Fully Automated Breast Density Segmentation and Classification Using Deep Learning
Abstract
1. Introduction
- Developing an effective conditional Generative Adversarial Network for segmenting the regions of dense tissues in a mammogram.
- Utilizing the ratio of the dense segmented regions (i.e., resulting in from the cGAN network) to the breasts total area. The computed percentage is used for classifying the mammogram into four different classes of the BI-RADS standard (i.e., fatty, scattered fibroglandular density, heterogeneously dense, and extremely dense).
- Developing a multi-class CNN architecture for breast density classification using the binary masks obtained from the cGAN.
2. Background Study
2.1. Traditional Computer Vision Methods
2.2. Deep Learning-Based Methods
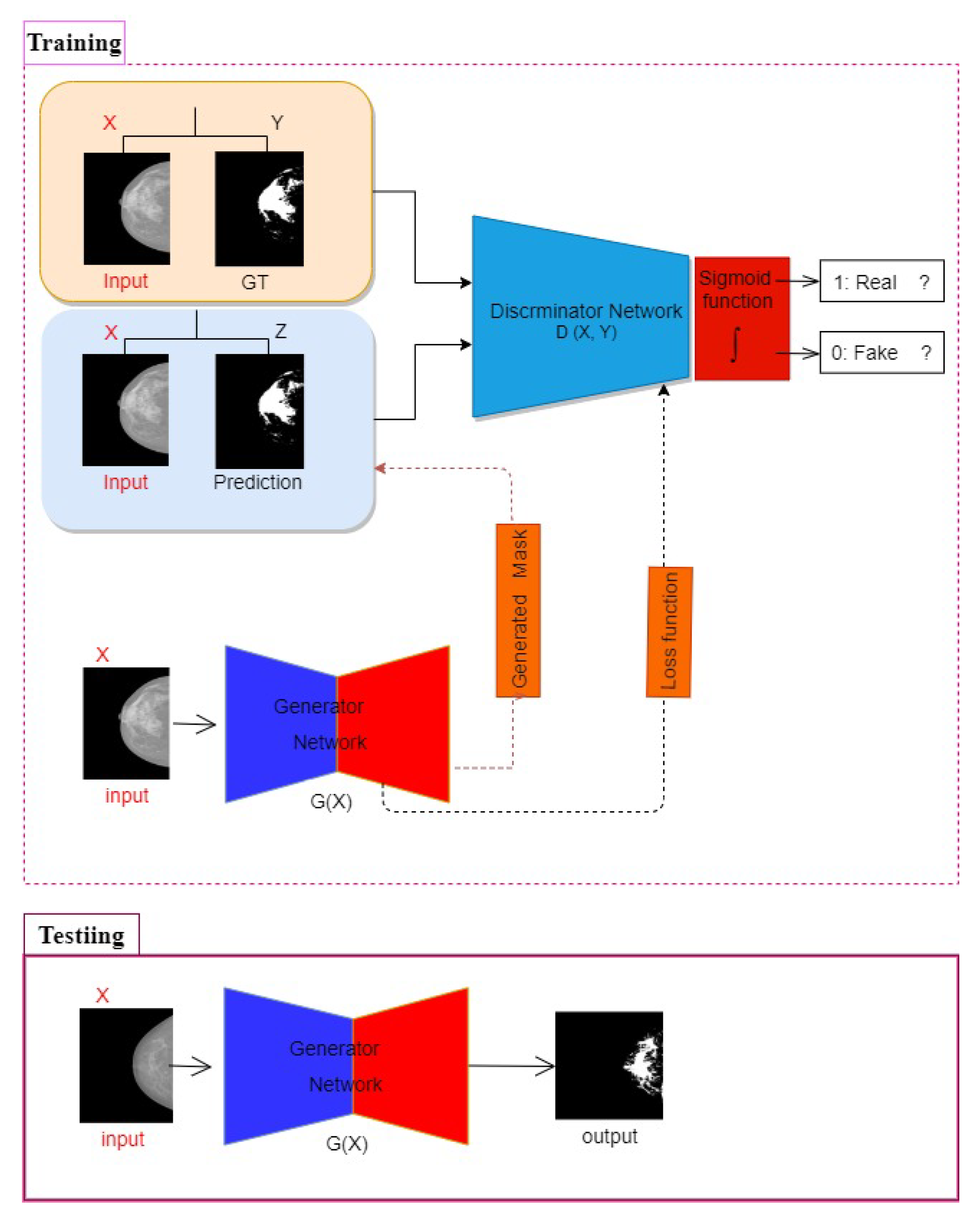
- The first adaptation of cGAN in the area of fully automated breast density segmentation in mammograms is developed,
- the breast density percentage classification by the developed multi-class CNN architecture correlated well with BI-RADS density ratings (BI-RADS I, BI-RADS II, BI-RADS III, and BI-RADS IV) using the binary mask segmented in the previous stage (cGAN output) by radiologists,
- a strong correspondence between the output of our automated algorithm and radiologist’s presented breast density measures can be obtained, and
- the proposed approach results in remarkably faster calculation while improving the classification efficiency compare to other methods in the literature
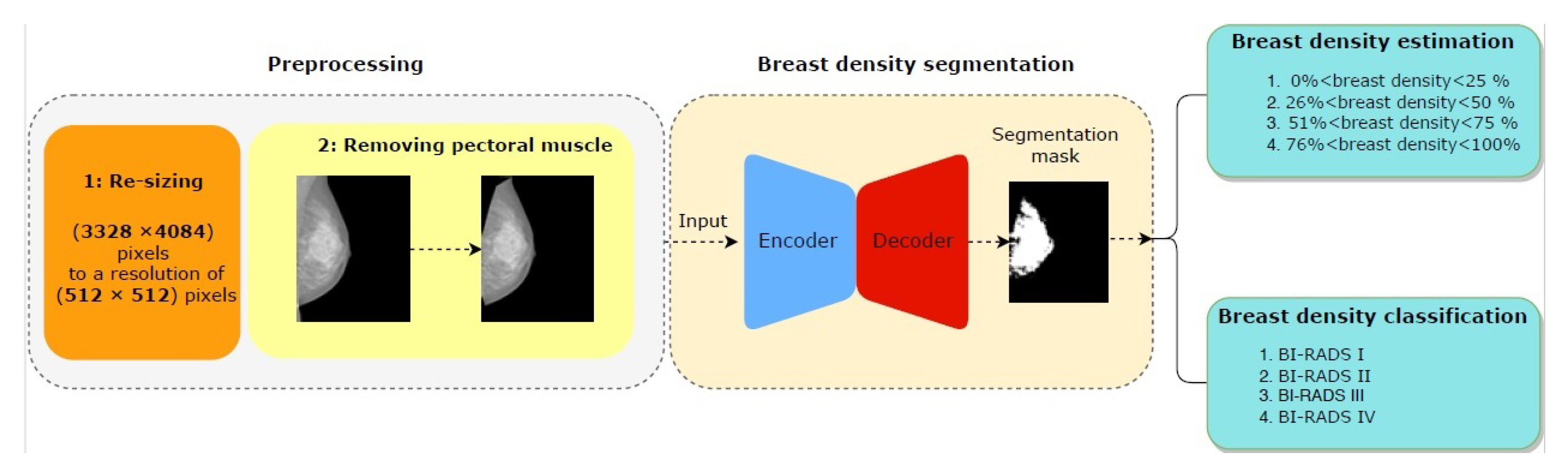
3. Methodology
3.1. Preprocessing
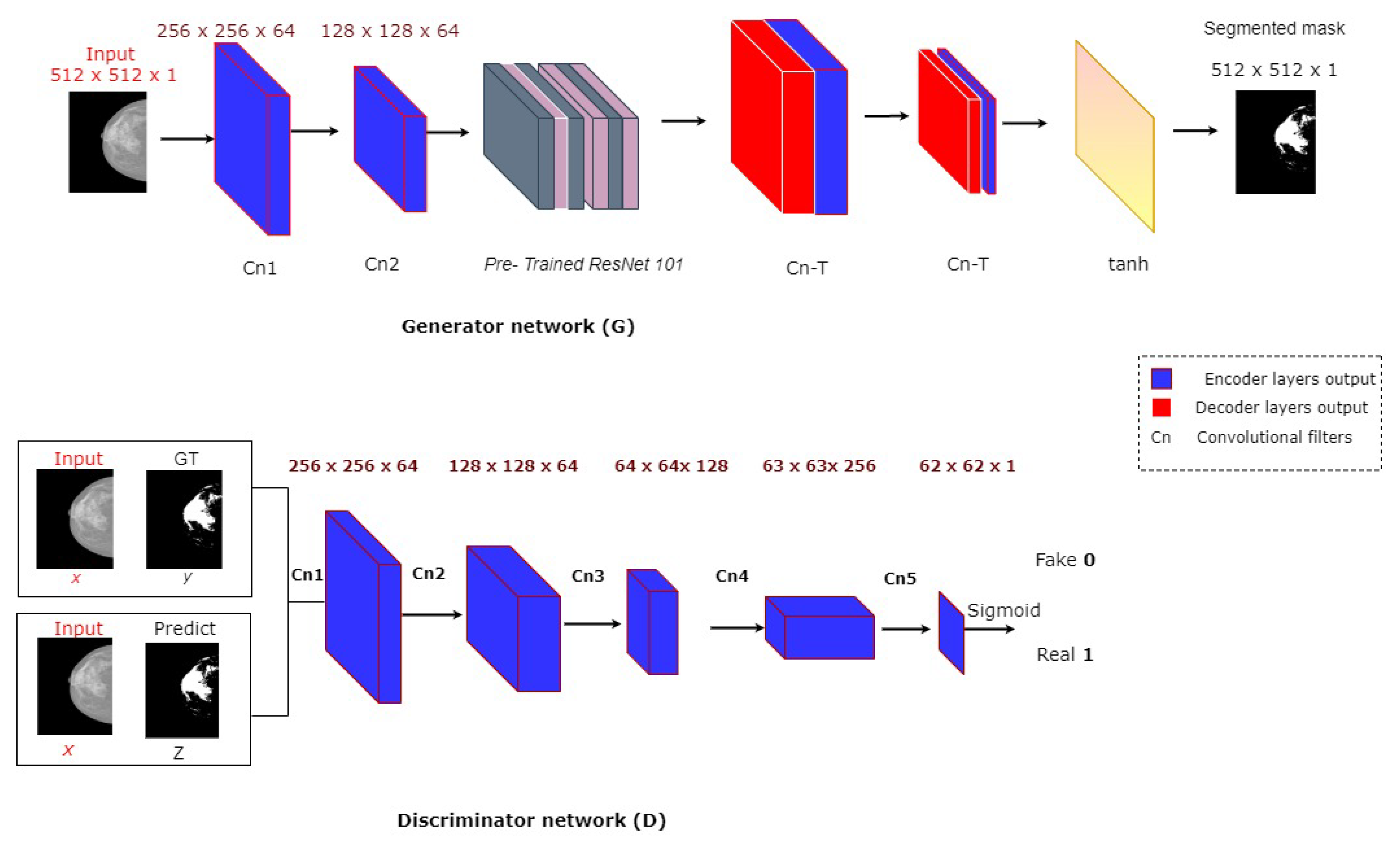
3.2. Breast Density Segmentation
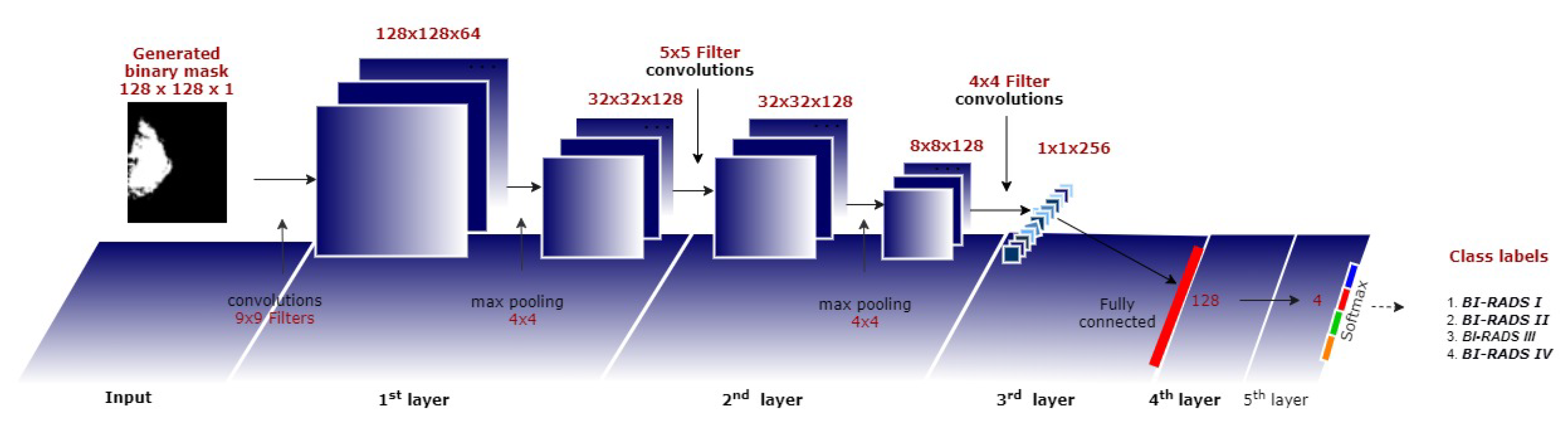
3.3. Breast Density Classification
3.3.1. Breast Density Percentage Estimation Based on Traditional Method
- First, we resize the generated mask images to the same resolution of the input mammography.
- We express the breast region area by the number of non-zero pixels in the mammogram images.
- We then count the non-zero pixels in the generated mask for expressing about the area dense tissues.
- Computing the ratio between the area of dense tissues and the area of the breast region to estimate the breast density in the input image.
- Finally by the thresholding rules defined in the BI-RADS density standard, we classify the breast density to 4 categories: ( < BDE <), ( < BDE < ), ( < BDE < ), ( < BDE < ).
3.3.2. Breast Density Classification Based on a CNN
4. Experimental Results
4.1. Dataset
4.2. Implementation Details
Evaluation Metrics of Breast Density
- True positive (TP) instances are gold standard class 1 predicted as class 1
- False-negative (FN) instances are gold standard class 1 predicted as class 2, 3, or 4
- False-positive (FP) instances are gold standard class 2, 3, or 4 predicted as class 1
- True negative (TN) instances are gold standard class 2,3 or 4 predicted as class 2, 3, or 4 (here errors do not matter as long as class 1 is not involved)
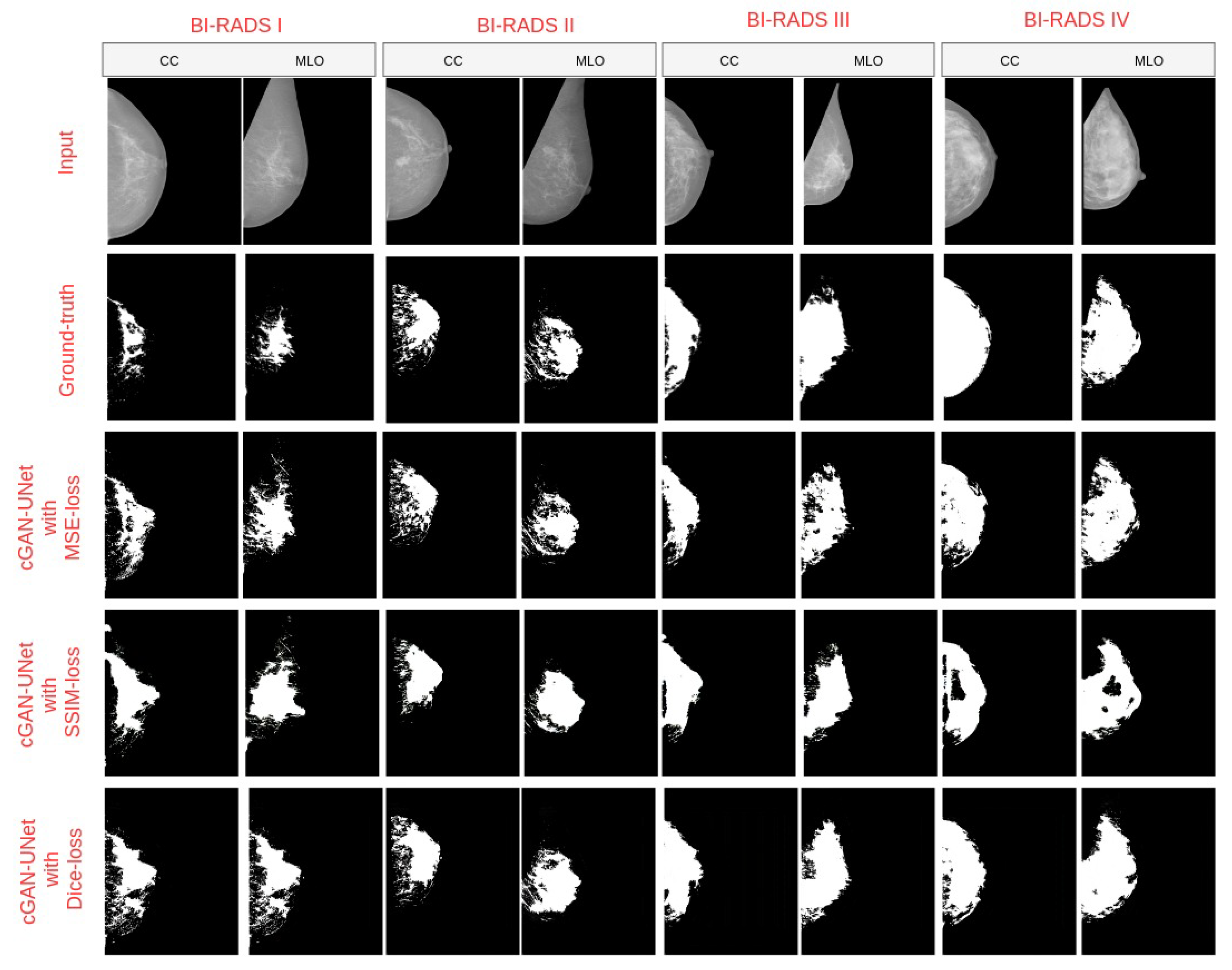
4.3. Breast Density Segmentation
4.4. Breast Density Classification
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abdel-Nasser, M.; Rashwan, H.A.; Puig, D.; Moreno, A. Analysis of tissue abnormality and breast density in mammographic images using a uniform local directional pattern. Expert Syst. Appl. 2015, 42, 9499–9511. [Google Scholar]
- Abbas, Q. DeepCAD: A computer-aided diagnosis system for mammographic masses using deep invariant features. Computers 2016, 5, 28. [Google Scholar]
- Astley, S.M.; Harkness, E.F.; Sergeant, J.C.; Warwick, J.; Stavrinos, P.; Warren, R.; Wilson, M.; Beetles, U.; Gadde, S.; Lim, Y.; et al. A comparison of five methods of measuring mammographic density: A case-control study. Breast Cancer Res. 2018, 20, 10. [Google Scholar] [PubMed]
- Keller, B.M.; Nathan, D.L.; Wang, Y.; Zheng, Y.; Gee, J.C.; Conant, E.F.; Kontos, D. Estimation of breast percent density in raw and processed full field digital mammography images via adaptive fuzzy c-means clustering and support vector machine segmentation. Med. Phys. 2012, 39, 4903–4917. [Google Scholar] [PubMed]
- Sprague, B.L.; Stout, N.K.; Schechter, C.; Van Ravesteyn, N.T.; Cevik, M.; Alagoz, O.; Lee, C.I.; Van Den Broek, J.J.; Miglioretti, D.L.; Mandelblatt, J.S.; et al. Benefits, harms, and cost-effectiveness of supplemental ultrasonography screening for women with dense breasts. Ann. Intern. Med. 2015, 162, 157–166. [Google Scholar] [PubMed]
- Wolfe, J.N. Breast patterns as an index of risk for developing breast cancer. Am. J. Roentgenol. 1976, 126, 1130–1137. [Google Scholar]
- Gram, I.T.; Funkhouser, E.; Tabár, L. The Tabar classification of mammographic parenchymal patterns. Eur. J. Radiol. 1997, 24, 131–136. [Google Scholar]
- McCormack, V.A.; dos Santos Silva, I. Breast density and parenchymal patterns as markers of breast cancer risk: A meta-analysis. Cancer Epidemiol. Prev. Biomarkers 2006, 15, 1159–1169. [Google Scholar]
- Boyd, N.F.; Lockwood, G.A.; Byng, J.W.; Tritchler, D.L.; Yaffe, M.J. Mammographic densities and breast cancer risk. Cancer Epidemiol. Prev. Biomarkers 1998, 7, 1133–1144. [Google Scholar]
- Youk, J.H.; Gweon, H.M.; Son, E.J.; Kim, J.A. Automated volumetric breast density measurements in the era of the BI-RADS fifth edition: A comparison with visual assessment. Am. J. Roentgenol. 2016, 206, 1056–1062. [Google Scholar]
- Lee, J.; Nishikawa, R.M. Automated mammographic breast density estimation using a fully convolutional network. Med. Phys. 2018, 45, 1178–1190. [Google Scholar] [PubMed]
- Kwok, S.M.; Chandrasekhar, R.; Attikiouzel, Y.; Rickard, M.T. Automatic pectoral muscle segmentation on mediolateral oblique view mammograms. IEEE Trans. Med. Imaging 2004, 23, 1129–1140. [Google Scholar] [PubMed]
- Tzikopoulos, S.D.; Mavroforakis, M.E.; Georgiou, H.V.; Dimitropoulos, N.; Theodoridis, S. A fully automated scheme for mammographic segmentation and classification based on breast density and asymmetry. Comput. Methods Programs Biomed. 2011, 102, 47–63. [Google Scholar] [PubMed]
- Nickson, C.; Arzhaeva, Y.; Aitken, Z.; Elgindy, T.; Buckley, M.; Li, M.; English, D.R.; Kavanagh, A.M. AutoDensity: An automated method to measure mammographic breast density that predicts breast cancer risk and screening outcomes. Breast Cancer Res. 2013, 15, R80. [Google Scholar]
- Kim, Y.; Kim, C.; Kim, J.H. Automated Estimation of Breast Density on Mammogram Using Combined Information of Histogram Statistics and Boundary Gradients; Medical Imaging 2010: Computer-Aided Diagnosis; International Society for Optics and Photonics: San Diego, CA, USA, 2010; Volume 7624, p. 76242F. [Google Scholar]
- Rouhi, R.; Jafari, M.; Kasaei, S.; Keshavarzian, P. Benign and malignant breast tumors classification based on region growing and CNN segmentation. Expert Syst. Appl. 2015, 42, 990–1002. [Google Scholar]
- Nagi, J.; Kareem, S.A.; Nagi, F.; Ahmed, S.K. Automated breast profile segmentation for ROI detection using digital mammograms. In Proceedings of the 2010 IEEE EMBS Conference on Biomedical Engineering and Sciences (IECBES), Kuala Lumpur, Malaysia, 30 November–2 December 2010; pp. 87–92. [Google Scholar]
- Zwiggelaar, R. Local greylevel appearance histogram based texture segmentation. In International Workshop on Digital Mammography; Springer: Berlin, Germany, 2010; pp. 175–182. [Google Scholar]
- Oliver, A.; Lladó, X.; Pérez, E.; Pont, J.; Denton, E.R.; Freixenet, J.; Martí, J. A statistical approach for breast density segmentation. J. Digit. Imaging 2010, 23, 527–537. [Google Scholar]
- Matsuyama, E.; Takehara, M.; Tsai, D.Y. Using a Wavelet-Based and Fine-Tuned Convolutional Neural Network for Classification of Breast Density in Mammographic Images. Open J. Med Imaging 2020, 10, 17. [Google Scholar]
- Gandomkar, Z.; Suleiman, M.E.; Demchig, D.; Brennan, P.C.; McEntee, M.F. BI-RADS density categorization using deep neural networks. In Medical Imaging 2019: Image Perception, Observer Performance, and Technology Assessment; International Society for Optics and Photonics: Bellingham WA, USA, 2019; Volume 10952, p. 109520N. [Google Scholar]
- Lehman, C.D.; Yala, A.; Schuster, T.; Dontchos, B.; Bahl, M.; Swanson, K.; Barzilay, R. Mammographic breast density assessment using deep learning: Clinical implementation. Radiology 2019, 290, 52–58. [Google Scholar]
- Chan, H.P.; Helvie, M.A. Deep learning for mammographic breast density assessment and beyond. Radiology 2019. [Google Scholar] [CrossRef]
- Byng, J.W.; Boyd, N.; Fishell, E.; Jong, R.; Yaffe, M.J. The quantitative analysis of mammographic densities. Phys. Med. Biol. 1994, 39, 1629. [Google Scholar]
- Sickles, E.A.; D’Orsi, C.J.; Bassett, L.W.; et al. ACR BI-RADS® Mammography. In ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar]
- Ciatto, S.; Bernardi, D.; Calabrese, M.; Durando, M.; Gentilini, M.A.; Mariscotti, G.; Monetti, F.; Moriconi, E.; Pesce, B.; Roselli, A.; et al. A first evaluation of breast radiological density assessment by QUANTRA software as compared to visual classification. Breast 2012, 21, 503–506. [Google Scholar] [PubMed]
- Highnam, R.; Brady, M.; Yaffe, M.J.; Karssemeijer, N.; Harvey, J. Robust breast composition measurement-Volpara TM. In International Workshop on Digital Mammography; Springer: Berlin, Germany, 2010; pp. 342–349. [Google Scholar]
- Seo, J.; Ko, E.; Han, B.K.; Ko, E.Y.; Shin, J.H.; Hahn, S.Y. Automated volumetric breast density estimation: A comparison with visual assessment. Clin. Radiol. 2013, 68, 690–695. [Google Scholar]
- Byng, J.; Boyd, N.; Fishell, E.; Jong, R.; Yaffe, M. Automated analysis of mammographic densities. Phys. Med. Biol. 1996, 41, 909. [Google Scholar] [PubMed]
- Boyd, N.F.; Martin, L.J.; Bronskill, M.; Yaffe, M.J.; Duric, N.; Minkin, S. Breast tissue composition and susceptibility to breast cancer. J. Natl. Cancer Inst. 2010, 102, 1224–1237. [Google Scholar] [PubMed]
- Kallenberg, M.; Petersen, K.; Nielsen, M.; Ng, A.Y.; Diao, P.; Igel, C.; Vachon, C.M.; Holland, K.; Winkel, R.R.; Karssemeijer, N.; et al. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans. Med. Imaging 2016, 35, 1322–1331. [Google Scholar]
- Dalmış, M.U.; Litjens, G.; Holland, K.; Setio, A.; Mann, R.; Karssemeijer, N.; Gubern-Mérida, A. Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Med. Phys. 2017, 44, 533–546. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Mohamed, A.A.; Luo, Y.; Peng, H.; Jankowitz, R.C.; Wu, S. Understanding clinical mammographic breast density assessment: A deep learning perspective. J. Digit. Imaging 2018, 31, 387–392. [Google Scholar]
- Mohamed, A.A.; Berg, W.A.; Peng, H.; Luo, Y.; Jankowitz, R.C.; Wu, S. A deep learning method for classifying mammographic breast density categories. Med. Phys. 2018, 45, 314–321. [Google Scholar]
- Li, S.; Wei, J.; Chan, H.P.; Helvie, M.A.; Roubidoux, M.A.; Lu, Y.; Zhou, C.; Hadjiiski, L.M.; Samala, R.K. Computer-aided assessment of breast density: Comparison of supervised deep learning and feature-based statistical learning. Phys. Med. Biol. 2018, 63, 025005. [Google Scholar] [PubMed]
- Dubrovina, A.; Kisilev, P.; Ginsburg, B.; Hashoul, S.; Kimmel, R. Computational mammography using deep neural networks. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2018, 6, 243–247. [Google Scholar]
- Ciritsis, A.; Rossi, C.; Vittoria De Martini, I.; Eberhard, M.; Marcon, M.; Becker, A.S.; Berger, N.; Boss, A. Determination of mammographic breast density using a deep convolutional neural network. Br. J. Radiol. 2019, 92, 20180691. [Google Scholar] [PubMed]
- Abdel-Nasser, M.; Moreno, A.; Puig, D. Temporal mammogram image registration using optimized curvilinear coordinates. Comput. Methods Programs Biomed. 2016, 127, 1–14. [Google Scholar] [PubMed]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing And Computer-Assisted Intervention; Springer: Berlin, Germany, 2015; pp. 234–241. [Google Scholar]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar]
- Moreira, I.C.; Amaral, I.; Domingues, I.; Cardoso, A.; Cardoso, M.J.; Cardoso, J.S. Inbreast: Toward a full-field digital mammographic database. Acad. Radiol. 2012, 19, 236–248. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar]
- Kerlikowske, K.; Zhu, W.; Tosteson, A.N.; Sprague, B.L.; Tice, J.A.; Lehman, C.D.; Miglioretti, D.L. Identifying women with dense breasts at high risk for interval cancer: A cohort study. Ann. Intern. Med. 2015, 162, 673–681. [Google Scholar]
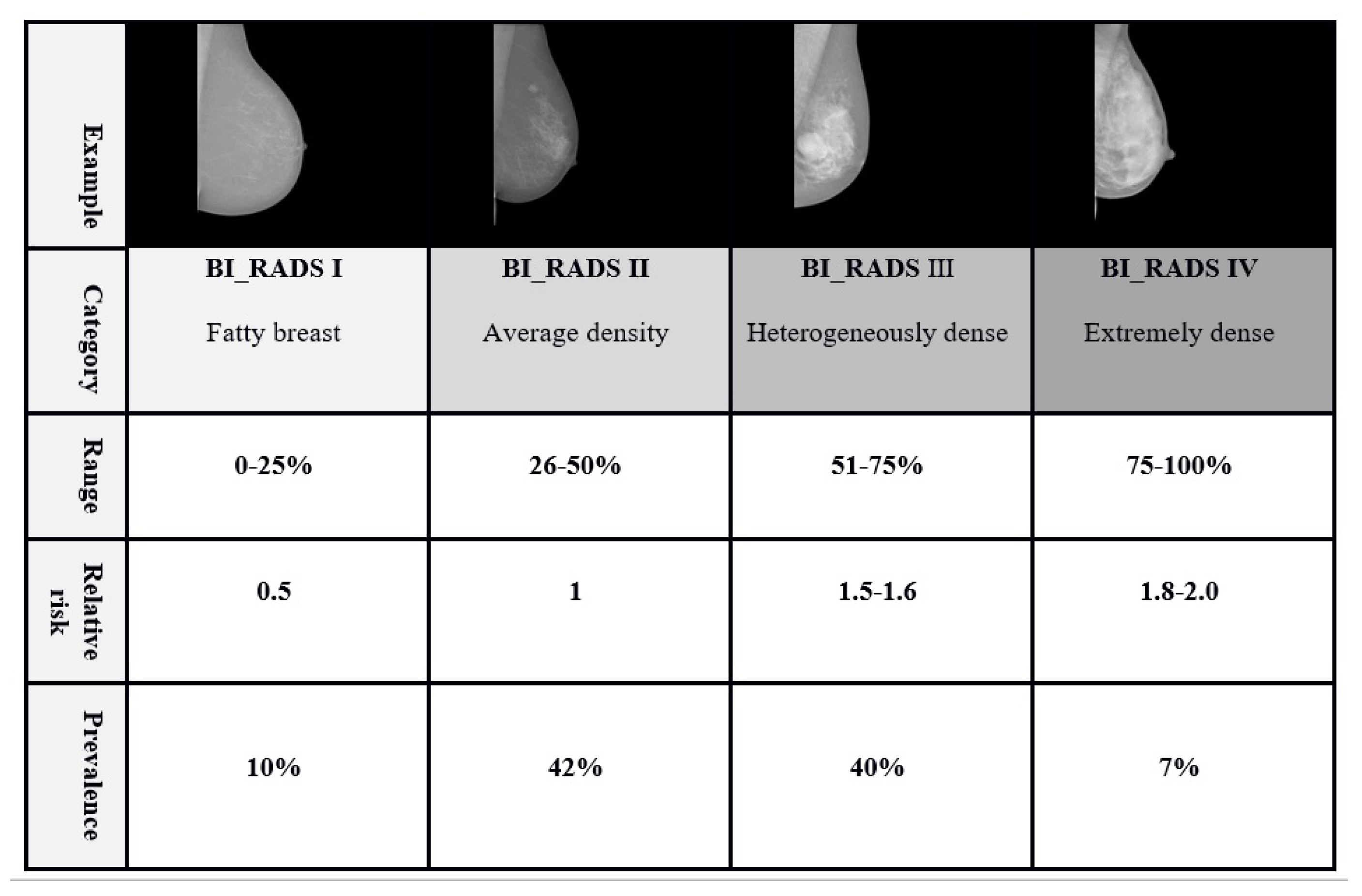
| I | II | III | IV | |
|---|---|---|---|---|
| Total dataset (410 images) | 33%, 136 images | 35%, 146 images | 25%, 100 images | 7%, 28 images |
| from 38 patients | from 41 patients | from 25 patients | from 8 patients | |
| Training and validation subset (328 images) | 33%, 108 images | 35%, 116 images | 25%, 80 images | 7%, 22 images |
| from 27 patients | from 29 patients | from 20 patients | from 6 patients | |
| Test subset | 33%, 27 images | 35%, 29 images | 25%, 20 images | 7%, 6 images |
| (82 images) | from 11 patients | from 12 patients | from 5 patients | from 2 patients |
| Model | Accuracy | DSC | J I | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | All | C1 | C2 | C3 | C4 | All | C1 | C2 | C3 | C4 | All | |
| cGAN-Unet with Dice-loss | 0.98 | 0.99 | 0.99 | 0.99 | 0.98 | 0.66 | 0.90 | 0.95 | 0.95 | 0.88 | 0.50 | 0.82 | 0.91 | 0.91 | 0.78 |
| cGAN-Unet with SSIM-loss | 0.94 | 0.98 | 0.98 | 0.96 | 0.96 | 0.54 | 0.86 | 0.91 | 0.85 | 0.79 | 0.37 | 0.75 | 0.83 | 0.74 | 0.65 |
| cGAN-Unet with MSE-loss | 0.68 | 0.84 | 0.93 | 0.95 | 0.80 | 0.53 | 0.85 | 0.94 | 0.97 | 0.81 | 0.36 | 0.74 | 0.90 | 0.94 | 0.67 |
| Methods | Sensitivity | Specificity | Precision | DSC |
|---|---|---|---|---|
| cGAN-UNet with Dice-loss | 0.957 | 0.985 | 0.81 | 0.88 |
| FCN-8 [44] | 0.748 | 0.997 | 0.69 | 0.72 |
| FCN-32 [44] | 0.5724 | 0.997 | 0.59 | 0.58 |
| Vgg-Segnet [45] | 0.832 | 0.996 | 0.66 | 0.73 |
| Class | Predicted Label | ||||
|---|---|---|---|---|---|
| Ground truth | I | II | III | IV | |
| I | 0.77 | 0.20 | 0.03 | 0 | |
| II | 0 | 0.76 | 0.24 | 0 | |
| III | 0 | 0 | 0.90 | 0.10 | |
| IV | 0 | 0 | 0.16 | 0.84 | |
| I | II | III | IV | |
|---|---|---|---|---|
| Total dataset (3192 images) | 25% (798 images) | 25% (798 images) | 25% (798 images) | 25% (798 images) |
| Training subset (2552 images) | 25% (638 images) | 25% (638 images ) | 25% (638 images) | 25% (638 images) |
| Validation subset (640 images) | 25% (160 images) | 25% (160 images) | 25% (160 images) | 25% (160 images) |
| Test subset | 27 images | 29 images | 20 images | 6 images |
| (82 images) | of 11 patients | of 12 patients | of 5 patients | of 2 patients |
| Size of Input Images | Accuracy | Precision | Sensitivity | Specificity | |
|---|---|---|---|---|---|
| Imbalanced dataset (410 images) | 128 × 128 | 90.29 | 90.29 | 90.29 | 96.76 |
| 64 × 64 | 94.95 | 94.17 | 94.17 | 98.05 | |
| Balanced dataset (3192 images) | 128 × 128 | 98.75 | 97.5 | 97.5 | 99.16 |
| 64 × 64 | 98.62 | 97.85 | 97.85 | 99.28 |
| Size of Input Images | Class | Predicted Label by CNN | |||||
|---|---|---|---|---|---|---|---|
| Ground truth | Imbalanced dataset (410 images) | I | II | III | IV | ||
| I | 1.0 | 0.0 | 0.0 | 0.0 | |||
| II | 0.03 | 0.93 | 0.03 | 0.0 | |||
| III | 0.0 | 0.12 | 0.77 | 0.12 | |||
| IV | 0.0 | 0.0 | 0.33 | 0.67 | |||
| I | 0.98 | 0.0 | 0.0 | 0.0 | |||
| II | 0.0 | 0.96 | 0.04 | 0.0 | |||
| III | 0.0 | 0.0 | 1.0 | 0.0 | |||
| IV | 0.0 | 0.0 | 0.57 | 0.43 | |||
| Balanced dataset (3192 images) | I | 1.0 | 0.0 | 0.0 | 0.0 | ||
| II | 0.0 | 0.97 | 0.03 | 0.0 | |||
| III | 0.05 | 0.0 | 0.95 | 0.0 | |||
| IV | 0.0 | 0.0 | 0.0 | 1.0 | |||
| I | 1.0 | 0.0 | 0.0 | 0.0 | |||
| II | 0.07 | 0.93 | 0.0 | 0.0 | |||
| III | 0.0 | 0.0 | 0.97 | 0.03 | |||
| IV | 0.0 | 0.0 | 0.0 | 1.0 | |||
| Study | Year | Method | No. of Images | No. of Density Category | Accuracy (%) |
|---|---|---|---|---|---|
| Volpara software [27] | 2010 | Hand-crafted | 2217 | Dense and Fatty | 94.0 |
| LIBRA software [4] | 2012 | Hand-crafted | 324 | 4 Classes | 81.0 |
| Lehman et al. [22] | 2018 | Deep Learning | 41479 | Dense and Non-dense | 87.0 |
| Lee and Nishikawa [11] | 2018 | Deep Learning | 455 | 4 Classes | 85.0 |
| Mohamed et al. [35] | 2018 | Deep Learning | 925 | BI-RADS II and BI-RADS III | 94.0 |
| Dubrovina et al. [37] | 2018 | Deep Learning | 40 | 4 Classes | 80.0 |
| Gandomkar et al. [21] | 2019 | Deep Learning | 3813 | Dense and Fatty | 92.0 |
| Our proposed CNN | 2020 | Deep Learning | 410 | 4 Classes | 98.75 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saffari, N.; Rashwan, H.A.; Abdel-Nasser, M.; Kumar Singh, V.; Arenas, M.; Mangina, E.; Herrera, B.; Puig, D. Fully Automated Breast Density Segmentation and Classification Using Deep Learning. Diagnostics 2020, 10, 988. https://doi.org/10.3390/diagnostics10110988
Saffari N, Rashwan HA, Abdel-Nasser M, Kumar Singh V, Arenas M, Mangina E, Herrera B, Puig D. Fully Automated Breast Density Segmentation and Classification Using Deep Learning. Diagnostics. 2020; 10(11):988. https://doi.org/10.3390/diagnostics10110988
Chicago/Turabian StyleSaffari, Nasibeh, Hatem A. Rashwan, Mohamed Abdel-Nasser, Vivek Kumar Singh, Meritxell Arenas, Eleni Mangina, Blas Herrera, and Domenec Puig. 2020. "Fully Automated Breast Density Segmentation and Classification Using Deep Learning" Diagnostics 10, no. 11: 988. https://doi.org/10.3390/diagnostics10110988
APA StyleSaffari, N., Rashwan, H. A., Abdel-Nasser, M., Kumar Singh, V., Arenas, M., Mangina, E., Herrera, B., & Puig, D. (2020). Fully Automated Breast Density Segmentation and Classification Using Deep Learning. Diagnostics, 10(11), 988. https://doi.org/10.3390/diagnostics10110988










