HLA Allele and Haplotype Frequencies in Three Urban Mexican Populations: Genetic Diversity for the Approach of Genomic Medicine
Abstract
1. Introduction
2. Materials and Methods
2.1. The Sample
2.2. HLA Typing
2.3. Statical Analysis and Data Visualization
3. Results
3.1. Allele Frequency
3.1.1. HLA-A
3.1.2. HLA-B
3.1.3. HLA-DRB1
3.1.4. HLA-DQB1
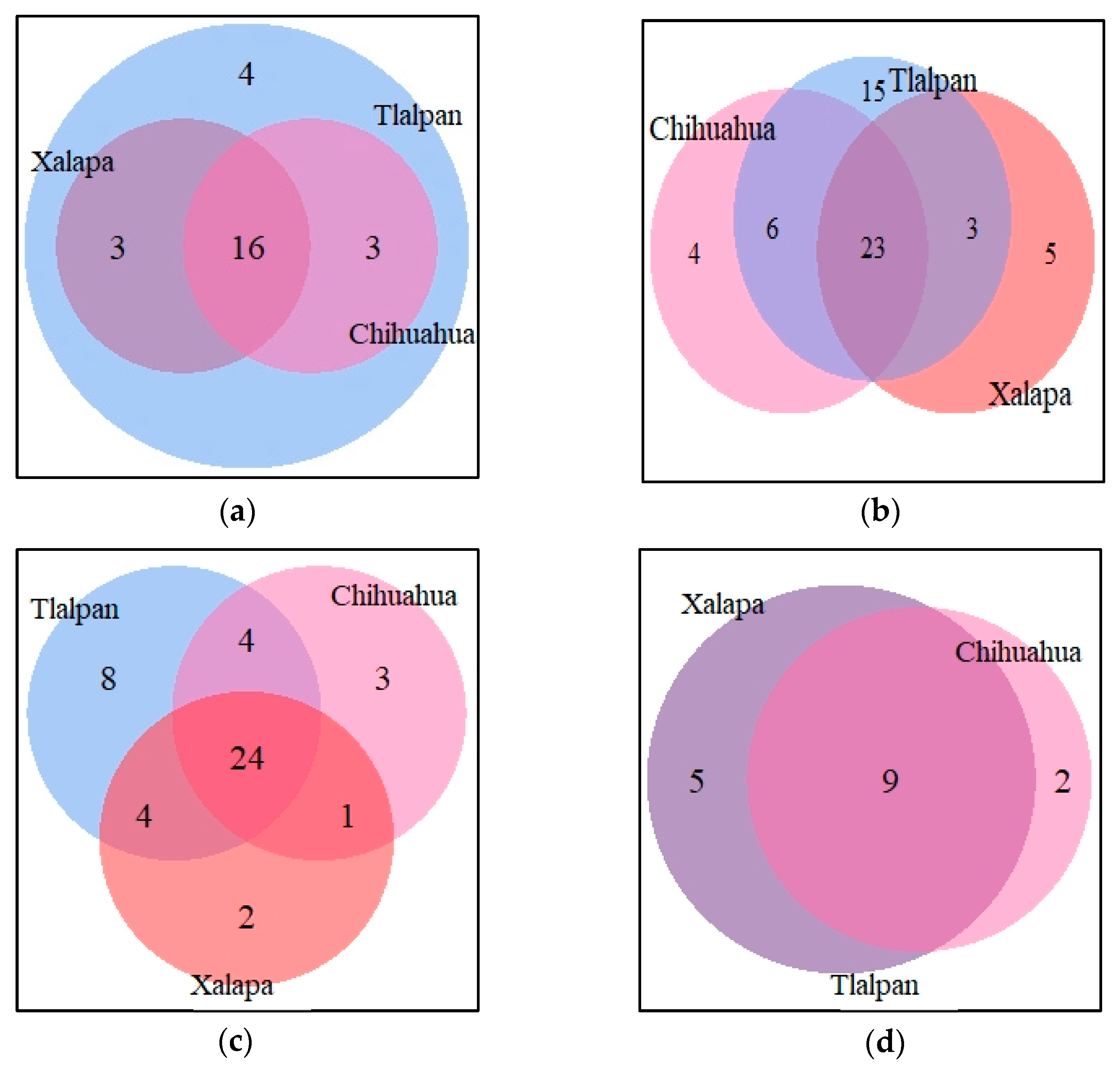
3.2. Haplotype Frequency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hughes, A.L.; Yeager, M. Natural selection at Major Histocompatibility Complex loci of vertebrates. Annu. Rev. Genet. 1998, 32, 415–435. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.C.; Plebanski, M.; Gupta, S.; Morris, J.; Cox, M.; Aidoo, M.; Kwiatkowski, D.; Greenwood, B.M.; Whittle, H.C.; Hill, A.V.S. Association of malaria parasite population structure, HLA, and immunological antagonism. Science 1998, 279, 1173–1177. [Google Scholar] [CrossRef] [PubMed]
- Falfán-Valencia, R.; Narayanankutty, A.; Reséndiz-Hernández, J.M.; Pérez-Rubio, G.; Ramírez-Venegas, A.; Nava-Quiroz, K.J.; Bautista-Félix, N.E.; Vargas-Alarcón, G.; Castillejos-López, M.D.J.; Hernández, A. An increased frequency in HLA class I alleles and haplotypes suggests genetic susceptibility to influenza A (H1N1) 2009 pandemic: A case-control study. J. Immunol. Res. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Mhc, H.; Horton, R.; Wilming, L.; Rand, V.; Lovering, R.C.; Bruford, E.A.; Wain, H.M.; Trowsdale, J.; Ziegler, A.; Beck, S. Gene map of the extended human MHC. Nat. Rev. Genet. 2004, 5, 889–899. [Google Scholar] [CrossRef]
- Gorodezky, C.; Alaez, C.; Vázquez-García, M.N.; de la Rosa, G.; Infante, E.; Balladares, S.; Toribio, R.; Pérez-Luque, E.; Muñoz, L. The Genetic structure of Mexican Mestizos of different locations: Tracking back their origins through MHC genes, blood group systems, and microsatellites. Hum. Immunol. 2001, 62, 979–991. [Google Scholar] [CrossRef]
- Instituto Nacional de Estadística y Geografía, Encuesta Intercensal 2015. INEGI. Available online: https://www.inegi.org.mx/ (accessed on 11 June 2019).
- Eberhard, D.M.; Gary, F.S.; Charles, D.F. Ethnologue: Languages of the World, Twenty-Second ed.; SIL Int.: Dallas, TX, USA, 2019; Available online: https://www.ethnologue.com/language/spa (accessed on 11 September 2019).
- Jimenez-Sanchez, G.; Silva-Zolezzi, I.; Hidalgo, A.; March, S. Genomic medicine in Mexico: Initial steps and the road ahead Genomic medicine in Mexico: Initial steps and the road ahead. Genome Res. 2008, 18, 1191–1198. [Google Scholar] [CrossRef]
- Burchard, E.G.; Borrell, L.N.; Choudhry, S.; Naqvi, M.; Tsai, H.-J.; Rodriguez-Santana, J.R.; Chapela, R.; Rogers, S.D.; Mei, R.; Rodriguez-Cintron, W.; et al. Latino populations: A unique opportunity for the study of race, genetics, and social environment in epidemiological research. Am. J. Public Health. 2005, 95, 2161–2168. [Google Scholar] [CrossRef]
- Instituto Nacional de Estadística y Geografía, México en Cifras: Ciudad de México (09). INEGI, 2015. Available online: https://www.inegi.org.mx/app/areasgeograficas/?ag=30 (accessed on 9 September 2019).
- Instituto Nacional de Lenguas Indígenas, Atlas de las Lenguas Indígenas de México. Secr. Cult. 2016. Available online: https://atlas.inali.gob.mx/inicio (accessed on 30 September 2019).
- Adams, R.E.W.; MacLeod, M.J. The Cambridge History of the Native Peoples of the Americas; Mesoamerica, Part 2; Cambridge University Press: Cambridge, UK, 2008; Volume II. [Google Scholar] [CrossRef]
- Schryer, F.J. Native Peoples of Central Mexico Since Independence. In The Cambridge History of the Native Peoples of the Americas; Mesoamerica, Part 2; Cambridge University Press: Cambridge, UK, 2008; Volume II, pp. 223–273. [Google Scholar] [CrossRef]
- Cline, S.L. Native Peoples of Colonial Central Mexico. In The Cambridge History of the Native Peoples of the Americas; Cambridge University Press: Cambridge, UK, 2008; pp. 187–222. [Google Scholar] [CrossRef]
- Instituto Nacional de Estadística y Geografía, México en Cifras: Chihuahua (08) INEGI. 2015. Available online: https://www.inegi.org.mx/app/areasgeograficas/?ag=30 (accessed on 9 September 2019).
- Frye, D. The Native Peoples of Northeastern Mexico. In The Cambridge History of the Native Peoples of the Americas; Mesoamerica, Part 2; Cambridge University Press: Cambridge, UK, 2008; Volume II, pp. 89–135. [Google Scholar] [CrossRef]
- Instituto Nacional de los Pueblos Indígenas, Atlas de los Pueblos Indígenas de México. Inst. Nac. Los Pueblos Indígenas. 2018. Available online: http://atlas.cdi.gob.mx/ (accessed on 12 June 2019).
- Instituto Nacional de Estadística y Geografía, México en Cifras: Veracruz de Ignacio de la Llave (30). INEGI, 2015. Available online: https://www.inegi.org.mx/app/areasgeograficas/?ag=30 (accessed on 9 September 2019).
- Deans-Smith, S. Native Peoples of the Gulf Coast from the Colonial Period to The Present. In The Cambridge History of the Native Peoples of the Americas; Mesoamerica, Part 2; Cambridge University Press: Cambridge, UK, 2008; Volume II, pp. 274–301. [Google Scholar] [CrossRef]
- Marsh, S.G.E.; Albert, E.D.; Bodmer, W.F.; Bontrop, R.E.; Dupont, B.; Erlich, H.A.; Fernández-Viña, M.; Geraghty, D.E.; Holdsworth, R.; Hurley, C.K.; et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens 2010, 75, 291–455. [Google Scholar] [CrossRef]
- Mullis, K.B. The unusual origin of the polymerase chain reaction. Sci. Am. 1990, 262, 56–65. [Google Scholar] [CrossRef]
- One Lambda, Micro SSPTM Generic Trays. A Thermo Fish. Sci. Brand. 2004, 1, 1–10. Available online: https://www.onelambda.com/en/product/micro-ssp-generic-trays.html (accessed on 2 January 2020).
- Lee, P.Y.; Costumbrado, J.; Hsu, C.Y.; Kim, Y.H. Agarose gel electrophoresis for the separation of DNA fragments. J. Vis. Exp. 2012, 62, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Slatkin, M. Incorporating genotypes of relatives into a test of linkage disequilibrium. Am. J. Hum. Genet. 1998, 62, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. 2007, 1, 47–50. [Google Scholar] [CrossRef]
- Lewontin, R.C. The Interaction of Selection and Linkage. II. Optimum Models. Genetics 1964, 50, 757–782. [Google Scholar] [PubMed]
- Haseman, J.K.; Elston, R.C. The investigation of linkage between a quantitative trait and a marker locus. Behav. Genet. 1972, 2, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Cedillo, T.; Zuñiga, J.; Acuña-Alonzo, V.; Pérez-Hernández, N.; Rodríguez-Pérez, J.M.; Barquera, R.; Gallardo, G.J.; Sánchez-Arenas, R.; García-Peña, M.D.; Granados, J.; et al. Genetic admixture and diversity estimations in the Mexican Mestizo population from Mexico City using 15 STR polymorphic markers. Forensic Sci. Int. Genet. 2008, 2, 37–39. [Google Scholar] [CrossRef]
- Arnaiz-Villena, A.; Vargas-Alarcon, G.; Granados, J.; Gomez-Casado, E.; Longas, J.; Gonzales-Hevilla, M.; Zuñiga, J.; Salgado, N.; Hernandez-Pacheco, G.; Guillen, J.; et al. HLA genes in Mexican Mazatecans, the peopling of the Americas and the uniqueness of Amerindians. Tissue Antigens. 2000, 56, 405–416. [Google Scholar] [CrossRef]
- Garcia-Ortiz, J.E.; Sandoval-Ramirez, L.; Rangel-Villalobos, H.; Maldonado-Torres, H.; Cox, S.; Garcia-Sepulveda, C.A.; Figuera, L.E.; Marsh, S.G.E.; Little, A.M.; Madrigal, J.A.; et al. High-resolution molecular characterization of the HLA class I and class II in the Tarahumara Amerindian population. Tissue Antigens. 2006, 68, 135–146. [Google Scholar] [CrossRef]
- Vargas-Alarcon, G.; Hernandez-Pacheco, G.; Zuñiga, J.; Rodriguez-Perez, J.M.; Perez-Hernandez, N.; Rangel, C.; Villarreal-Garza, C.; Martinez-Laso, J.; Granados, J.; Arnaiz-Villena, A. Distribution of HLA-B alleles in Mexican Amerindian populations. Immunogenetics 2003, 54, 756–760. [Google Scholar] [CrossRef]
- Arnaiz-Villena, A.; Moscoso, J.; Granados, J.; Serrano-Vela, J.I.; de la Peña, A.; Reguera, R.; Ferri, A.; Seclen, E.; Izaguirre, R.; Perez-Hernandez, N.; et al. HLA Genes in Mayos Population from Northeast Mexico. Curr. Genomics 2007, 8, 466–475. [Google Scholar] [CrossRef]
- Arnaiz-Villena, A.; Parga-Lozano, C.; Moreno, E.; Areces, C.; Rey, D.; Gomez-Prieto, P. The Origin of Amerindians and the Peopling of the Americas According to HLA Genes: Admixture with Asian and Pacific People. Curr. Genomics 2010, 11, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Silva-Zolezzi, I.; Hidalgo-Miranda, A.; Estrada-Gil, J.; Fernandez-Lopez, J.C.; Uribe-Figueroa, L.; Contreras, A.; Balam-Ortiz, E.; Bosque-Plata, L.; Velazquez-Fernandez, D.; Lara, C.; et al. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc. Natl. Acad. Sci. USA 2009, 106, 8611–8616. [Google Scholar] [CrossRef] [PubMed]
- Arnaiz-Villena, A.; Moscoso, J.; Serrano-Vela, I.; Martinez-Laso, J. The uniqueness of amerindians according to HLA genes and the peopling of the Americas. Inmunología 2006, 25, 13–24. [Google Scholar]
- Vargas-Alarcón, G.; Moscoso, J.; Laso, J.M.; Perez, J.R.; Dominguez, C.F.; Serrano-Vela, J.; Moreno, A.; Granados, J.; Arnaiz-Villena, A. Origin of Mexican Nahuas (Aztecs) according to HLA genes and their relationships with worldwide populations. Mol. Immunol. 2007, 44, 747–755. [Google Scholar] [CrossRef]
- Santaniello, A.; Salazar, G.; Lenna, S.; Antonioli, R.; Colombo, G.; Beretta, L.; Scorza, R. HLA-B35 upregulates the production of endothelin-1 in HLA-transfected cells: A possible pathogenetic role in pulmonary hypertension. Tissue Antigens. 2006, 68, 239–244. [Google Scholar] [CrossRef]
- Yang, M.; Kuang, X.; Li, J.; Pan, Y.; Tan, M.; Lu, B.; Cheng, Q.; Wu, L.; Pang, G. Meta-analysis of the association of HLA-DRB1 with rheumatoid arthritis in Chinese populations. BMC Musculoskelet. Disord. 2013, 14, 307. [Google Scholar] [CrossRef]
- Gregersen, P.K.; Silver, J.; Winchester, R.J. The shared epitope hypothesis. Arthritis Rheum. 1987, 30, 1205–1213. [Google Scholar] [CrossRef]
- Holoshitz, J. The Rheumatoid Arthritis HLA-DRB1 Share Epitope. Curr. Opin. Rheumatol. 2011, 22, 293–298. [Google Scholar] [CrossRef]
- Granados, J.; Vargas-Alarcón, G.; Andrade, F.; Melin-Aldana, H.; Alcocer-Varela, J.; Alarcón-Segovia, D. The role of HLA-DR alleles and complotypes through the ethnic barrier in systemic lupus erythematosus in Mexicans. Lupus 1996, 5, 184–189. [Google Scholar] [CrossRef]
- Morris, D.L.; Fernando, M.M.A.; Taylor, K.E.; Chung, S.A.; Nititham, J.; Alarcón-Riquelme, M.E.; Barcellos, L.F.; Behrens, T.W.; Cotsapas, C.; Gaffney, P.M.; et al. MHC associations with clinical and autoantibody manifestations in European SLE. Genes Immun. 2014, 15, 210–217. [Google Scholar] [CrossRef]
- Chen, J.; Yang, F.; Zhang, Y.; Fan, X.; Xiao, H.; Qian, W.; Chang, Y. HLA-A*01:01 in MHC is associated with psoriatic arthritis in Chinese Han population. Arch. Dermatol. Res. 2019, 311, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Bernal-Silva, S.; Granados, J.; Gorodezky, C.; Aláez, C.; Flores-Aguilar, H.; Cerda-Flores, R.M.; Guerrero-González, G.; Valdez-Chapa, L.D.; Morales-Casas, J.; González-Guerrero, J.F.; et al. HLA-DRB1 Class II antigen level alleles are associated with persistent HPV infection in Mexican women; A pilot study. Infect. Agents Cancer 2013, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Skinningsrud, B.; Lie, B.A.; Lavant, E.; Carlson, J.A.; Erlich, H.; Akselsen, H.E.; Gervin, K.; Wolff, A.B.; Erichsen, M.M.; Løvås, K.; et al. Multiple Loci in the HLA Complex Are Associated with Addison’s Disease. J. Clin. Endocrinol. Metab. 2015, 96, 1703–1708. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, A.; Abbasi, F.; Taghvaei, M.; Rabbani, B.; Moradi, B.; Shakiba, Y.; Rabbani, A. HLA-DRB,-DQA, and DQB alleles and haplotypes in Iranian patients with diabetes mellitus type I. Pediatr. Diabetes 2012, 14, 1–6. [Google Scholar] [CrossRef]
| Population | HLA-A | HLA-B | HLA-DRB1 | HLA-DQB1 | |
|---|---|---|---|---|---|
| MM | Obs. Het. | 0.8725 | 0.9383 | 0.9143 | 0.8327 |
| Exp. Het. | 0.8714 | 0.9326 | 0.9329 | 0.8186 | |
| p-value | 0.0127 * | 0.0069 * | 0.1043 | 0.6764 | |
| Tlalpan | Obs. Het. | 0.8636 | 0.9364 | 0.9273 | 0.8546 |
| Exp. Het. | 0.8712 | 0.9328 | 0.9324 | 0.8214 | |
| p-value | 0.0411 * | 0.0221 * | 0.1350 | 0.4558 | |
| Chihuahua | Obs. Het. | 0.9091 | 0.9546 | 0.9205 | 0.8296 |
| Exp. Het. | 0.8753 | 0.9371 | 0.9494 | 0.8510 | |
| p-value | 0.1074 | 0.0729 | 0.4980 | 0.5634 | |
| Xalapa | Obs. Het. | 0.8691 | 0.9286 | 0.8571 | 0.7500 |
| Exp. Het. | 0.8602 | 0.9146 | 0.8954 | 0.7457 | |
| p-value | 0.9130 | 0.3824 | 0.0939 | 0.6401 |
| Allele | MM | Tlalpan | Chihuahua | Xalapa | p-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tlalpan vs. | Chihuahua vs. Xalapa | ||||||||||
| n | AF | n | AF | n | AF | n | AF | Chihuahua | Xalapa | ||
| A*01:01 | 59 | 5.88 | 40 | 6.06 | 16 | 9.09 | 3 | 1.79 | 0.0300 | 0.0036 | |
| A*02:01 | 265 | 26.39 | 171 | 25.91 | 48 | 27.27 | 46 | 27.38 | |||
| A*02:02 | 20 | 1.99 | 16 | 2.42 | 1 | 0.57 | 3 | 1.79 | |||
| A*03:01 | 48 | 4.78 | 29 | 4.39 | 10 | 5.68 | 9 | 5.36 | |||
| A*11:01 | 27 | 2.69 | 18 | 2.73 | 6 | 3.41 | 3 | 1.79 | |||
| A*23:01 | 30 | 2.99 | 18 | 2.73 | 6 | 3.41 | 6 | 3.57 | |||
| A*24:02 | 165 | 16.43 | 113 | 17.12 | 28 | 15.91 | 24 | 14.29 | |||
| A*26:01 | 33 | 3.29 | 19 | 2.88 | 9 | 5.11 | 5 | 2.98 | |||
| A*29:01 | 29 | 2.89 | 18 | 2.73 | 8 | 4.55 | 3 | 1.79 | |||
| A*30:01 | 34 | 3.39 | 26 | 3.94 | 4 | 2.27 | 4 | 2.38 | |||
| A*31:01 | 65 | 6.47 | 34 | 5.15 | 15 | 8.52 | 16 | 9.52 | |||
| A*32:01 | 17 | 1.69 | 9 | 1.36 | 4 | 2.27 | 4 | 2.38 | |||
| A*33:01 | 19 | 1.89 | 13 | 1.97 | 3 | 1.7 | 3 | 1.79 | |||
| A*36 | 18 | 1.79 | 10 | 1.52 | 4 | 2.27 | 4 | 2.38 | |||
| A*68:01 | 129 | 12.85 | 91 | 13.79 | 8 | 4.55 | 30 | 17.86 | 0.0011 | 0.0002 | |
| A*68:03 | 10 | 1.00 | 9 | 1.36 | 0 | 0.00 | 1 | 0.60 | |||
| Others | 36 | 3.58 | 26 | 3.94 | 6 | 3.42 | 4 | 2.35 | |||
| Allele | MM | Tlalpan | Chihuahua | Xalapa | p-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tlalpan vs. | Chihuahua vs. Xalapa | ||||||||||
| n | AF | n | AF | n | AF | n | AF | Chihuahua | Xalapa | ||
| B*07:02 | 47 | 4.68 | 23 | 3.48 | 16 | 9.09 | 8 | 4.76 | 0.0336 | ||
| B*08:01 | 30 | 2.99 | 20 | 3.03 | 9 | 5.11 | 1 | 0.60 | 0.0200 | ||
| B*13:01 | 12 | 1.20 | 9 | 1.36 | 2 | 1.14 | 1 | 0.60 | |||
| B*14:01 | 24 | 2.39 | 12 | 1.82 | 9 | 5.11 | 3 | 1.79 | 0.0270 | ||
| B*14:02 | 25 | 2.49 | 22 | 3.33 | 3 | 1.70 | 0 | 0.00 | |||
| B*15:01 | 54 | 5.38 | 36 | 5.45 | 7 | 3.98 | 11 | 6.55 | |||
| B*15:02 | 10 | 1.00 | 10 | 1.52 | 0 | 0.00 | 0 | 0.00 | |||
| B*18:01 | 25 | 2.49 | 15 | 2.27 | 7 | 3.98 | 3 | 1.79 | |||
| B*35:01 | 161 | 16.04 | 109 | 16.52 | 19 | 10.80 | 33 | 19.64 | 0.0324 | ||
| B*35:02 | 43 | 4.28 | 28 | 4.24 | 5 | 2.84 | 10 | 5.95 | |||
| B*38:01 | 11 | 1.10 | 3 | 0.45 | 6 | 3.41 | 2 | 1.19 | |||
| B*39:01 | 131 | 13.05 | 86 | 13.03 | 20 | 11.36 | 25 | 14.88 | |||
| B*39:02 | 23 | 2.29 | 21 | 3.18 | 1 | 0.57 | 1 | 0.60 | |||
| B*39:06 | 16 | 1.59 | 14 | 2.12 | 1 | 0.57 | 1 | 0.60 | |||
| B*40:01 | 22 | 2.19 | 15 | 2.27 | 5 | 2.84 | 2 | 1.19 | |||
| B*40:02 | 65 | 6.47 | 41 | 6.21 | 7 | 3.98 | 17 | 10.12 | 0.0430 | ||
| B*44:02 | 62 | 6.18 | 39 | 5.91 | 14 | 7.95 | 9 | 5.36 | |||
| B*48:01 | 32 | 3.19 | 26 | 3.94 | 1 | 0.57 | 5 | 2.98 | 0.0274 | ||
| B*49:01 | 13 | 1.29 | 9 | 1.36 | 1 | 0.57 | 3 | 1.79 | |||
| B*50:01 | 11 | 1.10 | 8 | 1.21 | 3 | 1.70 | 0 | 0.00 | |||
| B*51:01 | 65 | 6.47 | 36 | 5.45 | 21 | 11.93 | 8 | 4.76 | 0.0042 | 0.0279 | |
| B*52:01 | 16 | 1.59 | 8 | 1.21 | 4 | 2.27 | 4 | 2.38 | |||
| B*53:01 | 11 | 1.10 | 7 | 1.06 | 0 | 0.00 | 4 | 2.38 | |||
| Others | 95 | 9.46 | 63 | 9.58 | 15 | 8.53 | 17 | 10.09 | |||
| Allele | MM | Tlalpan | Chihuahua | Xalapa | p-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tlalpan vs. | Chihuahua vs. Xalapa | ||||||||||
| n | AF | n | AF | n | AF | n | AF | Chihuahua | Xalapa | ||
| DRB1*01:01 | 35 | 3.49 | 23 | 3.48 | 9 | 5.11 | 3 | 1.79 | |||
| DRB1*01:02 | 29 | 2.89 | 22 | 3.33 | 6 | 3.41 | 1 | 0.60 | |||
| DRB1*03:01 | 49 | 4.88 | 36 | 5.45 | 11 | 6.25 | 2 | 1.19 | 0.0133 | 0.0205 | |
| DRB1*04:01 | 13 | 1.29 | 10 | 1.52 | 2 | 1.14 | 1 | 0.60 | |||
| DRB1*04:02 | 16 | 1.59 | 10 | 1.52 | 2 | 1.14 | 4 | 2.38 | |||
| DRB1*04:03 | 24 | 2.39 | 19 | 2.88 | 2 | 1.14 | 3 | 1.79 | |||
| DRB1*04:04 | 66 | 6.57 | 38 | 5.76 | 13 | 7.39 | 15 | 8.93 | |||
| DRB1*04:05 | 13 | 1.29 | 13 | 1.97 | 0 | 0.00 | 0 | 0.00 | |||
| DRB1*04:07 | 174 | 17.33 | 110 | 16.67 | 18 | 10.23 | 46 | 27.38 | 0.0466 | 0.0022 | 0.0001 |
| DRB1*04:11 | 17 | 1.69 | 8 | 1.21 | 2 | 1.14 | 7 | 4.17 | 0.0251 | ||
| DRB1*07:01 | 74 | 7.37 | 53 | 8.03 | 15 | 8.52 | 6 | 3.57 | |||
| DRB1*08:02 | 100 | 9.96 | 73 | 11.06 | 13 | 7.39 | 14 | 8.33 | |||
| DRB1*10:01 | 13 | 1.29 | 9 | 1.36 | 3 | 1.70 | 1 | 0.60 | |||
| DRB1*11:01 | 36 | 3.59 | 27 | 4.09 | 7 | 3.98 | 2 | 1.19 | |||
| DRB1*11:04 | 13 | 1.29 | 6 | 0.91 | 3 | 1.70 | 4 | 2.38 | |||
| DRB1*13:01 | 33 | 3.29 | 20 | 3.03 | 7 | 3.98 | 6 | 3.57 | |||
| DRB1*13:02 | 10 | 1.00 | 7 | 1.06 | 1 | 0.57 | 2 | 1.19 | |||
| DRB1*13:03 | 10 | 1.00 | 6 | 0.91 | 4 | 2.27 | 0 | 0.00 | |||
| DRB1*14:01 | 15 | 1.49 | 3 | 0.45 | 9 | 5.11 | 3 | 1.79 | <0.0001 | ||
| DRB1*14:02 | 49 | 4.88 | 29 | 4.39 | 9 | 5.11 | 11 | 6.55 | |||
| DRB1*14:06 | 51 | 5.08 | 37 | 5.61 | 11 | 6.25 | 3 | 1.79 | 0.0424 | ||
| DRB1*15:01 | 47 | 4.68 | 28 | 4.24 | 12 | 6.82 | 7 | 4.17 | |||
| DRB1*15:02 | 11 | 1.10 | 9 | 1.36 | 0 | 0.00 | 2 | 1.19 | |||
| DRB1*16:02 | 43 | 4.28 | 27 | 4.09 | 3 | 1.70 | 13 | 7.74 | |||
| Others | 63 | 6.27 | 37 | 5.62 | 14 | 7.95 | 12 | 7.11 | |||
| Allele | MM | Tlalpan | Chihuahua | Xalapa | p-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tlalpan vs. | Chihuahua vs. Xalapa | ||||||||||
| n | AF | n | AF | n | AF | n | AF | Chihuahua | Xalapa | ||
| DQB1*02:01 | 78 | 7.77 | 59 | 8.94 | 14 | 7.95 | 5 | 2.98 | 0.0088 | ||
| DQB1*02:02 | 55 | 5.48 | 36 | 5.45 | 16 | 9.09 | 3 | 1.79 | 0.0426 | 0.0036 | |
| DQB1*03:01 | 222 | 22.11 | 145 | 21.97 | 43 | 24.43 | 34 | 20.24 | |||
| DQB1*03:02 | 316 | 31.47 | 202 | 30.61 | 40 | 22.73 | 74 | 44.05 | 0.0013 | <0.0001 | |
| DQB1*04:02 | 118 | 11.75 | 82 | 12.42 | 14 | 7.95 | 22 | 13.10 | |||
| DQB1*05:01 | 91 | 9.06 | 62 | 9.39 | 21 | 11.93 | 8 | 4.76 | 0.0279 | ||
| DQB1*06:01 | 29 | 2.89 | 19 | 2.88 | 4 | 2.27 | 6 | 3.57 | |||
| DQB1*06:02 | 36 | 3.59 | 22 | 3.33 | 10 | 5.68 | 4 | 2.38 | |||
| DQB1*06:03 | 31 | 3.09 | 16 | 2.42 | 10 | 5.68 | 5 | 2.98 | 0.0491 | ||
| Others | 28 | 2.79 | 17 | 2.59 | 4 | 2.29 | 7 | 4.15 | |||
| Haplotype | n (2n = 1004) | HF | ∆′ |
|---|---|---|---|
| A*02:01-B*35:01-DRB1*08:02-DQB1*04:02 | 17 | 1.69 | 0.1889 |
| A*68:01-B*39:01-DRB1*04:07-DQB1*03:02 | 17 | 1.69 | 0.2429 |
| A*02:01-B*35:01-DRB1*04:07-DQB1*03:02 | 12 | 1.20 | 0.0242 |
| A*01:01-B*08:01-DRB1*03:01-DQB1*02:01 | 11 | 1.10 | 0.7747 |
| A*68:01-B*39:01-DRB1*08:02-DQB1*04:02 | 11 | 1.10 | 0.1596 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Del Angel-Pablo, A.D.; Juárez-Martín, A.I.; Pérez-Rubio, G.; Ambrocio-Ortiz, E.; López-Flores, L.A.; Camarena, A.E.; Falfán-Valencia, R. HLA Allele and Haplotype Frequencies in Three Urban Mexican Populations: Genetic Diversity for the Approach of Genomic Medicine. Diagnostics 2020, 10, 47. https://doi.org/10.3390/diagnostics10010047
Del Angel-Pablo AD, Juárez-Martín AI, Pérez-Rubio G, Ambrocio-Ortiz E, López-Flores LA, Camarena AE, Falfán-Valencia R. HLA Allele and Haplotype Frequencies in Three Urban Mexican Populations: Genetic Diversity for the Approach of Genomic Medicine. Diagnostics. 2020; 10(1):47. https://doi.org/10.3390/diagnostics10010047
Chicago/Turabian StyleDel Angel-Pablo, Alma D., Ana Itzel Juárez-Martín, Gloria Pérez-Rubio, Enrique Ambrocio-Ortiz, Luis A. López-Flores, Angel E. Camarena, and Ramcés Falfán-Valencia. 2020. "HLA Allele and Haplotype Frequencies in Three Urban Mexican Populations: Genetic Diversity for the Approach of Genomic Medicine" Diagnostics 10, no. 1: 47. https://doi.org/10.3390/diagnostics10010047
APA StyleDel Angel-Pablo, A. D., Juárez-Martín, A. I., Pérez-Rubio, G., Ambrocio-Ortiz, E., López-Flores, L. A., Camarena, A. E., & Falfán-Valencia, R. (2020). HLA Allele and Haplotype Frequencies in Three Urban Mexican Populations: Genetic Diversity for the Approach of Genomic Medicine. Diagnostics, 10(1), 47. https://doi.org/10.3390/diagnostics10010047







