Flavivirus Cross-Reactivity to Dengue Nonstructural Protein 1 Antigen Detection Assays
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Virus Preparation
2.2. Viral Infection and Harvesting
2.3. Quantification of Infectious Virus Particles and Viral RNA Copies
2.4. Evaluation of Commercial Dengue NS1 Rapid Assays and ELISAs
2.5. Sequence Analysis, Localization of Mutations and Prediction of Epitopes Targeted by Assays
2.6. Statistical Analysis
3. Results
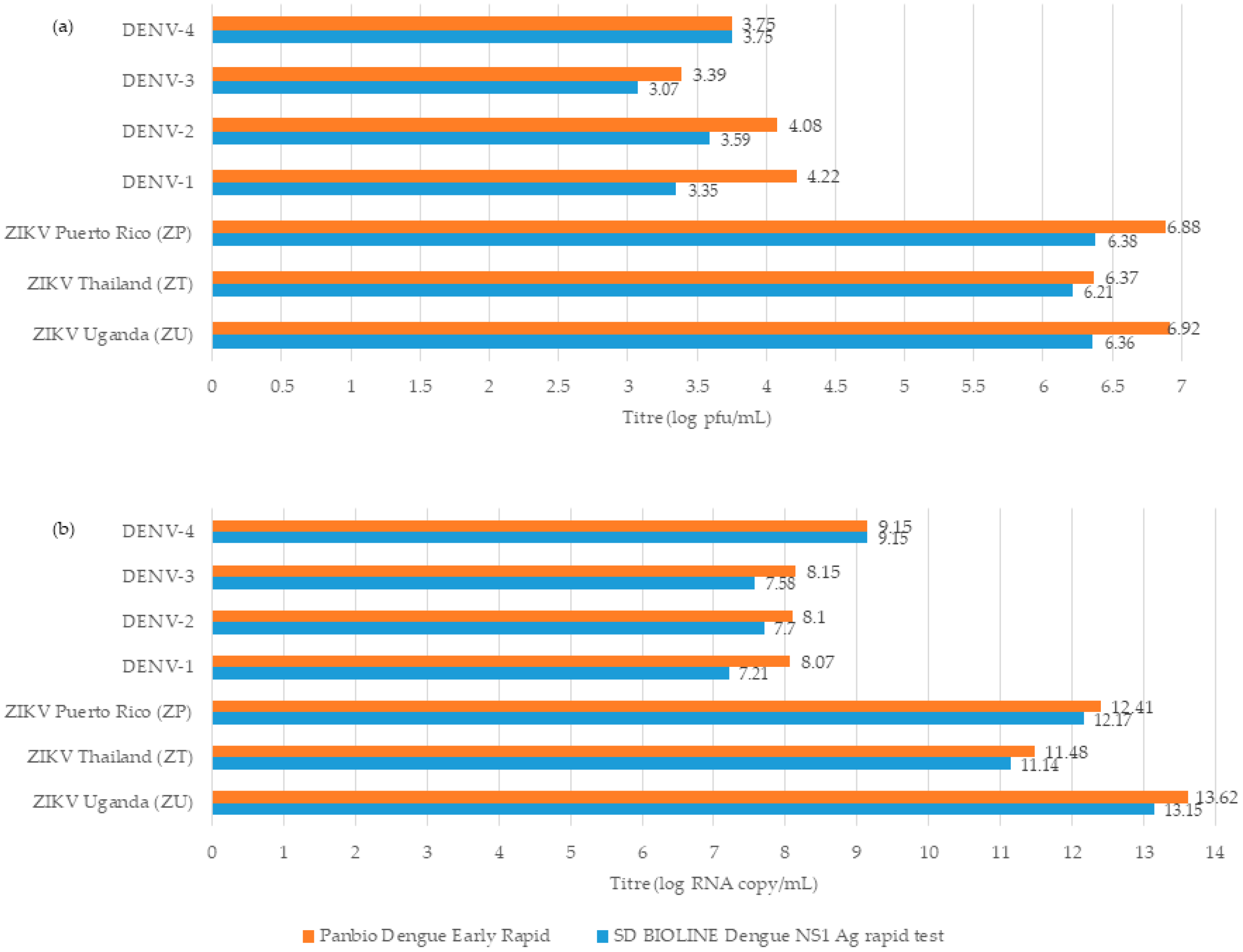
3.1. Dengue NS1 Ag Assays Detected DENV Early and at Low Titres
3.2. SD BIOLINE Dengue NS1 Ag Rapid Test and Panbio Dengue Early Rapid Assay Showed False-Positive Results for ZIKV at Significantly Higher Titres than DENV
3.3. SD BIOLINE Dengue NS1 Ag Rapid Test and Panbio Dengue Early Rapid Assay Showed Mixed False-Positive Results for KUNV and YFV at Higher Titres than DENV
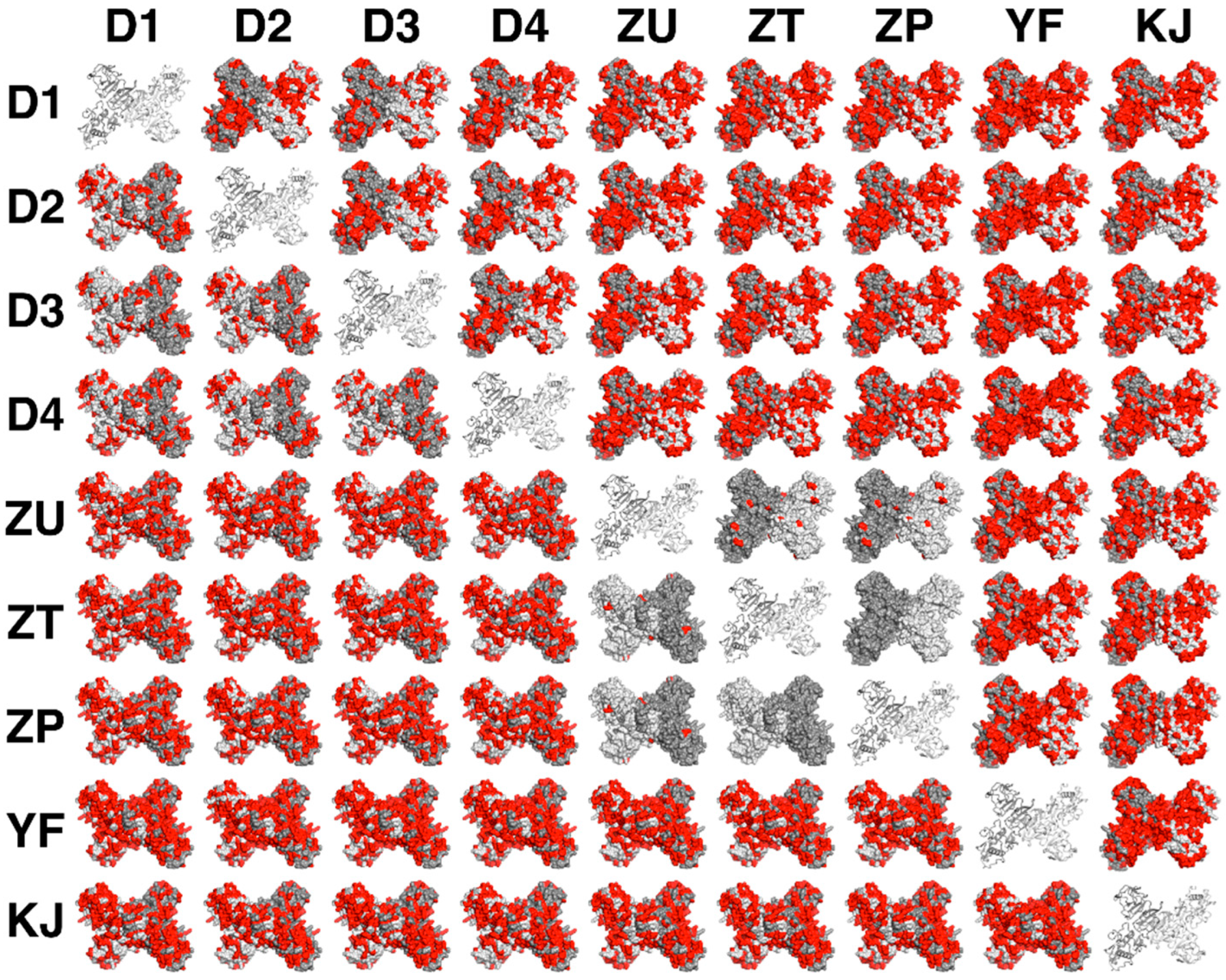
3.4. Similarity of Residues among Flavivirus NS1 Dimers Illustrated the Possibility of Cross-Reactivity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gubler, D.J.; Vasilakis, N.; Musso, D. History and Emergence of Zika Virus. J. Infect. Dis. 2017, 216, S860–S867. [Google Scholar] [CrossRef] [PubMed]
- Song, B.H.; Yun, S.I.; Woolley, M.; Lee, Y.M. Zika virus: History, epidemiology, transmission, and clinical presentation. J. Neuroimmunol. 2017, 308, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Landry, M.L.; St George, K. Laboratory Diagnosis of Zika Virus Infection. Arch. Pathol. Lab. Med. 2017, 141, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.A.; Depelsenaire, A.C.; Young, P.R. Clinical and Laboratory Diagnosis of Dengue Virus Infection. J. Infect. Dis. 2017, 215, S89–S95. [Google Scholar] [CrossRef] [PubMed]
- Winkler, G.; Maxwell, S.E.; Ruemmler, C.; Stollar, V. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology 1989, 171, 302–305. [Google Scholar] [CrossRef]
- Flamand, M.; Megret, F.; Mathieu, M.; Lepault, J.; Rey, F.A.; Deubel, V. Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J. Virol. 1999, 73, 6104–6110. [Google Scholar]
- Alcon, S.; Talarmin, A.; Debruyne, M.; Falconar, A.; Deubel, V.; Flamand, M. Enzyme-linked immunosorbent assay specific to Dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J. Clin. Microbiol. 2002, 40, 376–381. [Google Scholar] [CrossRef]
- Muller, D.A.; Young, P.R. The flavivirus NS1 protein: molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antiviral Res. 2013, 98, 192–208. [Google Scholar] [CrossRef]
- Ding, X.X.; Li, X.F.; Deng, Y.Q.; Guo, Y.H.; Hao, W.; Che, X.Y.; Qin, C.F.; Fu, N. Development of a double antibody sandwich ELISA for West Nile virus detection using monoclonal antibodies against non-structural protein 1. PLoS ONE 2014, 9, e108623. [Google Scholar] [CrossRef]
- Alcon-LePoder, S.; Sivard, P.; Drouet, M.T.; Talarmin, A.; Rice, C.; Flamand, M. Secretion of flaviviral non-structural protein NS1: from diagnosis to pathogenesis. Novartis Found. Symp 2006, 277, 233–247, discussion 247–253. [Google Scholar]
- Kumar, J.S.; Parida, M.; Rao, P.V. Monoclonal antibody-based antigen capture immunoassay for detection of circulating non-structural protein NS1: implications for early diagnosis of Japanese encephalitis virus infection. J. Med. Virol. 2011, 83, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Wee, S.; Alli-Shaik, A.; Kek, R.; Swa, H.L.F.; Tien, W.P.; Lim, V.W.; Leo, Y.S.; Ng, L.C.; Hapuarachchi, H.C.; Gunaratne, J. Multiplex targeted mass spectrometry assay for one-shot flavivirus diagnosis. Proc. Natl. Acad. Sci. USA 2019, 116, 6754–6759. [Google Scholar] [CrossRef] [PubMed]
- Gyurech, D.; Schilling, J.; Schmidt-Chanasit, J.; Cassinotti, P.; Kaeppeli, F.; Dobec, M. False positive dengue NS1 antigen test in a traveller with an acute Zika virus infection imported into Switzerland. Swiss Med. Wkly. 2016, 146, w14296. [Google Scholar] [CrossRef] [PubMed]
- Matheus, S.; Boukhari, R.; Labeau, B.; Ernault, V.; Bremand, L.; Kazanji, M.; Rousset, D. Specificity of Dengue NS1 Antigen in Differential Diagnosis of Dengue and Zika Virus Infection. Emerg. Infect. Dis. 2016, 22, 1691–1693. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.K.; Lam, S.; Low, S.L.; Tan, F.H.; Ng, L.C.; Teo, D. Evaluation of Pathogen Reduction Systems to Inactivate Dengue and Chikungunya Viruses in Apheresis Platelets Suspended in Plasma. Adv. Infect. Dis. 2013, 3, 9. [Google Scholar] [CrossRef]
- Zhang, L.; Wei, Q.; Mao, L.; Liu, W.; Mills, G.B.; Coombes, K. Serial dilution curve: a new method for analysis of reverse phase protein array data. Bioinformatics 2009, 25, 650–654. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232–1239. [Google Scholar] [CrossRef]
- Tan, C.H.; Wong, P.S.; Li, M.Z.; Vythilingam, I.; Ng, L.C. Evaluation of the Dengue NS1 Ag Strip(R) for detection of dengue virus antigen in Aedes aegypti (Diptera: Culicidae). Vector Borne Zoonotic Dis. 2011, 11, 789–792. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Brown, W.C.; Akey, D.L.; Konwerski, J.R.; Tarrasch, J.T.; Skiniotis, G.; Kuhn, R.J.; Smith, J.L. Extended surface for membrane association in Zika virus NS1 structure. Nat. Struct Mol. Biol. 2016, 23, 865–867. [Google Scholar] [CrossRef]
- Grell, L.; Parkin, C.; Slatest, L.; Craig, P.A. EZ-Viz, a tool for simplifying molecular viewing in PyMOL. Biochem. Mol. Biol. Educ 2006, 34, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Perkasa, A.; Yudhaputri, F.; Haryanto, S.; Hayati, R.F.; Ma’roef, C.N.; Antonjaya, U.; Yohan, B.; Myint, K.S.; Ledermann, J.P.; Rosenberg, R.; et al. Isolation of Zika Virus from Febrile Patient, Indonesia. Emerg. Infect. Dis. 2016, 22, 924–925. [Google Scholar] [CrossRef] [PubMed]
- Bonaldo, M.C.; Ribeiro, I.P.; Lima, N.S.; Dos Santos, A.A.; Menezes, L.S.; da Cruz, S.O.; de Mello, I.S.; Furtado, N.D.; de Moura, E.E.; Damasceno, L.; et al. Isolation of Infective Zika Virus from Urine and Saliva of Patients in Brazil. PLoS Negl. Trop. Dis. 2016, 10, e0004816. [Google Scholar] [CrossRef] [PubMed]
- Pessôa, R.; Patriota, J.V.; Lourdes de Souza, M.; Felix, A.C.; Mamede, N.; Sanabani, S.S. Investigation Into an Outbreak of Dengue-like Illness in Pernambuco, Brazil, Revealed a Cocirculation of Zika, Chikungunya, and Dengue Virus Type 1. Medicine (Baltimore) 2016, 95, e3201. [Google Scholar] [CrossRef]
- Waggoner, J.J.; Gresh, L.; Vargas, M.J.; Ballesteros, G.; Tellez, Y.; Soda, K.J.; Sahoo, M.K.; Nuñez, A.; Balmaseda, A.; Harris, E.; et al. Viremia and Clinical Presentation in Nicaraguan Patients Infected with Zika Virus, Chikungunya Virus, and Dengue Virus. Clin. Infect. Dis. 2016, 63, 1584–1590. [Google Scholar] [CrossRef]
- Tan, C.H.; Tan, L.K.; Hapuarachchi, H.C.; Lai, Y.L.; Wong, P.S.J.; Yap, G.; Mak, K.W.; Wong, W.Y.; Leo, Y.S.; Wong, M.C.; et al. Viral and Antibody Kinetics, and Mosquito Infectivity of an Imported Case of Zika Fever Due to Asian Genotype (American Strain) in Singapore. Viruses 2018, 10, 44. [Google Scholar] [CrossRef]
- Barzon, L.; Pacenti, M.; Franchin, E.; Squarzon, L.; Sinigaglia, A.; Ulbert, S.; Cusinato, R.; Palù, G. Isolation of West Nile virus from urine samples of patients with acute infection. J. Clin. Microbiol. 2014, 52, 3411–3413. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, L.; Lv, Y.; Zhang, W.; Li, J.; Zhang, Y.; Di, T.; Zhang, S.; Liu, J.; Qu, J.; et al. A fatal yellow fever virus infection in China: description and lessons. Emerg. Microbes Infect. 2016, 5, e69. [Google Scholar] [CrossRef]
- Scott, M.G. Monoclonal antibodies—Approaching adolescence in diagnostic immunoassays. Trends Biotechnol. 1985, 3, 170–175. [Google Scholar] [CrossRef]
- Frank, S.A. Molecular processes: Specificity and cross-reactivity. In Immunology and Evolution of Infectious Disease; Frank, S.A., Ed.; Princeton University Press: Princeton, NJ, USA, 2002; pp. 33–54. [Google Scholar]
- Rocha, L.B.; Alves, R.P.D.S.; Caetano, B.A.; Pereira, L.R.; Mitsunari, T.; Amorim, J.H.; Polatto, J.M.; Botosso, V.F.; Gallina, N.M.F.; Palacios, R.; et al. Epitope Sequences in Dengue Virus NS1 Protein Identified by Monoclonal Antibodies. Antibodies (Basel) 2017, 6, 14. [Google Scholar] [CrossRef]
- Andries, A.C.; Duong, V.; Ngan, C.; Ong, S.; Huy, R.; Sroin, K.K.; Te, V.; Y, B.; Try, P.L.; Buchy, P. Field evaluation and impact on clinical management of a rapid diagnostic kit that detects dengue NS1, IgM and IgG. PLoS Negl. Trop. Dis. 2012, 6, e1993. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.M.; Chua, T.H.; Sulaiman, W.Y.; Joanne, S.; Lim, Y.A.; Sekaran, S.D.; Chinna, K.; Venugopalan, B.; Vythilingam, I. A new paradigm for Aedes spp. surveillance using gravid ovipositing sticky trap and NS1 antigen test kit. Parasit Vectors 2017, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Stettler, K.; Beltramello, M.; Espinosa, D.A.; Graham, V.; Cassotta, A.; Bianchi, S.; Vanzetta, F.; Minola, A.; Jaconi, S.; Mele, F.; et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 2016, 353, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Qi, J.; Haywood, J.; Shi, Y.; Gao, G.F. Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses. Nat. Struct Mol. Biol. 2016, 23, 456–458. [Google Scholar] [CrossRef]
- Freire, M.; Pol-Fachin, L.; Coelho, D.F.; Viana, I.F.T.; Magalhaes, T.; Cordeiro, M.T.; Fischer, N.; Loeffler, F.F.; Jaenisch, T.; Franca, R.F.; et al. Mapping Putative B-Cell Zika Virus NS1 Epitopes Provides Molecular Basis for Anti-NS1 Antibody Discrimination between Zika and Dengue Viruses. ACS Omega 2017, 2, 3913–3920. [Google Scholar] [CrossRef]
- Lee, A.J.; Bhattacharya, R.; Scheuermann, R.H.; Pickett, B.E. Identification of diagnostic peptide regions that distinguish Zika virus from related mosquito-borne Flaviviruses. PLoS ONE 2017, 12, e0178199. [Google Scholar] [CrossRef]
- Chang, H.H.; Huber, R.G.; Bond, P.J.; Grad, Y.H.; Camerini, D.; Maurer-Stroh, S.; Lipsitch, M. Systematic analysis of protein identity between Zika virus and other arthropod-borne viruses. Bull. World Health Organ. 2017, 95, 517–525. [Google Scholar] [CrossRef]
- Duyen, H.T.; Ngoc, T.V.; Ha do, T.; Hang, V.T.; Kieu, N.T.; Young, P.R.; Farrar, J.J.; Simmons, C.P.; Wolbers, M.; Wills, B.A. Kinetics of plasma viremia and soluble nonstructural protein 1 concentrations in dengue: differential effects according to serotype and immune status. J. Infect. Dis. 2011, 203, 1292–1300. [Google Scholar] [CrossRef]
| Virus | Isolate | GenBank Accession No. | Lineage/Genotype | Isolation Location | Source |
|---|---|---|---|---|---|
| ZIKV Uganda (ZU) | MR766 | LC002520 | African | Uganda, 1947 | ATCC© VR-84™ |
| ZIKV Thailand (ZT) | PLCal_ZV | KF993678 | Asian | Thailand, 2013 | National Microbiology Laboratory, Canada |
| ZIKV Puerto Rico (ZPR) | PRVABC59 | KU501215 | Asian | Puerto Rico, 2015 | ATCC© VR-1843™ |
| DENV-1 | EHIE11986Y13 | KJ806943 | Genotype I | Singapore, 2013 | EHI, Singapore |
| DENV-2 | EHIE18944Y13 | KR779784 | Cosmopolitan Clade 1b | Singapore, 2013 | EHI, Singapore |
| DENV-3 | EHIE26592Y13 | KR685235 | Genotype III | Singapore, 2013 | EHI, Singapore |
| DENV-4 | EHIE2641Y08 | JN019830 | Genotype II | Singapore, 2008 | EHI, Singapore |
| CHIKV | EHICH06071Y13 | KP685237 | Asian | Singapore, 2013 | EHI, Singapore |
| KUNV | KUNV_EHI | MF289571 | ND | ND | ND |
| YFV | YFV_EHI | MF289572 | 17D vaccine | ND | ND |
| Dengue Virus (DENV) | Dengue NS1 Ag Rapid Assays | Dengue NS1 Ag ELISAs | |||
|---|---|---|---|---|---|
| SD BIOLINE Dengue NS1 Ag Rapid Test | Panbio Dengue Early Rapid | Bio-Rad Dengue NS1 Ag STRIP | SD Dengue NS1 Ag ELISA | Panbio ELISA | |
| DENV-1 | 3.35 ± 0.09 (7.21 ± 0.24) | 4.22 ± 0.14 (8.07 ± 0.07) | 4.22 ± 0.14 (8.07 ± 0.07) | 0 (5.41 ± 3.13) | 3.25 ± 0.02 (7.00 ± 0.12) |
| DENV-2 | 3.59 ± 0.04 (7.70 ± 0.14) | 4.08 ± 0.20 (8.10 ± 0.06) | 3.59 ± 0.04 (7.70 ± 0.14) | 0.57 ± 0.98 (2.03 ± 3.51) | 3.12 ± 0.07 (7.10 ± 0.21) |
| DENV-3 | 3.07 ± 0.18 (7.58 ± 0.10) | 3.39 ± 0.04 (8.15 ± 0.04) | 3.07 ± 0.18 (7.58 ± 0.10) | 0.57 ± 0.98 (5.40 ± 0.29) | 2.20 ± 0.17 (6.64 ± 0.22) |
| DENV-4 | 3.75 ± 0.18 (9.15 ± 0.13) | 4.22 ± 0.10 (9.83 ± 0.26) | 2.94 ± 0.10 (8.83 ± 0.12) | 0 (6.66 ± 0.15) | 2.43 ± 0.23 (8.17 ± 0.02) |
| Virus | SD BIOLINE Dengue NS1 Ag Rapid Test | Panbio Dengue Early Rapid | Bio-Rad Dengue NS1 Ag STRIP | SD Dengue NS1 Ag ELISA | Panbio ELISA |
|---|---|---|---|---|---|
| KUNV | Reactive 5.96 ± 0.23 | Reactive 6.50 ± 0.11 | Nonreactive ≤7.46 ± 0.06 | Nonreactive ≤7.93 ± 0.55 | Reactive 5.96 ± 0.23 |
| YFV | Reactive 7.14 ± 0.37 | Nonreactive ≤7.13 ± 0.46 | Nonreactive ≤7.13 ± 0.46 | Nonreactive ≤7.14 ± 0.37 | Nonreactive ≤7.43 ± 0.06 |
| CHIKV | Nonreactive ≤6.98 ± 0.27 | Nonreactive ≤6.98 ± 0.27 | Nonreactive ≤6.98 ± 0.27 | Nonreactive ≤6.98 ± 0.27 | Nonreactive ≤6.98 ± 0.27 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, L.K.; Wong, W.Y.; Yang, H.T.; Huber, R.G.; Bond, P.J.; Ng, L.C.; Maurer-Stroh, S.; Hapuarachchi, H.C. Flavivirus Cross-Reactivity to Dengue Nonstructural Protein 1 Antigen Detection Assays. Diagnostics 2020, 10, 11. https://doi.org/10.3390/diagnostics10010011
Tan LK, Wong WY, Yang HT, Huber RG, Bond PJ, Ng LC, Maurer-Stroh S, Hapuarachchi HC. Flavivirus Cross-Reactivity to Dengue Nonstructural Protein 1 Antigen Detection Assays. Diagnostics. 2020; 10(1):11. https://doi.org/10.3390/diagnostics10010011
Chicago/Turabian StyleTan, Li Kiang, Wing Yan Wong, Hui Ting Yang, Roland G. Huber, Peter J. Bond, Lee Ching Ng, Sebastian Maurer-Stroh, and Hapuarachchige Chanditha Hapuarachchi. 2020. "Flavivirus Cross-Reactivity to Dengue Nonstructural Protein 1 Antigen Detection Assays" Diagnostics 10, no. 1: 11. https://doi.org/10.3390/diagnostics10010011
APA StyleTan, L. K., Wong, W. Y., Yang, H. T., Huber, R. G., Bond, P. J., Ng, L. C., Maurer-Stroh, S., & Hapuarachchi, H. C. (2020). Flavivirus Cross-Reactivity to Dengue Nonstructural Protein 1 Antigen Detection Assays. Diagnostics, 10(1), 11. https://doi.org/10.3390/diagnostics10010011






