Identification and Characterization of Serum microRNAs as Biomarkers for Human Disc Degeneration: An RNA Sequencing Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Collection of Serum from Peripheral Blood
2.3. RNA Extraction
2.4. Filtering of Small RNAs
2.5. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.6. Purification of PCR Products, Circularization, Library Validation, and Sequencing
2.7. Data Processing and Differentially Expressed Gene (DEG) Screening
2.8. Differentially Expressed Analysis of miRNA and the Target Genes
2.9. Establishment and Analysis of miRNA Regulated Gene Networks
2.10. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
2.11. Statistical Analysis
3. Results
3.1. Quality Analysis
3.2. Differentially Expressed miRNA and Target DEG Analysis
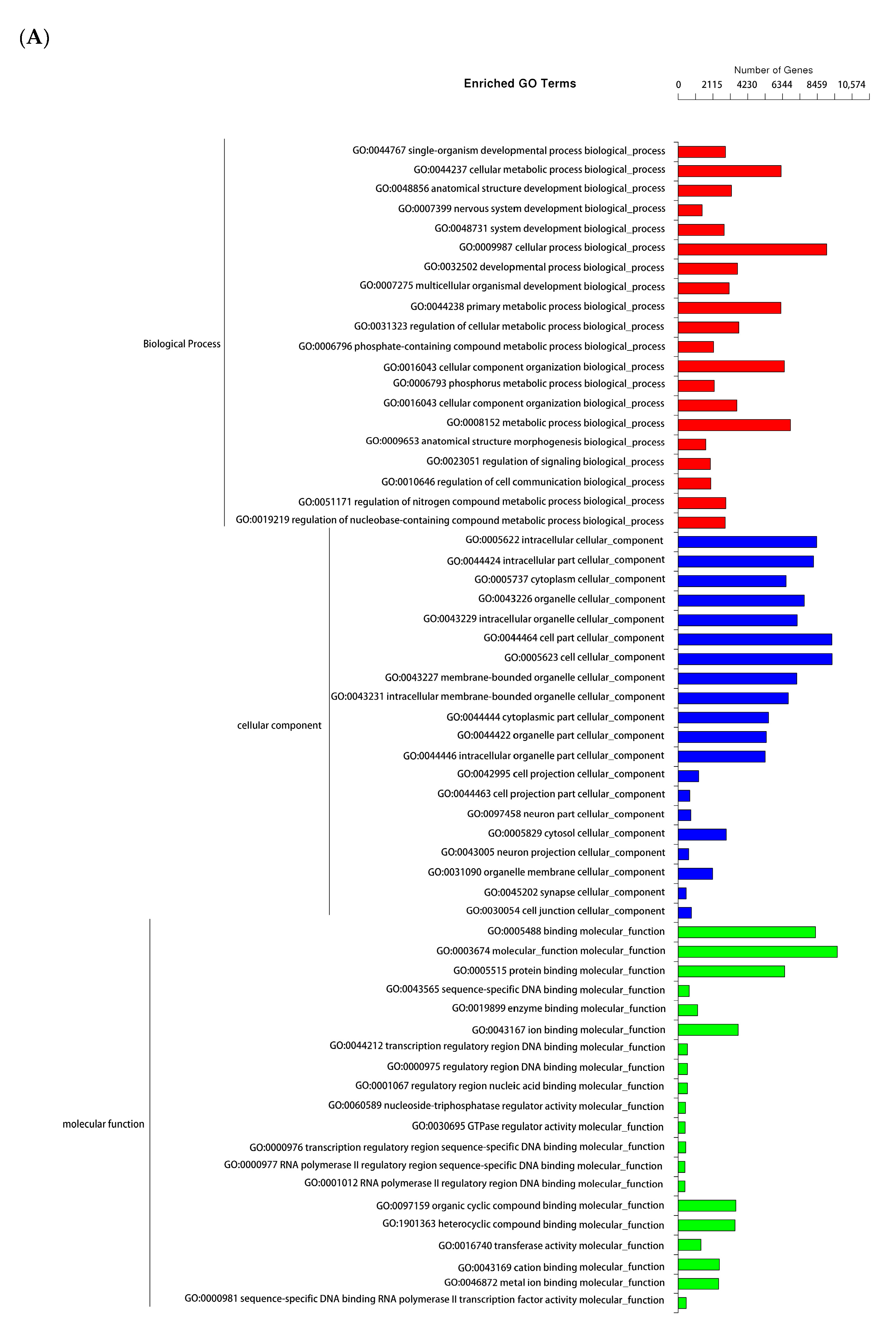
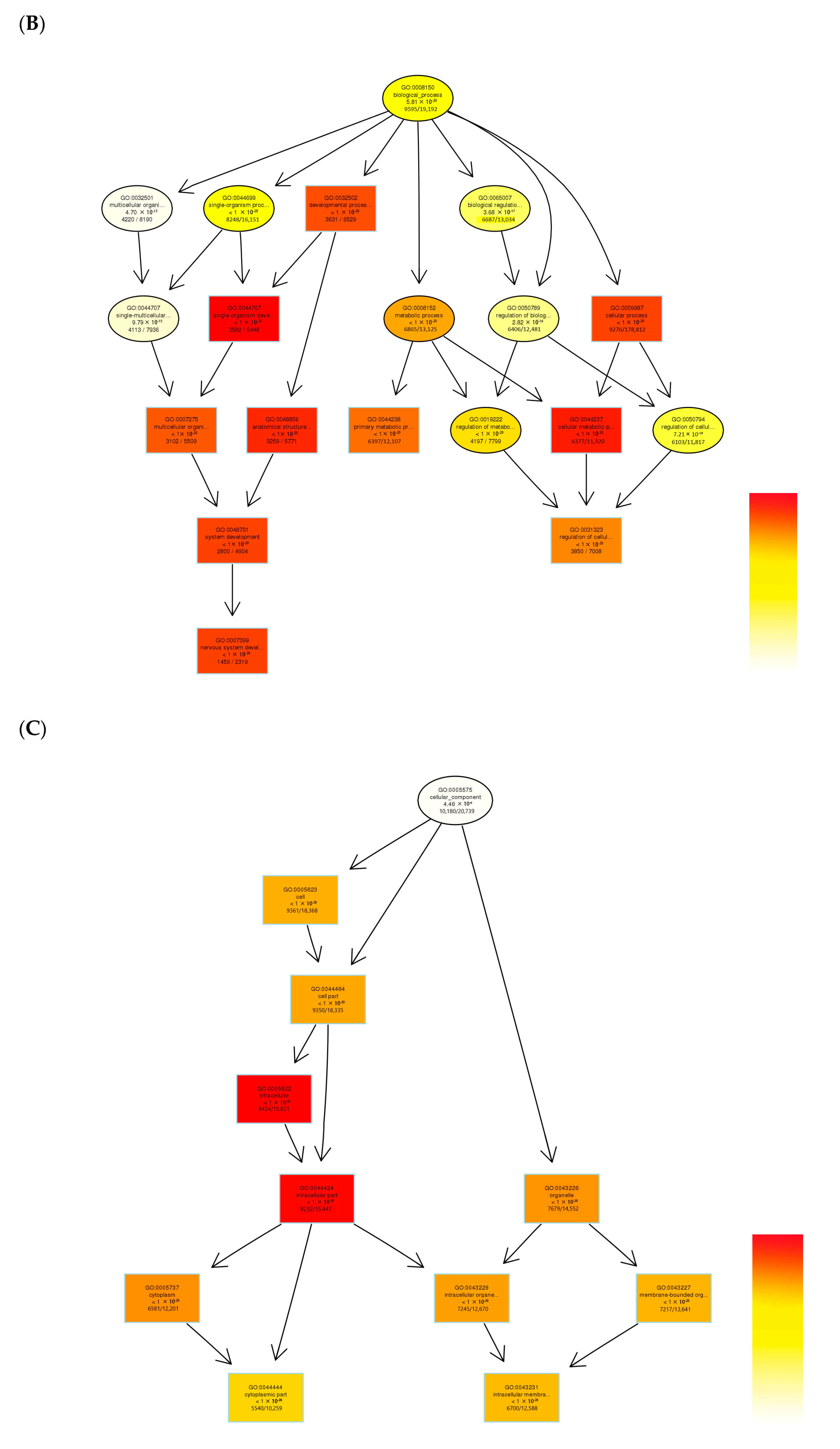
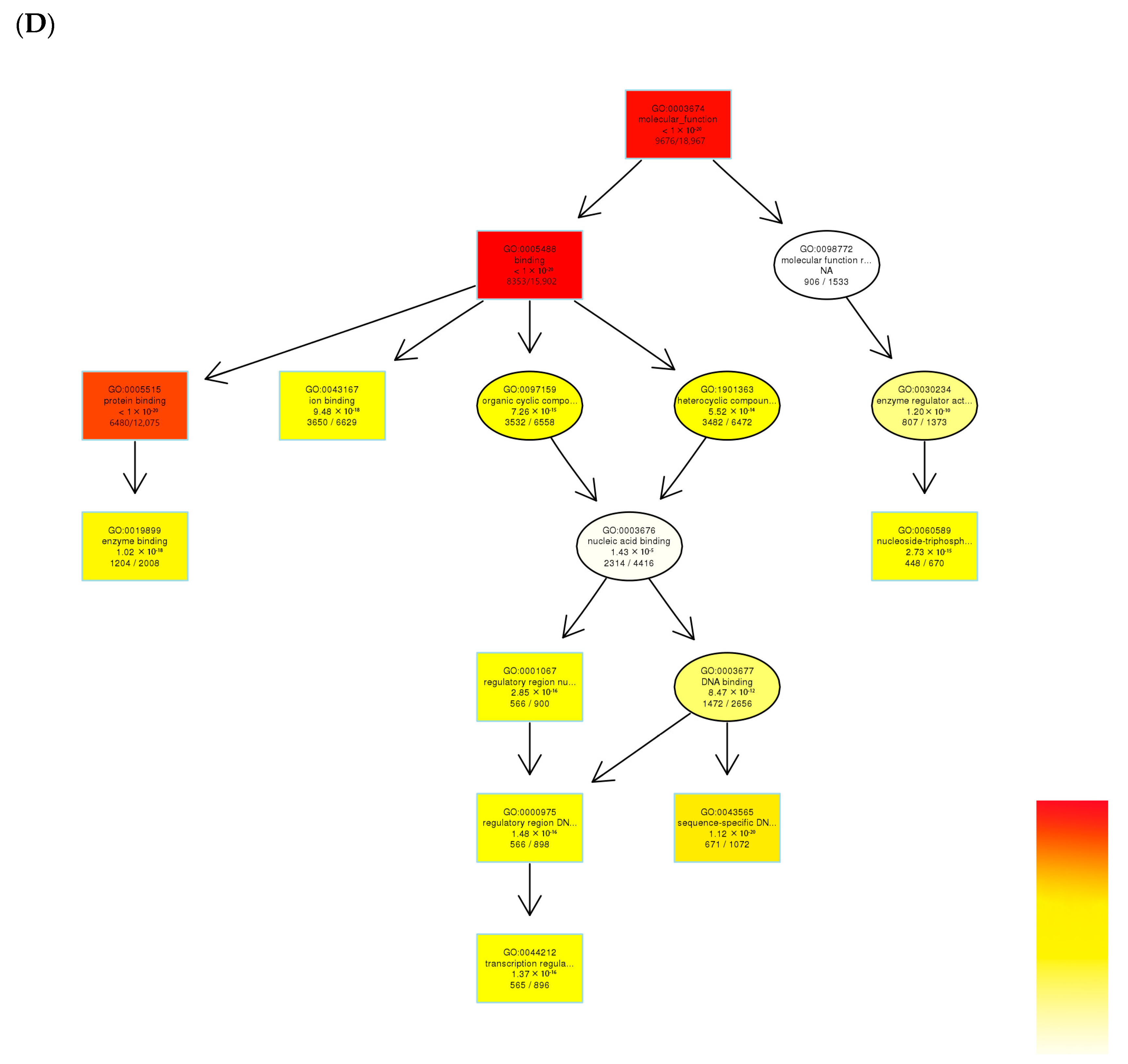
3.3. Gene Ontology (GO) Functional Enrichment Analysis
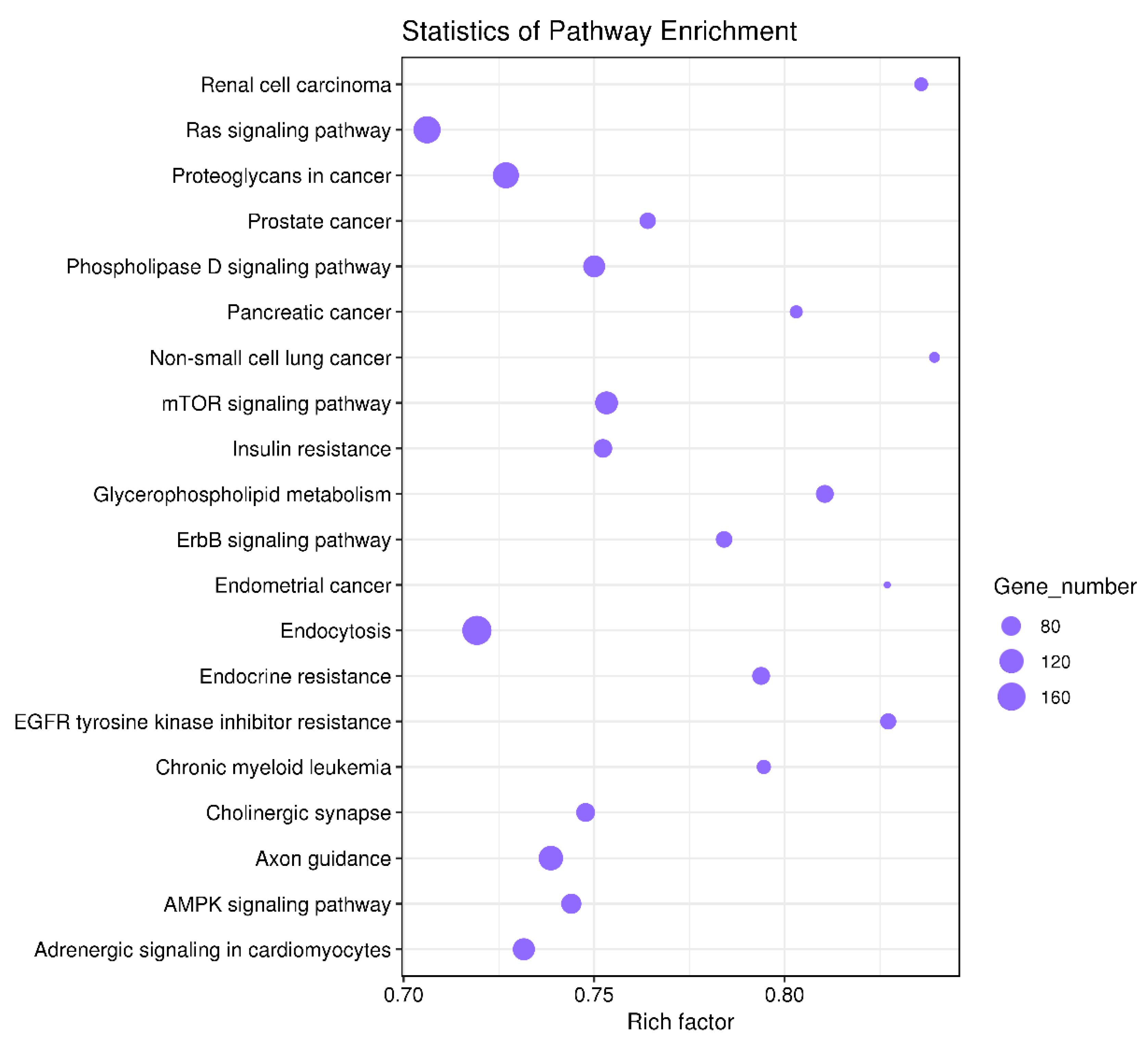
3.4. Enrichment Analysis
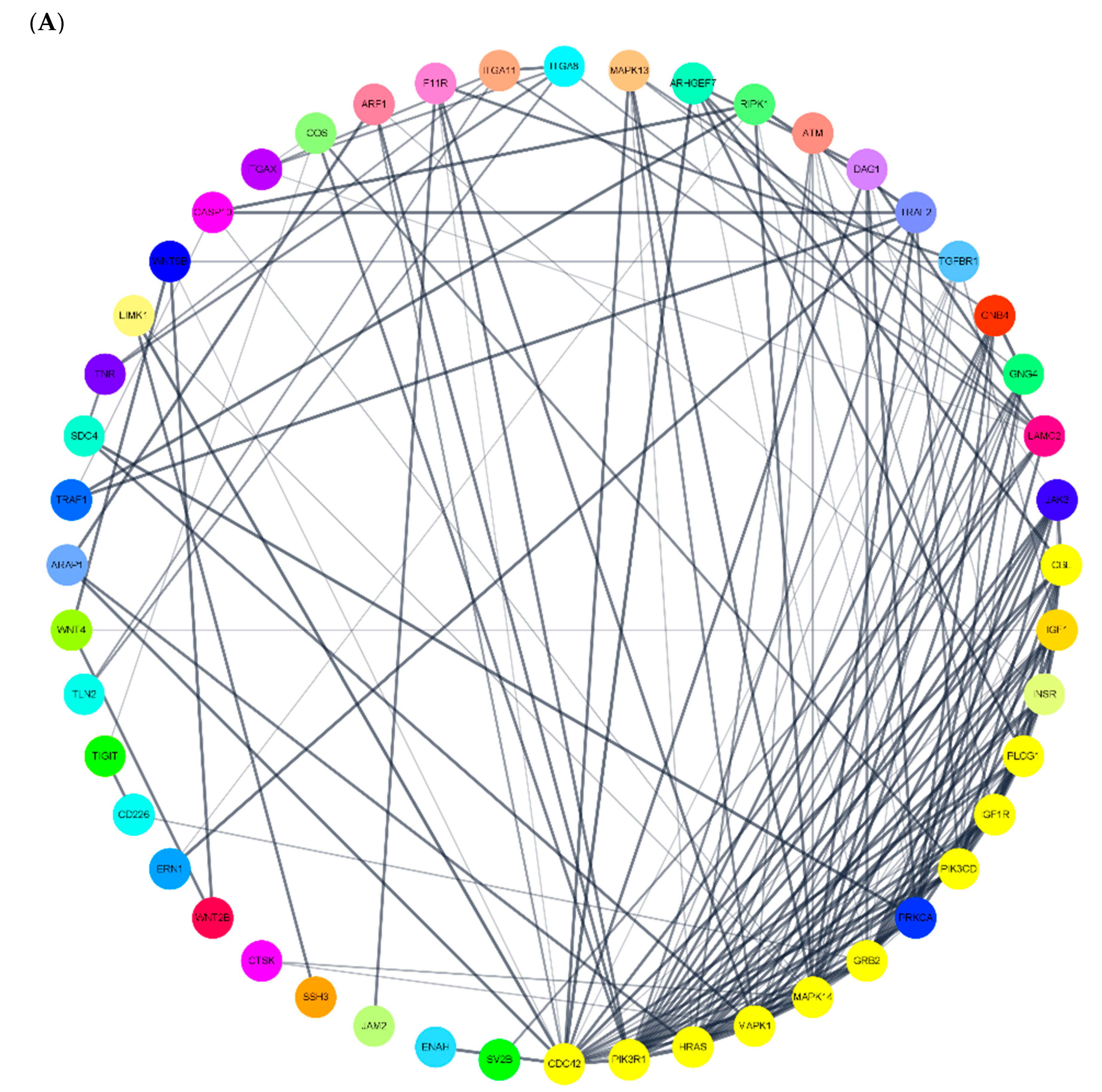
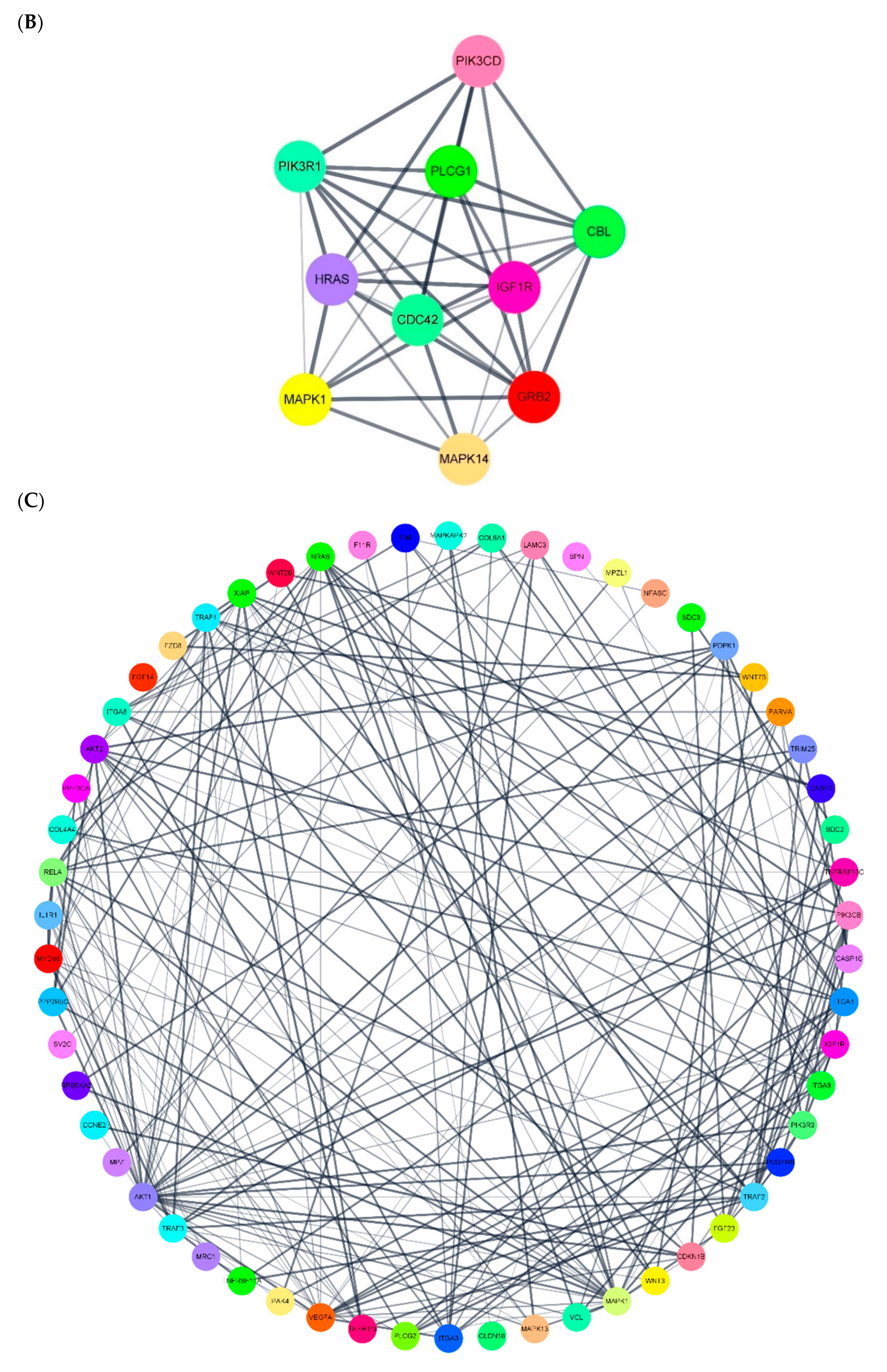
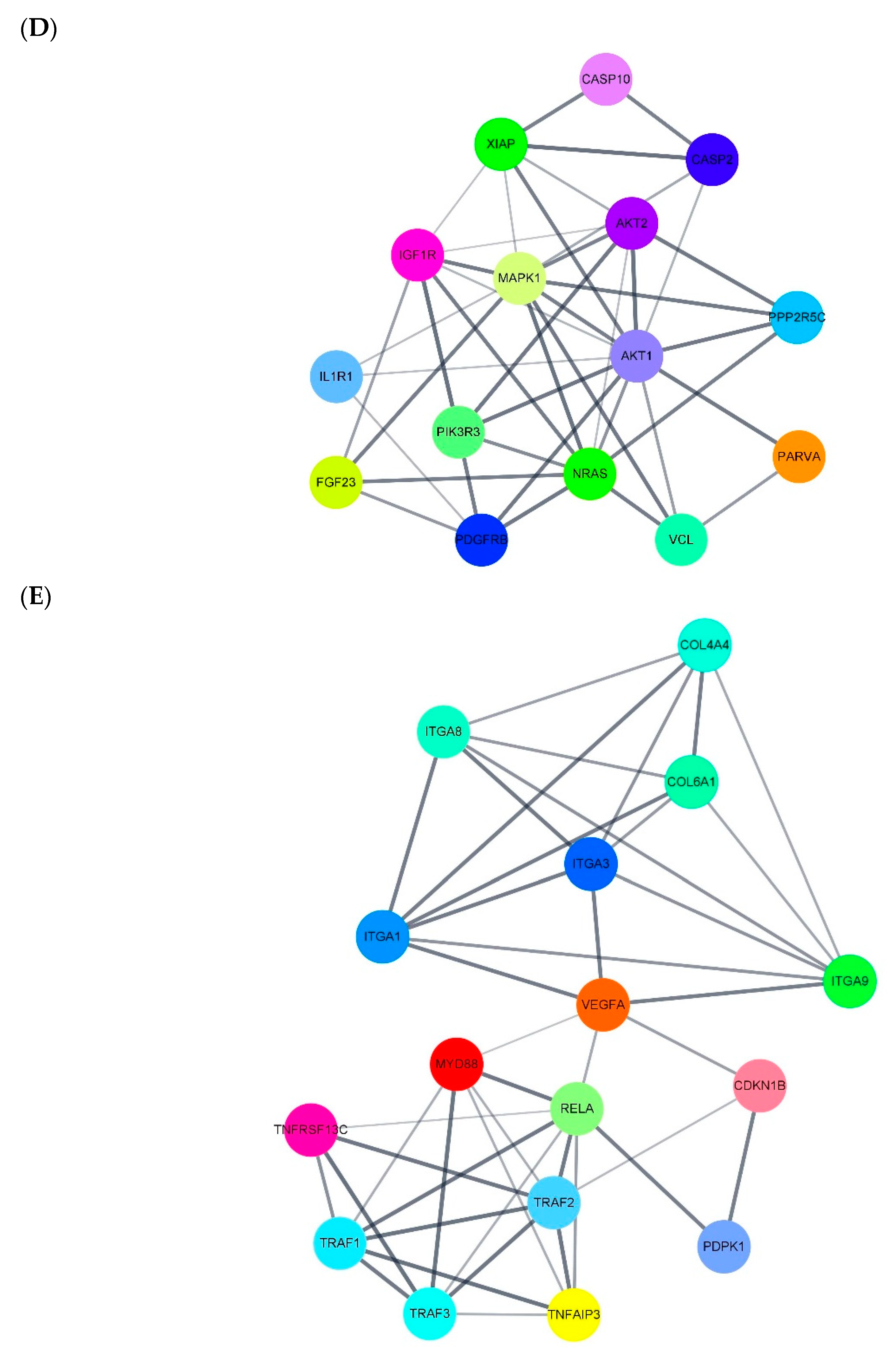
3.5. miRNA Target Gene Regulation Association and miRNA-DEG Regulation Network Construction
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Availability of Data and Material
References
- Disease, G.B.D.; Injury, I.; Prevalence, C. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1545–1602. [Google Scholar] [CrossRef]
- Rosenfeld, S.B.; Schroeder, K.; Watkins-Castillo, S.I. The economic burden of musculoskeletal disease in children and adolescents in the United States. J. Pediatr. Orthop. 2018, 38, e230–e236. [Google Scholar] [CrossRef] [PubMed]
- Boos, N.; Weissbach, S.; Rohrbach, H.; Weiler, C.; Spratt, K.F.; Nerlich, A.G. Classification of age-related changes in lumbar intervertebral discs: 2002 Volvo Award in basic science. Spine (Phila Pa 1976) 2002, 27, 2631–2644. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.A.; Roughley, P.J. What is intervertebral disc degeneration, and what causes it? Spine (Phila Pa 1976) 2006, 31, 2151–2161. [Google Scholar] [CrossRef] [PubMed]
- Battie, M.C.; Videman, T.; Parent, E. Lumbar disc degeneration: Epidemiology and genetic influences. Spine (Phila Pa 1976) 2004, 29, 2679–2690. [Google Scholar] [CrossRef]
- Kukurba, K.R.; Montgomery, S.B. RNA sequencing and analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef]
- Araldi, E.; Chamorro-Jorganes, A.; van Solingen, C.; Fernandez-Hernando, C.; Suarez, Y. Therapeutic potential of modulating microRNAs in atherosclerotic vascular disease. Curr. Vasc. Pharmacol. 2013, 13, 291–304. [Google Scholar] [CrossRef]
- Ivey, K.N.; Srivastava, D. microRNAs as developmental regulators. Cold Spring Harb. Perspect. Biol. 2015, 7, a008144. [Google Scholar] [CrossRef]
- Economou, E.K.; Oikonomou, E.; Siasos, G.; Papageorgiou, N.; Tsalamandris, S.; Mourouzis, K.; Papaioanou, S.; Tousoulis, D. The role of microRNAs in coronary artery disease: From pathophysiology to diagnosis and treatment. Atherosclerosis 2015, 241, 624–633. [Google Scholar] [CrossRef]
- Wang, C.; Wang, W.J.; Yan, Y.G.; Xiang, Y.X.; Zhang, J.; Tang, Z.H.; Jiang, Z.S. MicroRNAs: New players in intervertebral disc degeneration. Clin. Chim. Acta 2015, 450, 333–341. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, L.; Grad, S.; Alini, M.; Pan, H.; Yang, D.; Zhen, W.; Li, Z.; Huang, S.; Peng, S. The roles and perspectives of microRNAs as biomarkers for intervertebral disc degeneration. J. Tissue Eng. Regen. Med. 2017, 11, 3481–3487. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Hirai, T.; Ikeda, K.; Tsushima, H.; Fujishiro, M.; Hayakawa, K.; Yoshida, Y.; Morimoto, S.; Yamaji, K.; Takasaki, Y.; Takamori, K.; et al. Circulating plasma microRNA profiling in patients with polymyositis/dermatomyositis before and after treatment: miRNA may be associated with polymyositis/dermatomyositis. Inflamm. Regen. 2018, 38, 1. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wei, Y.; Yu, L.; Xiao, Y. Identification of altered circular RNA expression in serum exosomes from patients with papillary thyroid carcinoma by high-throughput sequencing. Med. Sci. Monit. 2019, 25, 2785–2791. [Google Scholar] [CrossRef]
- Lu, K.; Li, H.Y.; Yang, K.; Wu, J.L.; Cai, X.W.; Zhou, Y.; Li, C.Q. Exosomes as potential alternatives to stem cell therapy for intervertebral disc degeneration: In-vitro study on exosomes in interaction of nucleus pulposus cells and bone marrow mesenchymal stem cells. Stem Cell Res. Ther. 2017, 8, 108. [Google Scholar] [CrossRef]
- Ban, E.; Chae, D.K.; Yoo, Y.S.; Song, E.J. An improvement of miRNA extraction efficiency in human plasma. Anal. Bioanal. Chem. 2017, 409, 6397–6404. [Google Scholar] [CrossRef]
- Lekchnov, E.A.; Zaporozhchenko, I.A.; Morozkin, E.S.; Bryzgunova, O.E.; Vlassov, V.V.; Laktionov, P.P. Protocol for miRNA isolation from biofluids. Anal. Biochem. 2016, 499, 78–84. [Google Scholar] [CrossRef]
- Brzyski, D.; Peterson, C.B.; Sobczyk, P.; Candes, E.J.; Bogdan, M.; Sabatti, C. Controlling the rate of GWAS false discoveries. Genetics 2017, 205, 61–75. [Google Scholar] [CrossRef]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Zheng, X.; Yang, J.; Imamichi, T.; Stephens, R.; Lempicki, R.A. Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinform. 2009, 27, 13.11.1–13.11.13. [Google Scholar] [CrossRef] [PubMed]
- Sherafatian, M.; Abdollahpour, H.R.; Ghaffarpasand, F.; Yaghmaei, S.; Azadegan, M.; Heidari, M. MicroRNA expression profiles, target genes and pathways in intervertebral disc degeneration; A meta-analysis of three microarray studies. World Neurosurg. 2019. [Google Scholar] [CrossRef]
- Hu, P.; Feng, B.; Wang, G.; Ning, B.; Jia, T. Microarray based analysis of gene regulation by microRNA in intervertebral disc degeneration. Mol. Med. Rep. 2015, 12, 4925–4930. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Phillips, K.L.; Chiverton, N.; Michael, A.L.; Cole, A.A.; Breakwell, L.M.; Haddock, G.; Bunning, R.A.; Cross, A.K.; Le Maitre, C.L. The cytokine and chemokine expression profile of nucleus pulposus cells: Implications for degeneration and regeneration of the intervertebral disc. Arthritis Res. Ther. 2013, 15, R213. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wang, S.; Liu, Y.; Wang, X. Microarray analysis of genes and gene functions in disc degeneration. Exp. Ther. Med. 2014, 7, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Minogue, B.M.; Richardson, S.M.; Zeef, L.A.; Freemont, A.J.; Hoyland, J.A. Transcriptional profiling of bovine intervertebral disc cells: Implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res. Ther. 2010, 12, R22. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef]
- Li, M.; Zeringer, E.; Barta, T.; Schageman, J.; Cheng, A.; Vlassov, A.V. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef]
- Dowdell, J.; Erwin, M.; Choma, T.; Vaccaro, A.; Iatridis, J.; Cho, S.K. Intervertebral disk degeneration and repair. Neurosurgery 2017, 80, S46–S54. [Google Scholar] [CrossRef]
- Ma, K.; Chen, S.; Li, Z.; Deng, X.; Huang, D.; Xiong, L.; Shao, Z. Mechanisms of endogenous repair failure during intervertebral disc degeneration. Osteoarthr. Cartil. 2019, 27, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Yu, Q.; Li, H.; Guo, X.; He, X. Characterization of microRNA expression profiles in patients with intervertebral disc degeneration. Int. J. Mol. Med. 2014, 33, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Divi, S.N.; Markova, D.Z.; Fang, T.; Guzek, R.; Kurd, M.F.; Rihn, J.A.; Hilibrand, A.S.; Anderson, D.G.; Vaccaro, A.R.; Schroeder, G.D.; et al. Circulating miR-155-5p as a novel biomarker of lumbar degenerative disc disease. Spine (Phila Pa 1976) 2020, 45, E499–E507. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, Z.; Hao, J.; Shen, J. [Low-intensity pulsed ultrasound stimulates the extracellular matrix synthesis of human degenerative nucleus pulposus cells via activating PI3K/Akt pathway]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2013, 29, 34–38. [Google Scholar]
- Miao, D.; Zhang, L. Leptin modulates the expression of catabolic genes in rat nucleus pulposus cells through the mitogen-activated protein kinase and Janus kinase 2/signal transducer and activator of transcription 3 pathways. Mol. Med. Rep. 2015, 12, 1761–1768. [Google Scholar] [CrossRef]
- Hiyama, A.; Yokoyama, K.; Nukaga, T.; Sakai, D.; Mochida, J. Response to tumor necrosis factor-alpha mediated inflammation involving activation of prostaglandin E2 and Wnt signaling in nucleus pulposus cells. J. Orthop. Res. 2015, 33, 1756–1768. [Google Scholar] [CrossRef]
- Hiyama, A.; Yokoyama, K.; Nukaga, T.; Sakai, D.; Mochida, J. A complex interaction between Wnt signaling and TNF-alpha in nucleus pulposus cells. Arthritis Res. Ther. 2013, 15, R189. [Google Scholar] [CrossRef]
- Zhang, F.; Zhao, X.; Shen, H.; Zhang, C. Molecular mechanisms of cell death in intervertebral disc degeneration (Review). Int. J. Mol. Med. 2016, 37, 1439–1448. [Google Scholar] [CrossRef]
- Hummel, R.; Wang, T.; Watson, D.I.; Michael, M.Z.; Van der Hoek, M.; Haier, J.; Hussey, D.J. Chemotherapy-induced modification of microRNA expression in esophageal cancer. Oncol. Rep. 2011, 26, 1011–1017. [Google Scholar] [CrossRef]
- Sand, M.; Skrygan, M.; Georgas, D.; Sand, D.; Hahn, S.A.; Gambichler, T.; Altmeyer, P.; Bechara, F.G. Microarray analysis of microRNA expression in cutaneous squamous cell carcinoma. J. Dermatol. Sci. 2012, 68, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Li, C.F.; Chen, L.B.; Li, D.D.; Yang, L.; Jin, J.P.; Zhang, B. MicroRNA-766 targeting regulation of SOX6 expression promoted cell proliferation of human colorectal cancer. Onco Targets Ther. 2015, 8, 2981–2988. [Google Scholar] [CrossRef] [PubMed]
- Milagro, F.I.; Miranda, J.; Portillo, M.P.; Fernandez-Quintela, A.; Campion, J.; Martinez, J.A. High-throughput sequencing of microRNAs in peripheral blood mononuclear cells: Identification of potential weight loss biomarkers. PLoS ONE 2013, 8, e54319. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.; Akerstrom, T.; Rinnov, A.; Yfanti, C.; Scheele, C.; Pedersen, B.K.; Laye, M.J. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS ONE 2014, 9, e87308. [Google Scholar] [CrossRef]
- Liang, H.; Li, X.; Wang, L.; Yu, S.; Xu, Z.; Gu, Y.; Pan, Z.; Li, T.; Hu, M.; Cui, H.; et al. MicroRNAs contribute to promyelocyte apoptosis in As2O3-treated APL cells. Cell Physiol. Biochem. 2013, 32, 1818–1829. [Google Scholar] [CrossRef]
- Sharma, A.; Diecke, S.; Zhang, W.Y.; Lan, F.; He, C.; Mordwinkin, N.M.; Chua, K.F.; Wu, J.C. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. J. Biol. Chem. 2013, 288, 18439–18447. [Google Scholar] [CrossRef]
- Qian, Z.; Li, Y.; Chen, J.; Li, X.; Gou, D. miR-4632 mediates PDGF-BB-induced proliferation and antiapoptosis of human pulmonary artery smooth muscle cells via targeting cJUN. Am. J. Physiol. Cell Physiol. 2017, 313, C380–C391. [Google Scholar] [CrossRef]
- Hassanlou, M.; Soltani, B.M.; Medlej, A.; Kay, M.; Mowla, S.J. Hsa-miR-6165 downregulates insulin-like growth factor-1 receptor (IGF-1R) expression and enhances apoptosis in SW480 cells. Biol. Chem. 2020, 401, 477–485. [Google Scholar] [CrossRef]
- Walker, M.H.; Anderson, D.G. Molecular basis of intervertebral disc degeneration. Spine J. 2004, 4, 158S–166S. [Google Scholar] [CrossRef]
- Kepler, C.K.; Ponnappan, R.K.; Tannoury, C.A.; Risbud, M.V.; Anderson, D.G. The molecular basis of intervertebral disc degeneration. Spine J. 2013, 13, 318–330. [Google Scholar] [CrossRef]
- Bu, G.; Hou, S.; Ren, D.; Wu, Y.; Shang, W.; Huang, W. Increased expression of netrin-1 and its deleted in colorectal cancer receptor in human diseased lumbar intervertebral disc compared with autopsy control. Spine (Phila Pa 1976) 2012, 37, 2074–2081. [Google Scholar] [CrossRef]
- Hassanlou, M.; Soltani, B.M.; Mowla, S.J. Expression and function of hsa-miR-6165 in human cell lines and during the NT2 cell neural differentiation process. J. Mol. Neurosci. 2017, 63, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.K.; Hanson, D.A.; Schlaepfer, D.D. Focal adhesion kinase: In command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005, 6, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jia, X.; Duance, V.C.; Blain, E.J. The effects of cyclic tensile strain on the organisation and expression of cytoskeletal elements in bovine intervertebral disc cells: An in vitro study. Eur. Cells Mater. 2011, 21, 508–522. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, S.; Erickson, G.R.; Guilak, F. Hyperosmotically induced volume change and calcium signaling in intervertebral disk cells: The role of the actin cytoskeleton. Biophys. J. 2002, 83, 2502–2510. [Google Scholar] [CrossRef]
- Gao, B.; Li, S.; Tan, Z.; Ma, L.; Liu, J. ACTG1 and TLR3 are biomarkers for alcohol-associated hepatocellular carcinoma. Oncol. Lett. 2019, 17, 1714–1722. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Prado, S.; Cicione, C.; Muinos-Lopez, E.; Hermida-Gomez, T.; Oreiro, N.; Fernandez-Lopez, C.; Blanco, F.J. Characterization of microRNA expression profiles in normal and osteoarthritic human chondrocytes. BMC Musculoskelet. Disord. 2012, 13, 144. [Google Scholar] [CrossRef]
- Risbud, M.V.; Shapiro, I.M. Role of cytokines in intervertebral disc degeneration: Pain and disc content. Nat. Rev. Rheumatol. 2014, 10, 44–56. [Google Scholar] [CrossRef]
- Purmessur, D.; Walter, B.A.; Roughley, P.J.; Laudier, D.M.; Hecht, A.C.; Iatridis, J. A role for TNFalpha in intervertebral disc degeneration: A non-recoverable catabolic shift. Biochem. Biophys. Res. Commun. 2013, 433, 151–156. [Google Scholar] [CrossRef]
- Shen, C.; Yan, J.; Jiang, L.S.; Dai, L.Y. Autophagy in rat annulus fibrosus cells: Evidence and possible implications. Arthritis Res. Ther. 2011, 13, R132. [Google Scholar] [CrossRef]
- Zou, W.; Ding, F.; Niu, C.; Fu, Z.; Liu, S. Brg1 aggravates airway inflammation in asthma via inhibition of the PI3K/Akt/mTOR pathway. Biochem. Biophys. Res. Commun. 2018, 503, 3212–3218. [Google Scholar] [CrossRef]
- Du, C.; Zhang, T.; Xiao, X.; Shi, Y.; Duan, H.; Ren, Y. Protease-activated receptor-2 promotes kidney tubular epithelial inflammation by inhibiting autophagy via the PI3K/Akt/mTOR signalling pathway. Biochem. J. 2017, 474, 2733–2747. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shen, J.; Wu, W.K.; Yu, X.; Liang, J.; Qiu, G.; Liu, J. Leptin induces cyclin D1 expression and proliferation of human nucleus pulposus cells via JAK/STAT, PI3K/Akt and MEK/ERK pathways. PLoS ONE 2012, 7, e53176. [Google Scholar] [CrossRef] [PubMed]
- Krupkova, O.; Handa, J.; Hlavna, M.; Klasen, J.; Ospelt, C.; Ferguson, S.J.; Wuertz-Kozak, K. The natural polyphenol epigallocatechin gallate protects intervertebral disc cells from oxidative stress. Oxidative Med. Cell. Longev. 2016, 2016, 7031397. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.R.; Zhang, Z.; Gao, M.; Li, L.; Sun, P.Y.; Xu, L.N.; Qi, Y.; Yin, L.H.; Peng, J.Y. MicroRNA-27a-3p aggravates renal ischemia/reperfusion injury by promoting oxidative stress via targeting growth factor receptor-bound protein 2. Pharmacol. Res. 2020, 155, 104718. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Kawasaki, M.; Hirai, T.; Yoshida, Y.; Tsushima, H.; Fujishiro, M.; Ikeda, K.; Morimoto, S.; Takamori, K.; Sekigawa, I. MicroRNA-766-3p contributes to anti-inflammatory responses through the indirect inhibition of NF-kappaB signaling. Int. J. Mol. Sci. 2019, 20, 809. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Huang, Y.; Liang, Z.; Liu, D.; Lu, Y.; Dai, Y.; Feng, G.; Wang, C. Plasma miRNAs might be promising biomarkers of chronic obstructive pulmonary disease. Clin. Respir. J. 2016, 10, 104–111. [Google Scholar] [CrossRef]
- Cazzanelli, P.; Wuertz-Kozak, K. MicroRNAs in intervertebral disc degeneration, apoptosis, inflammation, and mechanobiology. Int. J. Mol. Sci. 2020, 21, 3601. [Google Scholar] [CrossRef]

| Sample | Gender | Age (Years) | Clinical Diagnosis |
|---|---|---|---|
| DD 1 | male | 58 | Lumbar disc herniation |
| DD 2 | male | 83 | Lumbar disc herniation |
| DD 3 | female | 67 | Lumbar disc herniation |
| DD 4 | male | 43 | Lumbar disc herniation |
| DD 5 | male | 32 | Lumbar disc herniation |
| DD 6 | male | 32 | Lumbar disc herniation |
| DD 7 | female | 20 | Lumbar disc herniation |
| DD 8 | male | 27 | Lumbar disc herniation |
| DD 9 | male | 56 | Lumbar disc herniation |
| DD 10 | female | 63 | Lumbar disc herniation |
| Control 1 | female | 15 | Idiopathic scoliosis |
| Control 2 | female | 26 | C2 Hangman fracture |
| Control 3 | female | 14 | Idiopathic scoliosis |
| Control 4 | male | 31 | Lumbar spine tuberculosis infection |
| Control 5 | female | 49 | T3 vertebrae body fracture |
| Control 6 | male | 11 | Idiopathic scoliosis |
| Control 7 | female | 22 | Idiopathic scoliosis |
| Control 8 | female | 26 | Idiopathic scoliosis |
| Control 9 | male | 12 | Idiopathic scoliosis |
| Control 10 | male | 26 | Spine hemangioma |
| Sample | No. of Total Reads | No. of Clean Reads | Q20 | Q30 | Mapped Known Total Small RNA |
|---|---|---|---|---|---|
| DD 1 | 29,947,800 | 18,808,543 (62.80%) | 98.72% | 93.50% | 1,428,828 |
| DD 2 | 24,083,779 | 216,430,52 (89.87%) | 98.94% | 94.67% | 426,634 |
| DD 3 | 48,583,019 | 45,634,320 (93.93%) | 98.91% | 94.62% | 1,468,324 |
| DD 4 | 47,625,147 | 41,735,829 (87.63%) | 98.91% | 94.02% | 12,845,203 |
| DD 5 | 50,936,612 | 46,158,268 (90.62%) | 98.69% | 93.32% | 7,900,045 |
| DD 6 | 28,051,208 | 16,648,215 (59.35%) | 98.81% | 94.20% | 1,448,967 |
| DD 7 | 81,098,971 | 67,786,634 (83.59%) | 98.98% | 95.11% | 2,177,393 |
| DD 8 | 50,374,765 | 44,617,225 (88.57%) | 98.91% | 93.87% | 1,746,933 |
| DD 9 | 54,017,791 | 51,321,862 (95.01%) | 99.12% | 97.56% | 4,259,498 |
| DD 10 | 38,095,238 | 31,881,721 (83.69%) | 98.64% | 96.35% | 7,543,771 |
| Control 1 | 55,492,708 | 47,926,094 (86.36%) | 99.06% | 94.58% | 681,164 |
| Control 2 | 50,594,530 | 46,373,575 (91.66%) | 98.83% | 93.58% | 3,380,139 |
| Control 3 | 37,387,520 | 25,946,219 (69.40%) | 98.91% | 94.44% | 856,419 |
| Control 4 | 44,852,156 | 37,645,238 (83.93%) | 98.98% | 94.14% | 1,284,731 |
| Control 5 | 20,785,656 | 45,634,320 (93.93%) | 98.91% | 94.48% | 234,324 |
| Control 6 | 38,977,001 | 35,108,194 (90.07%) | 98.89% | 97.31% | 2,494,358 |
| Control 7 | 25,745,004 | 19,298,221 (74.96%) | 99.04% | 94.28% | 299,844 |
| Control 8 | 33,928,731 | 30,275,015 (89.23%) | 98.99% | 93.80% | 6,523,253 |
| Control 9 | 55,606,502 | 49,864,823 (89.67%) | 99.12% | 94.07% | 3,003,336 |
| Control 10 | 33,322,179 | 27,434,382 (82.33%) | 99.01% | 94.17% | 399,765 |
| Upregulated miRNA | log2 Fold Change | p-Value | p-adj |
|---|---|---|---|
| hsa-miR-4685-3p | 4.3478 | 7.24 × 10−7 | 0.000241 |
| hsa-miR-766-3p | 3.0624 | 4.42 × 10−6 | 0.000458 |
| hsa-miR-3605-3p | 3.1164 | 7.75 × 10−6 | 0.000574 |
| hsa-miR-6749-3p | 4.667 | 0.000152 | 0.005617 |
| hsa-miR-1227-3p | 4.4573 | 0.0002 | 0.00666 |
| hsa-miR-6726-3p | 6.0323 | 0.000518 | 0.011509 |
| hsa-miR-877-3p | 4.2815 | 0.000493 | 0.011509 |
| hsa-miR-197-3p | 2.5895 | 0.000699 | 0.013684 |
| hsa-miR-6819-3p | 2.9766 | 0.001757 | 0.02489 |
| hsa-miR-3620-3p | 4.9605 | 0.00228 | 0.028649 |
| hsa-miR-1908-3p | 3.0883 | 0.002361 | 0.029119 |
| hsa-miR-211-5p | 4.4374 | 0.002771 | 0.03296 |
| hsa-miR-342-3p | 2.0702 | 0.003209 | 0.034472 |
| hsa-miR-130b-5p | 2.2154 | 0.003278 | 0.034655 |
| hsa-miR-654-5p | 2.223 | 0.003642 | 0.037897 |
| hsa-miR-6894-3p | 4.6656 | 0.004032 | 0.03892 |
| hsa-miR-543 | 3.4698 | 0.005086 | 0.046401 |
| Downregulated miRNA | log2 Fold Change | p-Value | p-adj |
| hsa-miR-4452 | −7.819 | 2.07 × 10−7 | 0.000138 |
| hsa-miR-1-3p | −4.3578 | 1.81 × 10−6 | 0.000304 |
| hsa-miR-5006-5p | −6.8699 | 1.83 × 10−6 | 0.000304 |
| hsa-miR-6165 | −9.193 | 2.44 × 10−6 | 0.000325 |
| hsa-miR-449a | −9.3266 | 5.5 × 10−6 | 0.000458 |
| hsa-miR-4632-5p | −7.6058 | 4.86 × 10−6 | 0.000458 |
| hsa-miR-1303 | −4.4186 | 3.37 × 10−5 | 0.002007 |
| hsa-miR-219a-2-3p | −5.8958 | 3.31 × 10−5 | 0.002007 |
| hsa-miR-4301 | −5.618 | 3.62 × 10−5 | 0.002007 |
| hsa-miR-27a-3p | −3.0627 | 5.02 × 10−5 | 0.002572 |
| hsa-miR-15b-5p | −3.7083 | 6.13 × 10−5 | 0.002722 |
| hsa-miR-566 | −6.0829 | 5.77 × 10−5 | 0.002722 |
| hsa-miR-5096 | −2.8231 | 9.08 × 10−5 | 0.003781 |
| hsa-miR-1285-5p | −3.0624 | 0.000109 | 0.004272 |
| hsa-miR-6859-3p | −5.8857 | 0.000181 | 0.006354 |
| hsa-miR-181c-3p | −5.4625 | 0.000238 | 0.006678 |
| hsa-miR-200b-3p | −4.8407 | 0.000211 | 0.006678 |
| hsa-miR-320c | −2.6456 | 0.000241 | 0.006678 |
| hsa-miR-4762-3p | −6.4726 | 0.000228 | 0.006678 |
| hsa-miR-5197-5p | −7.0974 | 0.000325 | 0.008651 |
| hsa-miR-320d | −3.0007 | 0.000338 | 0.008669 |
| hsa-miR-7847-3p | −5.6968 | 0.000396 | 0.009769 |
| hsa-miR-4261 | −6.8466 | 0.000514 | 0.011509 |
| hsa-miR-125b-5p | −3.7142 | 0.000542 | 0.011643 |
| hsa-miR-1290 | −2.4541 | 0.000669 | 0.013494 |
| hsa-miR-4488 | −4.5272 | 0.000667 | 0.013494 |
| hsa-miR-762 | −5.9577 | 0.00074 | 0.014083 |
| hsa-miR-5591-3p | −7.439 | 0.00087 | 0.015669 |
| hsa-miR-5591-5p | −7.439 | 0.00087 | 0.015669 |
| hsa-miR-7704 | −3.2961 | 0.000975 | 0.017088 |
| hsa-miR-548ba | −6.598 | 0.001137 | 0.018927 |
| hsa-miR-548d-5p | −6.3677 | 0.001123 | 0.018927 |
| hsa-miR-99a-5p | −1.7454 | 0.001213 | 0.019711 |
| hsa-miR-3178 | −4.214 | 0.001295 | 0.020535 |
| hsa-miR-29a-3p | −3.5301 | 0.001448 | 0.02242 |
| hsa-miR-1273c | −3.6329 | 0.001542 | 0.023337 |
| hsa-miR-195-5p | −3.8204 | 0.001641 | 0.024281 |
| hsa-miR-16-5p | −1.9705 | 0.001687 | 0.024423 |
| hsa-miR-1246 | −2.304 | 0.002231 | 0.028649 |
| hsa-miR-320b | −2.0358 | 0.002273 | 0.028649 |
| hsa-miR-3653-3p | −4.9585 | 0.002241 | 0.028649 |
| hsa-miR-4721 | −7.0703 | 0.002176 | 0.028649 |
| hsa-miR-6849-3p | −6.8278 | 0.002274 | 0.028649 |
| hsa-miR-1181 | −4.7855 | 0.002671 | 0.03234 |
| hsa-miR-141-3p | −3.2258 | 0.003021 | 0.034104 |
| hsa-miR-26b-5p | −2.6011 | 0.002971 | 0.034104 |
| hsa-miR-616-3p | −3.7777 | 0.002987 | 0.034104 |
| hsa-miR-199a-3p | −1.9663 | 0.003127 | 0.03414 |
| hsa-miR-6728-3p | −6.8316 | 0.003115 | 0.03414 |
| hsa-miR-4484 | −6.6985 | 0.003787 | 0.038356 |
| hsa-miR-450a-5p | −3.6107 | 0.003801 | 0.038356 |
| hsa-miR-143-3p | −2.002 | 0.004012 | 0.03892 |
| hsa-miR-4516 | −4.2535 | 0.003987 | 0.03892 |
| hsa-miR-144-5p | −5.0596 | 0.00481 | 0.045767 |
| hsa-miR-340-5p | −5.3597 | 0.00503 | 0.046401 |
| hsa-miR-5010-5p | −3.1516 | 0.005051 | 0.046401 |
| Pathway | miRNAs Upregulated | Target Genes | miRNAs Downregulated | Target Genes | ||
|---|---|---|---|---|---|---|
| endocytosis | miR-766-3p | miR-4685-3p | ARAP1, ZFYVE27, HSPA6, PSD4, ARF1, VPS37C, TGFBR1, IGF1R, PSD3, CBL | miR-4632-5p | miR-5006-5p | MARC1 |
| apoptosis | miR-766-3p | miR-6749-3p | CTSK, RIPK1, HRK, ERN1, CASP10, PIK3CD, ATM, TRAF2, MAPK1, TRAF1 | miR-4632-5p | miR-6165 | PDPK1, NRAS, RELA, CASP2, PIK3CB, CASP10, AKT2, HRK, TRAF2, XIAP |
| axon guidance | miR-766-3p | miR-6749-3p | Clorf220, SLIT3, SSH3, WNT4, HRAS, LIMK1, ENAH, C15orf37, PIK3R1, NTN1 | miR-4632-5p | miR-5006-5p | NRAS, PAK4, SEMA4F, RYK, PIK3CB, PLXNA3, ABL1, PAK3, SEMA4A, PLCG1 |
| VEGF | miR-766-3p | miR-6749-3p | Clorf220, MAPK13, HRAS, C15orf37, PIK3R1, MAPK14, PLCG1, SHPK, CDC42, PRKCA | miR-4632-5p | miR-1303 | MAPKAPK2, NRAS, AKT2, PPP3CA, PLCG2, PIK3R3, MAPK1, MAPK13, AKT1, MARC1 |
| regulation of cytoskeleton | miR-766-3p | miR-6749-3p | Clorf220, SSH3, HRAS, ARHGEF7, ITGA8, LIMK1, ENAH, ITGA11, C15orf37, ITGAX | miR-4452 | miR-1303 | COLGA6L9, MARCH1, MRC1 |
| cell adhesion molecular | miR-766-3p | miR-6749-3p | Clorf220, CD226, F11R, ICOS, ITGA8, C15orf37, TIGIT, NRXN2, JAM2, SDC4 | miR-4632-5p | miR-5006-5p | F11R, MPZ, PVRL1, SDC3, CLDN18, SDC2, NFASC, MPZL1, SPN, ITGA9 |
| focal adhesion | miR-766-3p | miR-6749-3p | Clorf220, HRAS, IGF1, ITGA8, TLN2, ITGA11, TNR, C15orf37, PIK3R1, LAMC2 | miR-4632-5p | miR-6165 | IGF1R, PDGFRB, PDPK1, BAB2, VCL, PAK4, PARVA, PIK3CB, VEGFA, ITGA9 |
| ECM-receptor interaction | miR-766-3p | miR-6749-3p | Clorf220, DAG1, ITGA8, ITGA11, TNR, C15orf37, LAMC2, SV2B, SDC4, SHPK | miR-4632-5p | miR-5006-5p | COL6A1, TNR, COL4A4, ITGA1, ITGA3, LAMC3, ITGA8, ITGA9, Sv2c |
| PI3K-AKT | miR-766-3p | miR-6749-3p | Clorf220, HRAS, IGF1, JAK3, ITGA8, INSR, GNB4, GNG4, ITGA11, TNR | miR-4632-5p | miR-1303 | FGF23, NRAS, RELA, CDKN1B, CCNE2, PPP2R5C, FGF14, ITGA9, COL4A4, AKT2 |
| mTOR | miR-766-3p | miR-6749-3p | Clorf220, WNT9B, WNT4, HRAS, IGF1, WNT2B, INSR, C15orf37, PIK3R1, GRB2 | miR-5006-5p | miR-6165 | WNT2B, PDPK1, PIK3CB, MAPK1, FZD8, GOLGA6L9, WNT7B, RPS6KA2, NPRL3, WNT3 |
| NF-Kb | miR-766-3p | miR-6749-3p | PLCG1, TMEM236 | miR-4632-5p | miR-5006-5p | MYD88, TNFRSF13C, TRIM25, RELA, IL1R1, TNFAIP3, TNFRSF11A, TRAF3, PLCG2, TRAF1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, S.; Zhou, Z.; Liu, X.; Richards, R.G.; Alini, M.; Peng, S.; Liu, S.; Zou, X.; Li, Z.; Grad, S. Identification and Characterization of Serum microRNAs as Biomarkers for Human Disc Degeneration: An RNA Sequencing Analysis. Diagnostics 2020, 10, 1063. https://doi.org/10.3390/diagnostics10121063
Cui S, Zhou Z, Liu X, Richards RG, Alini M, Peng S, Liu S, Zou X, Li Z, Grad S. Identification and Characterization of Serum microRNAs as Biomarkers for Human Disc Degeneration: An RNA Sequencing Analysis. Diagnostics. 2020; 10(12):1063. https://doi.org/10.3390/diagnostics10121063
Chicago/Turabian StyleCui, Shangbin, Zhiyu Zhou, Xizhe Liu, Robert Geoff Richards, Mauro Alini, Songlin Peng, Shaoyu Liu, Xuenong Zou, Zhen Li, and Sibylle Grad. 2020. "Identification and Characterization of Serum microRNAs as Biomarkers for Human Disc Degeneration: An RNA Sequencing Analysis" Diagnostics 10, no. 12: 1063. https://doi.org/10.3390/diagnostics10121063
APA StyleCui, S., Zhou, Z., Liu, X., Richards, R. G., Alini, M., Peng, S., Liu, S., Zou, X., Li, Z., & Grad, S. (2020). Identification and Characterization of Serum microRNAs as Biomarkers for Human Disc Degeneration: An RNA Sequencing Analysis. Diagnostics, 10(12), 1063. https://doi.org/10.3390/diagnostics10121063







