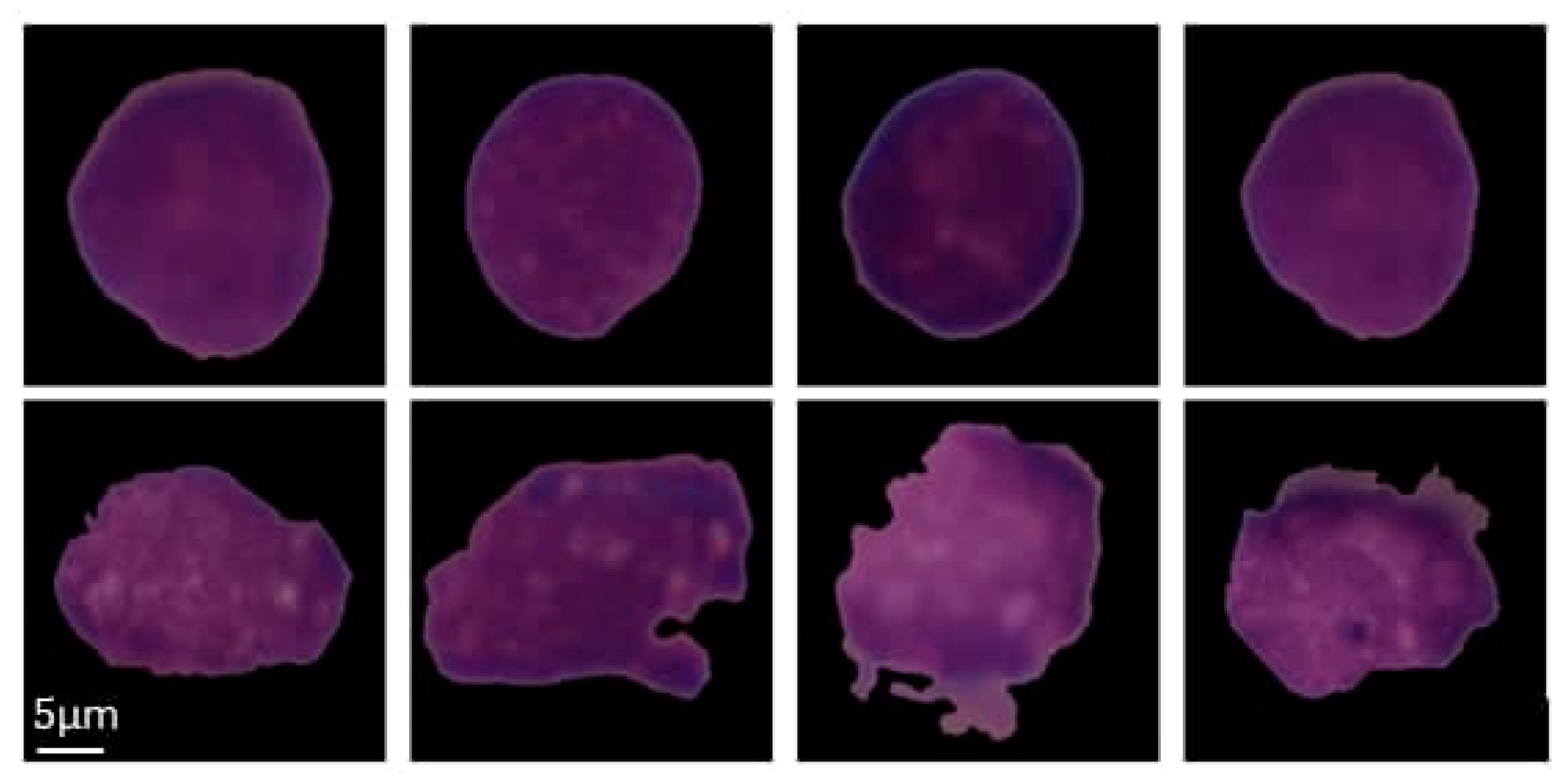
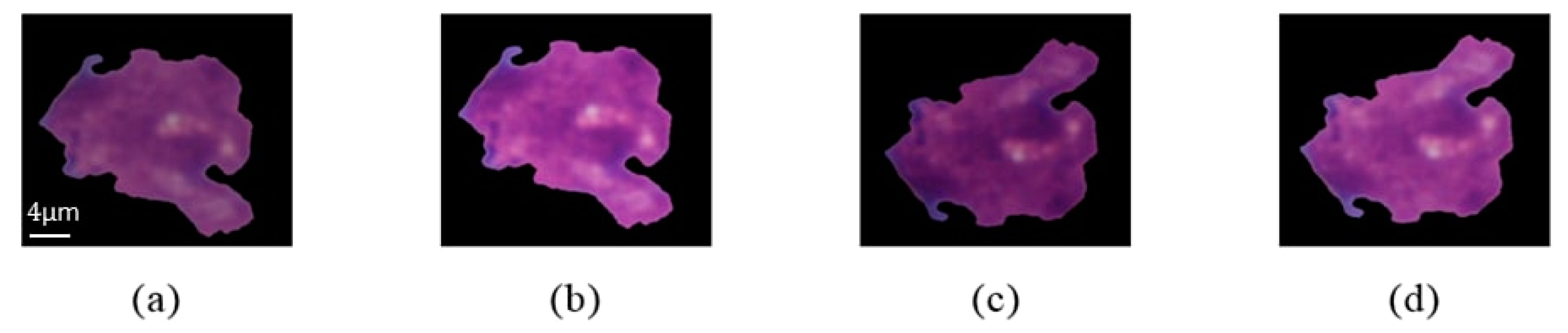

| Cell Type | Before Augmentation | After Augmentation |
|---|---|---|
| Healthy cells | 3389 | 24,616 |
| ALL cells | 7272 | 52,938 |
| Models | Fold1 (%) | Fold2 (%) | Fold3 (%) | Fold4 (%) | Fold5 (%) | Mean (%) | Std |
|---|---|---|---|---|---|---|---|
| KNN | 78.86 | 89.69 | 89.11 | 74.61 | 55.44 | 77.54 | ± 0.13 |
| Naive Bayes | 55.67 | 85.05 | 66.83 | 90.67 | 79.27 | 75.50 | ± 0.12 |
| Random Forest | 76.28 | 88.65 | 92.22 | 78.23 | 54.40 | 77.96 | ± 0.13 |
| Gradient Boosting | 72.68 | 87.62 | 90.15 | 78.23 | 56.47 | 77.03 | ± 0.12 |
| XGBoost | 74.74 | 91.75 | 95.33 | 77.72 | 58.54 | 79.62 | ± 0.13 |
| Models | Dataset Mean | Image Mean | Image Net Mean |
|---|---|---|---|
| AlexNet | 87.80% | 87.49% | 86.45% |
| NASNetLarge | 94.55% | 91.73% | 92.24% |
| NASNetMobile | 89.66% | 89.35% | 87.18% |
| InceptionV3 | 93.28% | 89.97% | 92.14% |
| VGG19 | 94.24% | 32.26% | 91.31% |
| VGG16 | 93.28% | 67.74% | 86.35% |
| Xception | 92.97% | 91.11% | 92.14% |
| MobileNet | 88.52% | 90.49% | 87.90% |
| ShuffleNet | 80.35% | 80.04% | 77.56% |
| DenseNet201 | 93.69% | 92.76% | 94.93% |
| Models | Adam | SGD | RMSProp |
|---|---|---|---|
| AlexNet | 87.80% | 85.73% | 88.11% |
| NASNetLarge | 95.55% | 92.14% | 32.26% |
| DenseNet201 | 93.69% | 91.86% | 91.68% |
| NASNetMobile | 89.66% | 88.93% | 60.70% |
| InceptionV3 | 93.28% | 92.66% | 92.86% |
| VGG19 | 95.24% | 88.24% | 93.90% |
| VGG16 | 93.10% | 90.49% | 91.49% |
| Xception | 92.97% | 88.13% | 91.31% |
| MobileNet | 88.52% | 92.45% | 92.24% |
| ShuffleNet | 80.35% | 84.49% | 80.66% |
| Average | 90.82% | 89.25% | 81.52% |
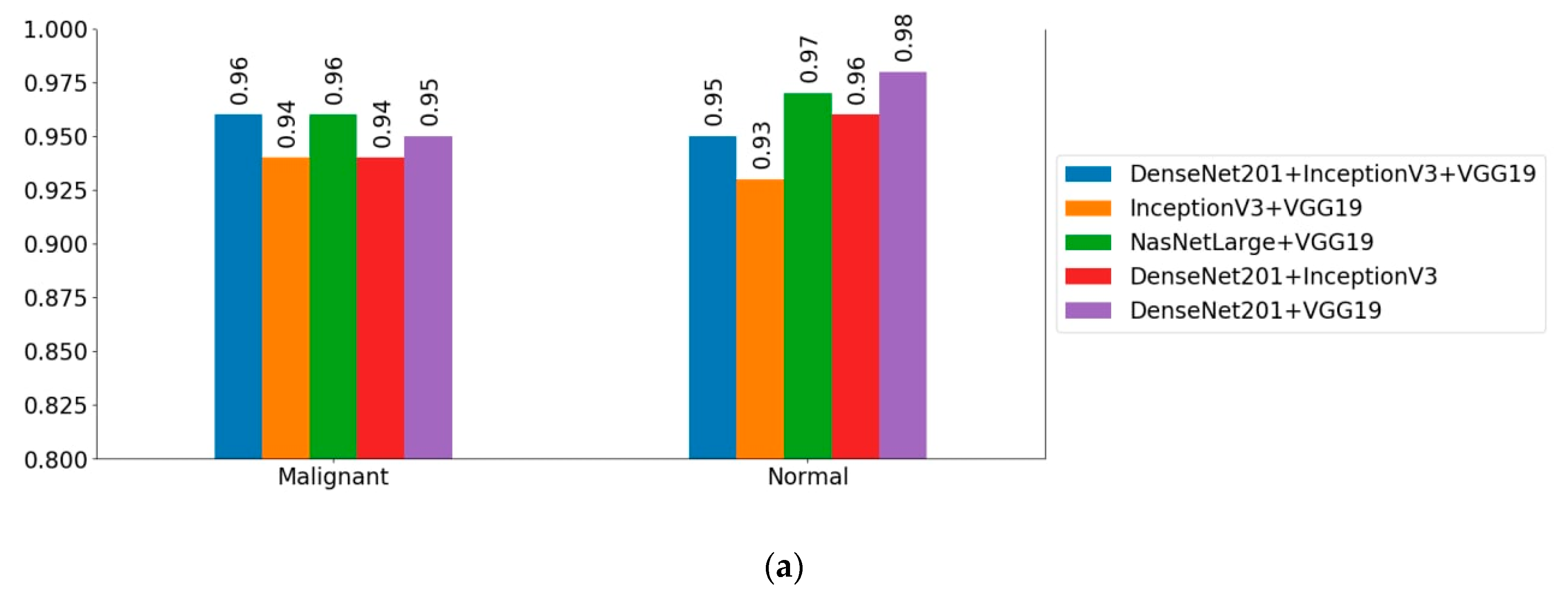
| Ensemble Models | Accuracy (%) | Precision (%) | Recall (%) | F1-score (%) |
|---|---|---|---|---|
| NASNetLarge + VGG19 * | 96.58 ± 1.09 | 96.94 | 91.75 | 94.67 |
| DenseNet201 + InceptionV3 | 94.73 ± 1.27 | 96.11 | 87.18 | 91.43 |
| DenseNet201 + VGG19 $ | 95.55 ± 1.10 | 92.43 | 93.91 | 93.16 |
| InceptionV3 + VGG19 | 93.80 ± 1.31 | 93.45 | 86.86 | 90.03 |
| DenseNet201 + VGG19 + InceptionV3 | 95.45 ± 1.29 | 95.30 | 90.38 | 92.76 |
| Models | Sensitivity (%) | Specificity (%) |
|---|---|---|
| NASNetLarge | 94.45 | 94.13 |
| DenseNet201 | 92.89 | 94.90 |
| VGG19 | 96.24 | 91.88 |
| InceptionV3 | 92.53 | 91.43 |
| DenseNet201 + VGG19 + InceptionV3 | 95.52 | 95.27 |
| NASNetLarge + VGG19 | 95.98 | 96.93 |
| DenseNet201 + InceptionV3 | 94.15 | 96.11 |
| DenseNet201 + VGG19 | 97.07 | 92.42 |
| InceptionV3 + VGG19 | 93.94 | 93.44 |
| Model | Total Parameters |
|---|---|
| NASNetLarge | 90 M |
| InceptionV3 | 24 M |
| AlexNet | 22 M |
| DenseNet201 | 21 M |
| VGG19 | 21 M |
| Xception | 20 M |
| VGG16 | 16 M |
| NASNetMobile | 6 M |
| MobileNet | 5 M |
| ShuffleNet | 1 M |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasani, P.H.; Park, S.-W.; Jang, J.-W. An Aggregated-Based Deep Learning Method for Leukemic B-lymphoblast Classification. Diagnostics 2020, 10, 1064. https://doi.org/10.3390/diagnostics10121064
Kasani PH, Park S-W, Jang J-W. An Aggregated-Based Deep Learning Method for Leukemic B-lymphoblast Classification. Diagnostics. 2020; 10(12):1064. https://doi.org/10.3390/diagnostics10121064
Chicago/Turabian StyleKasani, Payam Hosseinzadeh, Sang-Won Park, and Jae-Won Jang. 2020. "An Aggregated-Based Deep Learning Method for Leukemic B-lymphoblast Classification" Diagnostics 10, no. 12: 1064. https://doi.org/10.3390/diagnostics10121064
APA StyleKasani, P. H., Park, S.-W., & Jang, J.-W. (2020). An Aggregated-Based Deep Learning Method for Leukemic B-lymphoblast Classification. Diagnostics, 10(12), 1064. https://doi.org/10.3390/diagnostics10121064






