Membrane Lipid Composition and Amino Acid Excretion Patterns of Methanothermococcus okinawensis Grown in the Presence of Inhibitors Detected in the Enceladian Plume
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth Experiments
2.1.1. Design of Experiment (DoE)
2.1.2. Extreme Value Experiments
2.2. Lipid Extraction and Analysis
2.3. Amino Acid Analysis
2.4. Statistical Analyses
3. Results
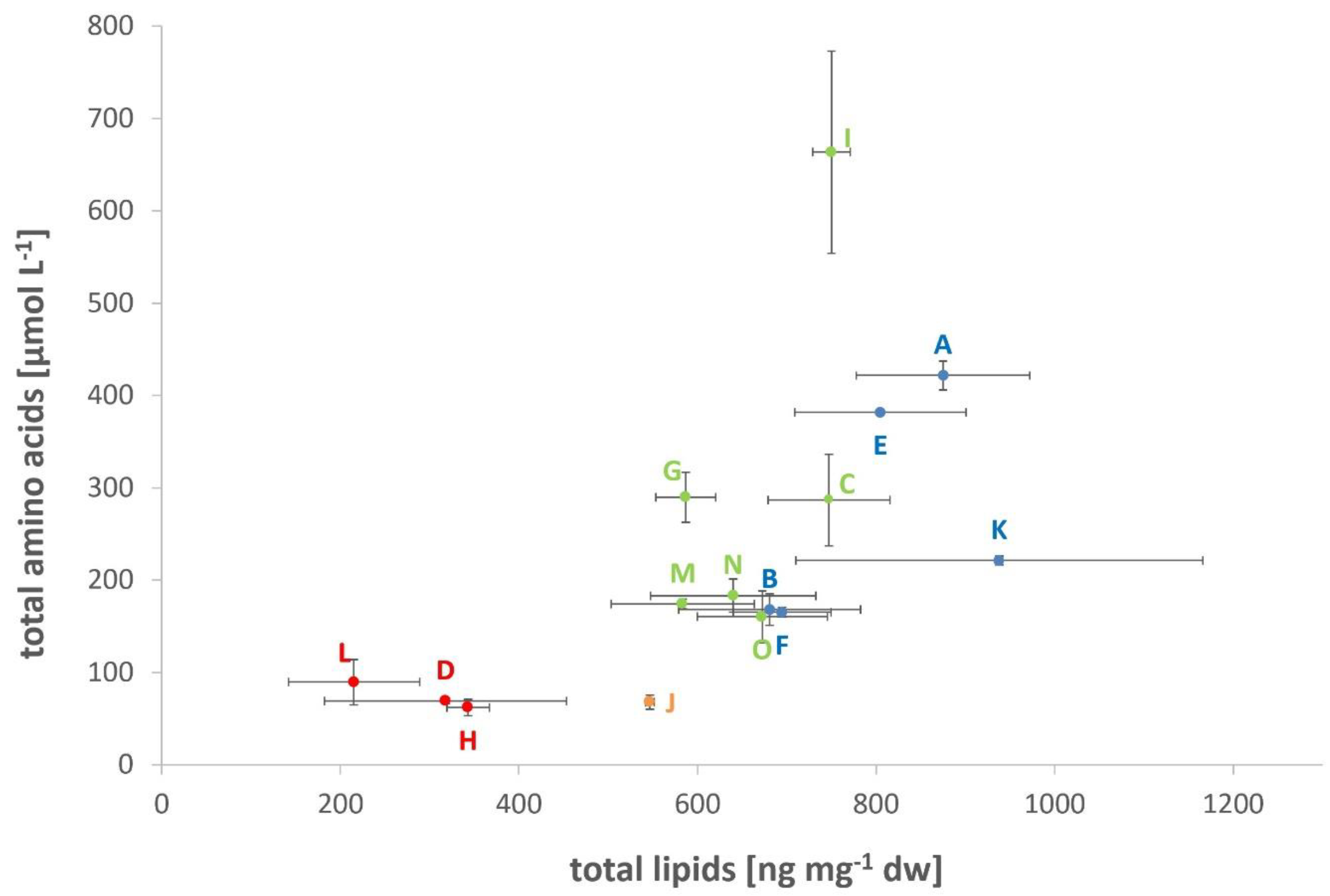
3.1. Lipid and Amino Acid Production under the DoE Setting
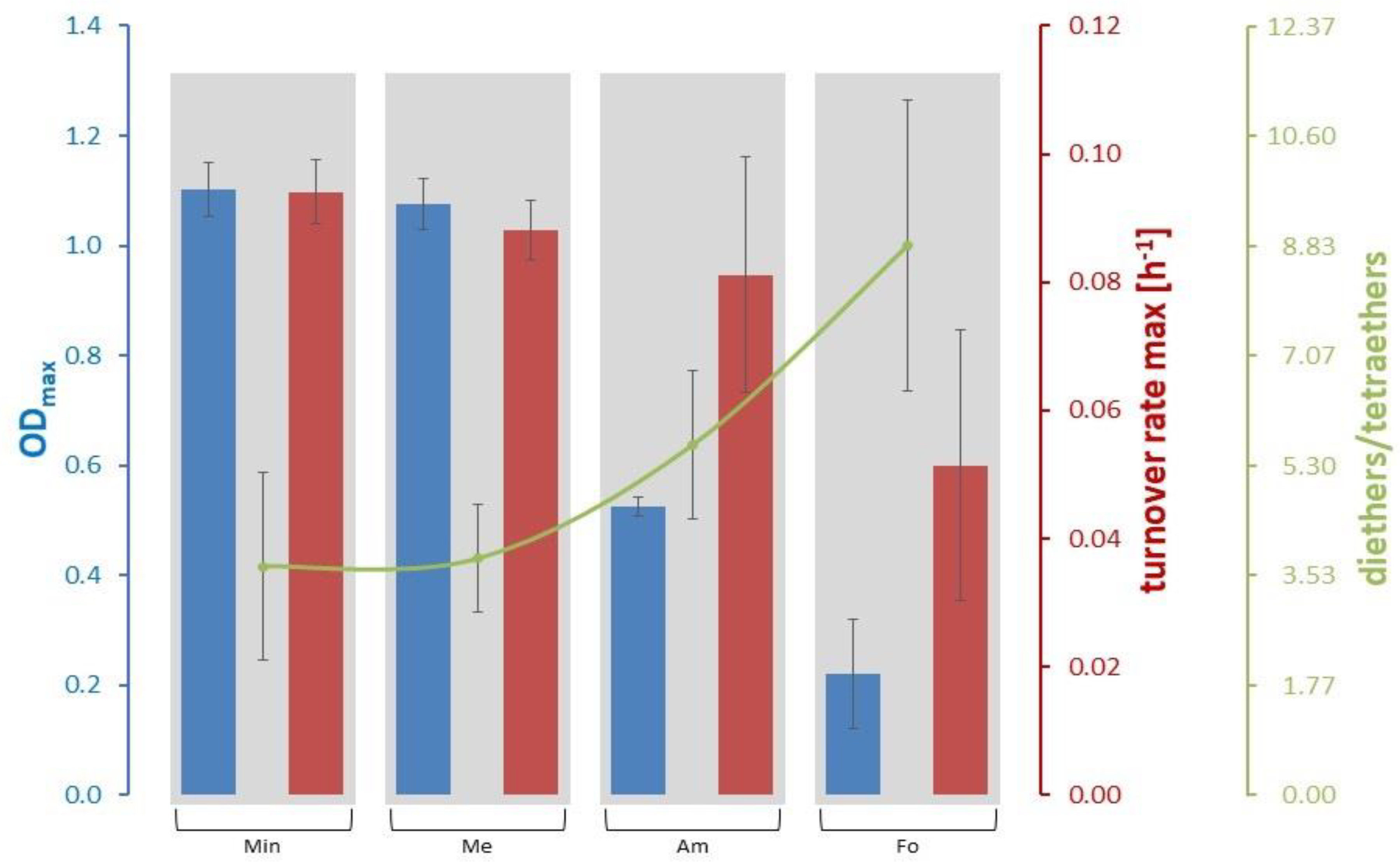
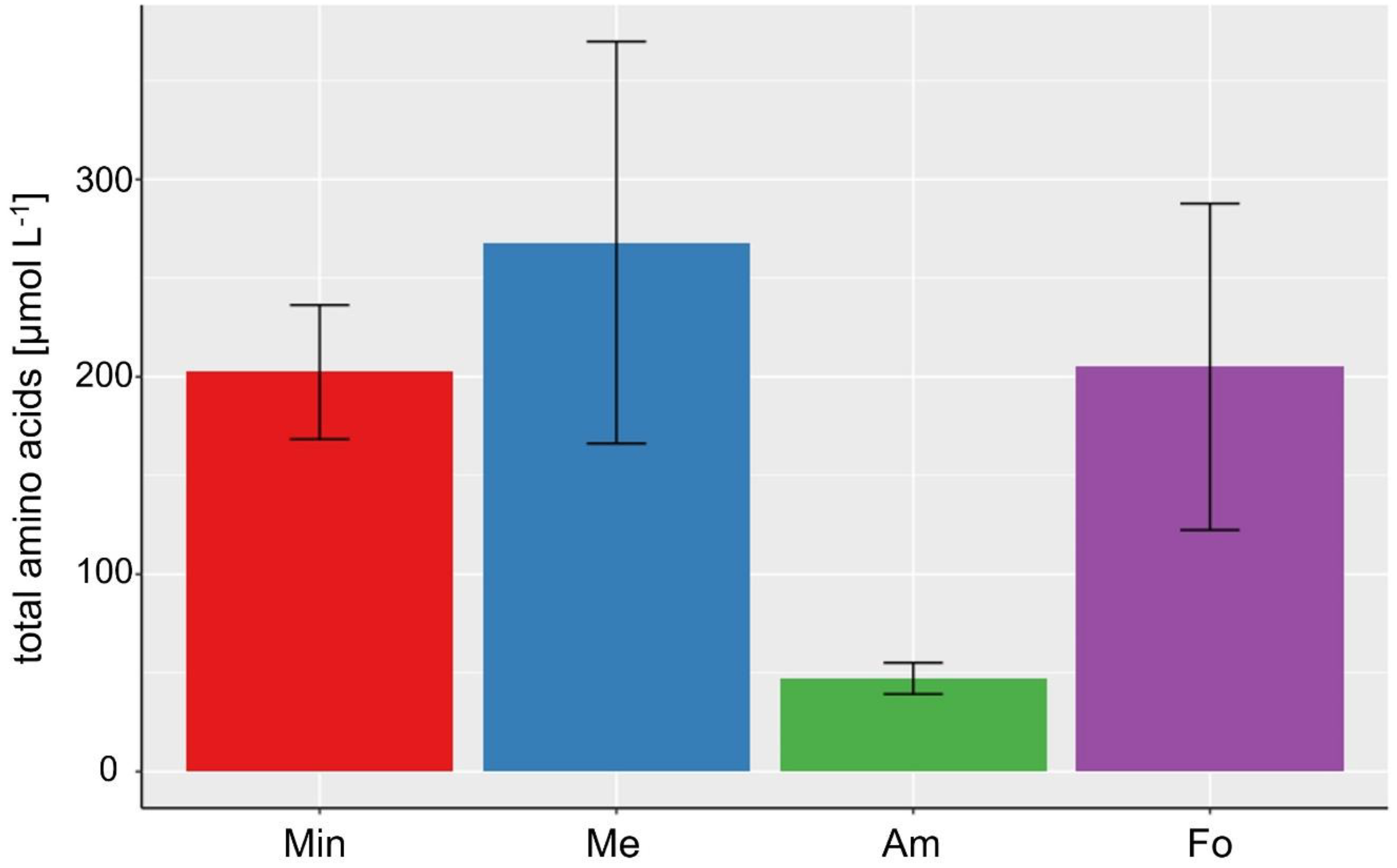
3.2. Lipid and Amino Acid Production in the Extreme Value Setting
4. Discussion
4.1. Effect of Varying Physical Conditions and Inhibitors on the Lipid Inventory of M. okinawensis
4.2. Amino Acids and Lipids as Biomarkers for Potential Extraterrestrial Methanogenic Life
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lombard, J.; López-García, P.; Moreira, D. The early evolution of lipid membranes and the three domains of life. Nat. Rev. Microbiol. 2012, 10, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Caforio, A.; Yang, Q.; Sun, B.; Yu, F.; Zhu, X.; Wang, J.; Dou, C.; Fu, Q.; Huang, N.; et al. Structural and mechanistic insights into the biosynthesis of CDP-archaeol in membranes. Cell Res. 2017, 27, 1378. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.H.; Percivalle, C.; Ritson, D.J.; Duffy, C.D.; Sutherland, J.D. Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat. Chem. 2015, 7, 301. [Google Scholar] [CrossRef] [PubMed]
- Oro, J. Chemical synthesis of lipids and the origin of life. J. Biol. Phys. 1995, 20, 135–147. [Google Scholar] [CrossRef]
- Segré, D.; Ben-Eli, D.; Deamer, D.W.; Lancet, D. The lipid world. Orig. Life Evol. Biosph. 2001, 31, 119–145. [Google Scholar] [CrossRef]
- Segré, D.; Lancet, D.; Kedem, O.; Pilpel, Y. Graded Autocatalysis Replication Domain (GARD): Kinetic analysis of self-replication in mutually catalytic sets. Orig. Life Evol. Biosph. 1998, 28, 501–514. [Google Scholar] [CrossRef]
- Lancet, D.; Zidovetzki, R.; Markovitch, O. Systems protobiology: Origin of life in lipid catalytic networks. J. Royal Soc. Interface 2018, 15, 20180159. [Google Scholar] [CrossRef]
- Lai, J.C.-Y.; Pearce, B.K.D.; Pudritz, R.E.; Lee, D. Meteoritic abundances of fatty acids and potential reaction pathways in planetesimals. Icarus 2019, 319, 685–700. [Google Scholar] [CrossRef]
- Cornell, C.E.; Black, R.A.; Xue, M.; Litz, H.E.; Ramsay, A.; Gordon, M.; Mileant, A.; Cohen, Z.R.; Williams, J.A.; Lee, K.K.; et al. Prebiotic amino acids bind to and stabilize prebiotic fatty acid membranes. Proc. Natl. Acad. Sci. USA 2019, 116, 17239–17244. [Google Scholar] [CrossRef]
- Cole, L.; Kramer, P.R. Chapter 1.4—Amino acid metabolism. In Human Physiology, Biochemistry and Basic Medicine; Cole, L., Kramer, P.R., Eds.; Academic Press: Boston, MA, USA, 2016; pp. 31–38. ISBN 978-0-12-803699-0. [Google Scholar]
- Ambrogelly, A.; Palioura, S.; Söll, D. Natural expansion of the genetic code. Nat. Chem. Biol. 2006, 3, 29. [Google Scholar] [CrossRef]
- Miller, S.L. A production of amino acids under possible primitive earth conditions. Science 1953, 117, 528–529. [Google Scholar] [CrossRef] [PubMed]
- Kitadai, N.; Maruyama, S. Origins of building blocks of life: A review. Geosci. Front. 2018, 9, 1117–1153. [Google Scholar] [CrossRef]
- Ménez, B.; Pisapia, C.; Andreani, M.; Jamme, F.; Vanbellingen, Q.P.; Brunelle, A.; Richard, L.; Dumas, P.; Réfrégiers, M. Abiotic synthesis of amino acids in the recesses of the oceanic lithosphere. Nature 2018, 564, 59–63. [Google Scholar]
- Schouten, S.; Van Der Maarel, M.J.E.C.; Huber, R.; Sinninghe Damsté, J.S. 2,6,10,15,19-Pentamethylicosenes in Methanolobus bombayensis, a marine methanogenic archaeon. Org. Geochem. 1997, 26, 409–414. [Google Scholar] [CrossRef]
- Londry, K.L.; Dawson, K.G.; Grover, H.D.; Bradley, A.S. Stable carbon isotope fractionation between substrates and products of Methanosarcina barkeri. Org. Geochem. 2008, 39, 608–621. [Google Scholar] [CrossRef]
- Koga, Y.; Nishihara, M.; Morii, H.; Akagawa-Matsushita, M. Ether polar lipids of methanogenic bacteria: Structures, comparative aspects, and biosyntheses. Microbiol. Mol. Biol. Rev. 1993, 57, 164–182. [Google Scholar]
- Langworthy, T.A. Long-chain diglycerol tetraethers from Thermoplasma acidophilum. Biochim. Biophys. Acta Lipids Lipid Metab. 1977, 487, 37–50. [Google Scholar] [CrossRef]
- De Rosa, M.; Esposito, E.; Gambacorta, A.; Nicolaus, B.; Bu’Lock, J.D. Effects of temperature on ether lipid composition of Caldariella acidophila. Phytochemistry 1980, 19, 827–831. [Google Scholar] [CrossRef]
- Koga, Y.; Morii, H.; Akagawa-Matsushita, M.; Ohga, M. Correlation of polar lipid composition with 16S rRNA phylogeny in methanogens. Further analysis of lipid component parts. Biosci. Biotechnol. Biochem. 1998, 62, 230–236. [Google Scholar] [CrossRef]
- Schouten, S.; Hopmans, E.C.; Sinninghe Damsté, J.S. The organic geochemistry of glycerol dialkyl glycerol tetraether lipids: A review. Org. Geochem. 2013, 54, 19–61. [Google Scholar] [CrossRef]
- Bauersachs, T.; Weidenbach, K.; Schmitz, R.A.; Schwark, L. Distribution of glycerol ether lipids in halophilic, methanogenic and hyperthermophilic archaea. Org. Geochem. 2015, 83–84, 101–108. [Google Scholar] [CrossRef]
- Robertson, D.E.; Noll, D.; Roberts, M.F. Free amino acid dynamics in marine methanogens. Beta-amino acids as compatible solutes. J. Biol. Chem. 1992, 267, 14893–14901. [Google Scholar] [PubMed]
- Robertson, D.E.; Roberts, M.F.; Belay, N.; Stetter, K.O.; Boone, D.R. Occurrence of beta-glutamate, a novel osmolyte, in marine methanogenic bacteria. Appl. Environ. Microbiol. 1990, 56, 1504–1508. [Google Scholar] [PubMed]
- Moissl-Eichinger, C.; Cockell, C.; Rettberg, P. Venturing into new realms? Microorganisms in space. FEMS Microbiol. Rev. 2016, 40, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Kral, T.A.; Goodhart, T.H.; Harpool, J.D.; Hearnsberger, C.E.; McCracken, G.L.; McSpadden, S.W. Sensitivity and adaptability of methanogens to perchlorates: Implications for life on Mars. Planet. Space Sci. 2016, 120, 87–95. [Google Scholar] [CrossRef]
- Taubner, R.S.; Pappenreiter, P.; Zwicker, J.; Smrzka, D.; Pruckner, C.; Kolar, P.; Bernacchi, S.; Seifert, A.H.; Krajete, A.; Bach, W.; et al. Biological methane production under putative Enceladus-like conditions. Nat. Commun. 2018, 9, 748. [Google Scholar] [CrossRef]
- Tan, J.; Lewis, J.M.T.; Sephton, M.A. The fate of lipid biosignatures in a Mars-analogue sulfur stream. Sci. Rep. 2018, 8, 7586. [Google Scholar] [CrossRef]
- Wilhelm, M.B.; Davila, A.F.; Eigenbrode, J.L.; Parenteau, M.N.; Jahnke, L.L.; Liu, X.-L.; Summons, R.E.; Wray, J.J.; Stamos, B.N.; O’Reilly, S.S.; et al. Xeropreservation of functionalized lipid biomarkers in hyperarid soils in the Atacama Desert. Org. Geochem. 2017, 103, 97–104. [Google Scholar] [CrossRef]
- Postberg, F.; Khawaja, N.; Abel, B.; Choblet, G.; Glein, C.R.; Gudipati, M.S.; Henderson, B.L.; Hsu, H.-W.; Kempf, S.; Klenner, F.; et al. Macromolecular organic compounds from the depths of Enceladus. Nature 2018, 558, 564–568. [Google Scholar] [CrossRef]
- Hsu, H.-W.; Postberg, F.; Sekine, Y.; Shibuya, T.; Kempf, S.; Horányi, M.; Juhász, A.; Altobelli, N.; Suzuki, K.; Masaki, Y.; et al. Ongoing hydrothermal activities within Enceladus. Nature 2015, 519, 207–210. [Google Scholar] [CrossRef]
- Waite, J.H.; Lewis, W.S.; Magee, B.A.; Lunine, J.I.; McKinnon, W.B.; Glein, C.R.; Mousis, O.; Young, D.T.; Brockwell, T.; Westlake, J.; et al. Liquid water on Enceladus from observations of ammonia and 40Ar in the plume. Nature 2009, 460, 487–490. [Google Scholar] [CrossRef]
- Cheung, A.C.; Rank, D.M.; Townes, C.H.; Thornton, D.D.; Welch, W.J. Detection of NH3 molecules in the interstellar medium by their microwave emission. Phys. Rev. Lett. 1968, 21, 1701–1705. [Google Scholar] [CrossRef]
- Snyder, L.E.; Buhl, D.; Zuckerman, B.; Palmer, P. Microwave detection of interstellar formaldehyde. Phys. Rev. Lett. 1969, 22, 679–681. [Google Scholar] [CrossRef]
- Kvenvolden, K.; Lawless, J.; Pering, K.; Peterson, E.; Flores, J.; Ponnamperuma, C.; Kaplan, I.R.; Moore, C. Evidence for extraterrestrial amino-acids and hydrocarbons in the murchison meteorite. Nature 1970, 228, 923–926. [Google Scholar] [CrossRef] [PubMed]
- Lees, R.M.; Haque, S.S. Microwave double resonance study of collision induced population transfer between levels of interstellar methanol lines. Can. J. Phys. 1974, 52, 2250–2271. [Google Scholar] [CrossRef]
- Ehrenfreund, P.; Charnley, S.B. Organic molecules in the interstellar medium, comets, and meteorites: A voyage from dark clouds to the early Earth. Annu. Rev. Astron. Astrophys. 2000, 38, 427–483. [Google Scholar] [CrossRef]
- Cooper, G.; Kimmich, N.; Belisle, W.; Sarinana, J.; Brabham, K.; Garrel, L. Carbonaceous meteorites as a source of sugar-related organic compounds for the early Earth. Nature 2001, 414, 879–883. [Google Scholar] [CrossRef]
- Gibb, E.L.; Whittet, D.C.B.; Chiar, J.E. Searching for ammonia in grain mantles toward massive young stellar objects. Astrophys. J. 2001, 558, 702–716. [Google Scholar] [CrossRef]
- Gibb, E.L.; Whittet, D.C.B.; Boogert, A.C.A.; Tielens, A.G.G.M. Interstellar ice: The infrared space observatory legacy. Astrophys. J. Suppl. Ser. 2004, 151, 35–73. [Google Scholar] [CrossRef]
- Ho, P.T.P.; Townes, C.H. Interstellar ammonia. Annu. Rev. Astron. Astrophys. 1983, 21, 239–270. [Google Scholar] [CrossRef]
- Woodman, J.H.; Trafton, L.; Owen, T. The abundances of ammonia in the atmospheres of Jupiter, Saturn, and Titan. Icarus 1977, 32, 314–320. [Google Scholar] [CrossRef]
- Bottke, W.F.; Norman, M.D. The late heavy bombardment. Annu. Rev. Earth Planet. Sci. 2017, 45, 619–647. [Google Scholar] [CrossRef]
- Deienno, R.; Morbidelli, A.; Gomes, R.S.; Nesvorný, D. Constraining the giant planets’ initial configuration from their evolution: Implications for the timing of the planetary instability. Astron. J. 2017, 153, 153. [Google Scholar] [CrossRef]
- Schlesinger, G.; Miller, S.L. Prebiotic synthesis in atmospheres containing CH4, CO, and CO2. J. Mol. Evol. 1983, 19, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J.D. The origin of life—Out of the blue. Angew. Chemie Int. Ed. 2016, 55, 104–121. [Google Scholar] [CrossRef] [PubMed]
- Taubner, R.S.; Rittmann, S.K.M.R. Method for indirect quantification of CH4 production via H2O production using hydrogenotrophic methanogens. Front. Microbiol. 2016, 7, 532. [Google Scholar] [CrossRef] [PubMed]
- Baumann, L.M.F.; Taubner, R.-S.; Bauersachs, T.; Steiner, M.; Schleper, C.; Peckmann, J.; Rittmann, S.K.-M.R.; Birgel, D. Intact polar lipid and core lipid inventory of the hydrothermal vent methanogens Methanocaldococcus villosus and Methanothermococcus okinawensis. Org. Geochem. 2018, 126, 33–42. [Google Scholar] [CrossRef]
- Krömer, J.O.; Fritz, M.; Heinzle, E.; Wittmann, C. In vivo quantification of intracellular amino acids and intermediates of the methionine pathway in Corynebacterium glutamicum. Anal. Biochem. 2005, 340, 171–173. [Google Scholar] [CrossRef]
- Clifford, E.L.; Hansell, D.A.; Varela, M.M.; Nieto-Cid, M.; Herndl, G.J.; Sintes, E. Crustacean zooplankton release copious amounts of dissolved organic matter as taurine in the ocean. Limnol. Oceanogr. 2017, 62, 2745–2758. [Google Scholar] [CrossRef]
- Hui, W.; Gel, Y.R.; Gastwirth, J.L. Lawstat: An R Package for Law, Public Policy and Biostatistics. J. Stat. Software 2008, 28. [Google Scholar] [CrossRef]
- Yoshinaga, M.Y.; Gagen, E.J.; Wörmer, L.; Broda, N.K.; Meador, T.B.; Wendt, J.; Thomm, M.; Hinrichs, K.-U. Methanothermobacter thermautotrophicus modulates its membrane lipids in response to hydrogen and nutrient availability. Front. Microbiol. 2015, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Siliakus, M.F.; van der Oost, J.; Kengen, S.W.M. Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 2017, 21, 651–670. [Google Scholar] [CrossRef] [PubMed]
- Sprott, G.D.; Meloche, M.; Richards, J.C. Proportions of diether, macrocyclic diether, and tetraether lipids in Methanococcus jannaschii grown at different temperatures. J. Bacteriol. 1991, 173, 3907–3910. [Google Scholar] [CrossRef] [PubMed]
- Kaneshiro, S.M.; Clark, D.S. Pressure effects on the composition and thermal behavior of lipids from the deep-sea thermophile Methanococcus jannaschii. J. Bacteriol. 1995, 177, 3668–3672. [Google Scholar] [CrossRef]
- Knappy, C.; Barillà, D.; Chong, J.; Hodgson, D.; Morgan, H.; Suleman, M.; Tan, C.; Yao, P.; Keely, B. Mono-, di- and trimethylated homologues of isoprenoid tetraether lipid cores in archaea and environmental samples: Mass spectrometric identification and significance. J. Mass Spectrom. 2015, 50, 1420–1432. [Google Scholar] [CrossRef]
- Knappy, C.S. Mass Spectrometric Studies of Ether Lipids in Archaea and Sediments. Ph.D. Thesis, University of York, Hearlington, York, UK, 2010. [Google Scholar]
- Kramer, J.K.G.; Sauer, F.D. Changes in the diether-to-tetraether-lipid ratio during cell growth in Methanobacterium thermoautotrophicum. FEMS Microbiol. Lett. 1991, 83, 45–50. [Google Scholar] [CrossRef]
- Morii, H.; Koga, Y. Tetraether type polar lipids increase after logarithmic growth phase of Methanobacterium thermoautotrophicum in compensation for the decrease of diether lipids. FEMS Microbiol. Lett. 1993, 109, 283–287. [Google Scholar] [CrossRef]
- Gräther, O.W. Zur Struktur und Biosynthese der Tetraetherlipide der Archea. Ph.D. Thesis, ETH Zürich, Zürich, Switzerland, 1994. [Google Scholar]
- De-Bashan, L.E.; Bashan, Y.; Moreno, M.; Lebsky, V.K.; Bustillos, J.J. Increased pigment and lipid content, lipid variety, and cell and population size of the microalgae Chlorella spp. when co-immobilized in alginate beads with the microalgae-growth-promoting bacterium Azospirillum brasilense. Can. J. Microbiol. 2002, 48, 514–521. [Google Scholar] [CrossRef]
- Naafs, B.D.A.; McCormick, D.; Inglis, G.N.; Pancost, R.D. Archaeal and bacterial H-GDGTs are abundant in peat and their relative abundance is positively correlated with temperature. Geochim. Cosmochim. Acta 2018, 227, 156–170. [Google Scholar] [CrossRef]
- McCartney, C.A.; Dewhurst, R.J.; Bull, I.D. Changes in the ratio of tetraether to diether lipids in cattle feces in response to altered dietary ratio of grass silage and concentrates. J. Anim. Sci. 2014, 92, 4095–4098. [Google Scholar] [CrossRef]
- van de Vossenberg, J.L.C.M.; Driessen, A.J.M.; Konings, W.N. The essence of being extremophilic: The role of the unique archaeal membrane lipids. Extremophiles 1998, 2, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Hegemann, W. Anaerobic toxicity and biodegradation of formaldehyde in batch cultures. Water Res. 1998, 32, 209–215. [Google Scholar] [CrossRef]
- Kato, S.; Kosaka, T.; Watanabe, K. Comparative transcriptome analysis of responses of Methanothermobacter thermautotrophicus to different environmental stimuli. Environ. Microbiol. 2008, 10, 893–905. [Google Scholar] [CrossRef]
- Bang, C.; Schilhabel, A.; Weidenbach, K.; Kopp, A.; Goldmann, T.; Gutsmann, T.; Schmitz, R.A. Effects of antimicrobial peptides on methanogenic archaea. Antimicrob. Agents Chemother. 2012, 56, 4123–4130. [Google Scholar] [CrossRef] [PubMed]
- Koga, Y.; Morii, H. Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol. Mol. Biol. Rev. 2007, 71, 97–120. [Google Scholar] [CrossRef] [PubMed]
- Caforio, A.; Driessen, A.J.M. Archaeal phospholipids: Structural properties and biosynthesis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1325–1339. [Google Scholar] [CrossRef]
- Kushwaha, S.C.; Kates, M.; Sprott, G.D.; Smith, I.C.P. Novel polar lipids from the methanogen Methanospirillum hungatei GP1. Biochim. Biophys. Acta Lipids Lipid Metab. 1981, 664, 156–173. [Google Scholar] [CrossRef]
- Morii, H.; Koga, Y. Asymmetrical topology of diether- and tetraether-type polar lipids in membranes of Methanobacterium thermoautotrophicum cells. J. Biol. Chem. 1994, 269, 10492–10497. [Google Scholar]
- Kon, T.; Nemoto, N.; Oshima, T.; Yamagishi, A. Effects of a squalene epoxidase inhibitor, terbinafine, on ether lipid biosyntheses in a thermoacidophilic archaeon, Thermoplasma acidophilum. J. Bacteriol. 2002, 184, 1395–1401. [Google Scholar] [CrossRef]
- Nemoto, N.; Shida, Y.; Shimada, H.; Oshima, T.; Yamagishi, A. Characterization of the precursor of tetraether lipid biosynthesis in the thermoacidophilic archaeon Thermoplasma acidophilum. Extremophiles 2003, 7, 235–243. [Google Scholar] [CrossRef]
- Kuan, Y.; Charnley, S.B.; Huang, H.; Tseng, W.; Kisiel, Z. Interstellar glycine. Astrophys. J. 2003, 593, 848–867. [Google Scholar] [CrossRef]
- Snyder, L.E.; Lovas, F.J.; Hollis, J.M.; Friedel, D.N.; Jewell, P.R.; Remijan, A.; Ilyushin, V.V.; Alekseev, E.A.; Dyubko, S.F. A rigorous attempt to verify interstellar glycine. Astrophys. J. 2005, 619, 914–930. [Google Scholar] [CrossRef]
- Elsila, J.E.; Glavin, D.P.; Dworkin, J.P. Cometary glycine detected in samples returned by Stardust. Meteorit. Planet. Sci. 2009, 44, 1323–1330. [Google Scholar] [CrossRef]
- Altwegg, K.; Balsiger, H.; Bar-Nun, A.; Berthelier, J.-J.; Bieler, A.; Bochsler, P.; Briois, C.; Calmonte, U.; Combi, M.R.; Cottin, H.; et al. Prebiotic chemicals—amino acid and phosphorus—in the coma of comet 67P/Churyumov-Gerasimenko. Sci. Adv. 2016, 2, e1600285. [Google Scholar] [CrossRef]
- Zaia, D.A.M.; Zaia, C.T.B.V.; De Santana, H. Which amino acids should be used in prebiotic chemistry studies? Orig. Life Evol. Biosph. 2008, 38, 469–488. [Google Scholar] [CrossRef]
- Koga, T.; Naraoka, H. A new family of extraterrestrial amino acids in the Murchison meteorite. Sci. Rep. 2017, 7, 636. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Y.; Alexandre, M.R.; Lee, T.; Rose-Petruck, C.; Fuller, M.; Pizzarello, S. Molecular and compound-specific isotopic characterization of monocarboxylic acids in carbonaceous meteorites. Geochim. Cosmochim. Acta 2005, 69, 1073–1084. [Google Scholar] [CrossRef]
- Hilts, R.W.; Herd, C.D.K.; Simkus, D.N.; Slater, G.F. Soluble organic compounds in the Tagish Lake meteorite. Meteorit. Planet. Sci. 2014, 49, 526–549. [Google Scholar] [CrossRef]
- Chyba, C.; Sagan, C. Endogenous production, exogenous delivery and impact-shock synthesis of organic molecules: An inventory for the origins of life. Nature 1992, 355, 125–132. [Google Scholar] [CrossRef]
- Steel, E.L.; Davila, A.; McKay, C.P. Abiotic and biotic formation of amino acids in the Enceladus ocean. Astrobiology 2017, 17, 862–875. [Google Scholar] [CrossRef]
- McKay, C.P.; Davila, A.; Glein, C.R.; Hand, K.; Stockton, A.M. Enceladus astrobiology, habitability, and the origin of life. In Enceladus and the Icy Moons of Saturn; Schenk, P.M., Clark, R.N., Howett, C.J.A., Verbiscer, A.J., Waite, J.H., Eds.; University of Arizona Press: Tucson, AZ, USA, 2018; pp. 437–452. [Google Scholar]
- Chyba, C.F.; Phillips, C.B. Possible ecosystems and the search for life on Europa. Proc. Natl. Acad. Sci. USA 2001, 98, 801–804. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Savageau, M.A.; Alves, R. Relative amino acid composition signatures of organisms and environments. PLoS ONE 2013, 8, e77319. [Google Scholar] [CrossRef] [PubMed]
- Cronin, J.R.; Moore, C.B. Amino acid analyses of the Murchison, Murray, and Allende carbonaceous chondrites. Science 1971, 172, 1327–1329. [Google Scholar] [CrossRef] [PubMed]
- Davila, A.F.; McKay, C.P. Chance and necessity in biochemistry: Implications for the search for extraterrestrial biomarkers in Earth-like environments. Astrobiology 2014, 14, 534–540. [Google Scholar] [CrossRef]
- Guzman, M.; Lorenz, R.; Hurley, D.; Farrell, W.; Spencer, J.; Hansen, C.; Hurford, T.; Ibea, J.; Carlson, P.; McKay, C.P. Collecting amino acids in the Enceladus plume. Int. J. Astrobiol. 2018, 18, 47–59. [Google Scholar] [CrossRef]
- Georgiou, C.D. Functional properties of amino acid side chains as biomarkers of extraterrestrial life. Astrobiology 2018, 18, 1479–1496. [Google Scholar] [CrossRef]
- Truong, N.; Monroe, A.A.; Glein, C.R.; Anbar, A.D.; Lunine, J.I. Decomposition of amino acids in water with application to in-situ measurements of Enceladus, Europa and other hydrothermally active icy ocean worlds. Icarus 2019, 329, 140–147. [Google Scholar] [CrossRef]
- Summons, R.E.; Powell, T.G.; Boreham, C.J. Petroleum geology and geochemistry of the Middle Proterozoic McArthur Basin, Northern Australia: III. Composition of extractable hydrocarbons. Geochim. Cosmochim. Acta 1988, 52, 1747–1763. [Google Scholar] [CrossRef]
- Summons, R.E.; Albrecht, P.; McDonald, G.; Moldowan, J.M. Molecular biosignatures. Space Sci. Rev. 2008, 135, 133–159. [Google Scholar] [CrossRef]
- Georgiou, C.D.; Deamer, D.W. Lipids as universal biomarkers of extraterrestrial life. Astrobiology 2014, 14, 541–549. [Google Scholar] [CrossRef]
- Bradley, A.S.; Hayes, J.M.; Summons, R.E. Extraordinary 13C enrichment of diether lipids at the Lost City Hydrothermal Field indicates a carbon-limited ecosystem. Geochim. Cosmochim. Acta 2009, 73, 102–118. [Google Scholar] [CrossRef]
- Blumenberg, M.; Seifert, R.; Petersen, S.; Michaelis, W. Biosignatures present in a hydrothermal massive sulfide from the Mid-Atlantic Ridge. Geobiology 2007, 5, 435–450. [Google Scholar] [CrossRef]
- Blumenberg, M.; Seifert, R.; Buschmann, B.; Kiel, S.; Thiel, V. Biomarkers reveal diverse microbial communities in black smoker sulfides from Turtle Pits (Mid-Atlantic ridge, recent) and Yaman Kasy (Russia, Silurian). Geomicrobiol. J. 2012, 29, 66–75. [Google Scholar] [CrossRef]
- Jaeschke, A.; Eickmann, B.; Lang, S.Q.; Bernasconi, S.M.; Strauss, H.; Früh-Green, G.L. Biosignatures in chimney structures and sediment from the Loki’s Castle low-temperature hydrothermal vent field at the Arctic Mid-Ocean Ridge. Extremophiles 2014, 18, 545–560. [Google Scholar] [CrossRef]
- Zwicker, J.; Birgel, D.; Bach, W.; Richoz, S.; Smrzka, D.; Grasemann, B.; Gier, S.; Schleper, C.; Rittmann, S.K.-M.R.; Koşun, E.; et al. Evidence for archaeal methanogenesis within veins at the onshore serpentinite-hosted Chimaera seeps, Turkey. Chem. Geol. 2018, 483, 567–580. [Google Scholar] [CrossRef]
- Birgel, D.; Himmler, T.; Freiwald, A.; Peckmann, J. A new constraint on the antiquity of anaerobic oxidation of methane: Late Pennsylvanian seep limestones from southern Namibia. Geology 2008, 36, 543–546. [Google Scholar] [CrossRef]
- Schinteie, R.; Brocks, J.J. Paleoecology of Neoproterozoic hypersaline environments: Biomarker evidence for haloarchaea, methanogens, and cyanobacteria. Geobiology 2017, 15, 641–663. [Google Scholar] [CrossRef]
- Judge, P. A Novel strategy to seek biosignatures at Enceladus and Europa. Astrobiology 2017, 17, 852–861. [Google Scholar] [CrossRef]
- Taubner, R. An interdisciplinary approach: On the habitability of Enceladus’ potential subsurface water reservoir. Ph.D. Thesis, University of Vienna, Wien, Austria, 2018. [Google Scholar]
- Lunine, J.I.; Coustenis, A.; Mitri, G.; Tobie, G.; Tosi, F. Future exploration of Enceladus and other Saturnian moons. In Enceladus and the Icy Moons of Saturn; Schenk, P.M., Clark, R.N., Howett, C.J.A., Verbiscer, A.J., Waite, J.H., Eds.; University of Arizona Press: Tucson, AZ, USA, 2018; pp. 453–468. [Google Scholar]
- Taubner, R.-S.; Olsson-Francis, K.; Vance, S.; Ramkissoon, N.; Postberg, F.; de Vera, J.-P.; Antunes, A.; Camprubi Casas, E.; Sekine, Y.; Noack, L.; et al. Experimental and simulation efforts in the astrobiological exploration of exooceans. Space Sci. Rev. 2019. (submitted). [Google Scholar]
- Deamer, D.; Damer, B. Can life begin on Enceladus? A perspective from hydrothermal chemistry. Astrobiology 2017, 17, 834–839. [Google Scholar] [CrossRef]
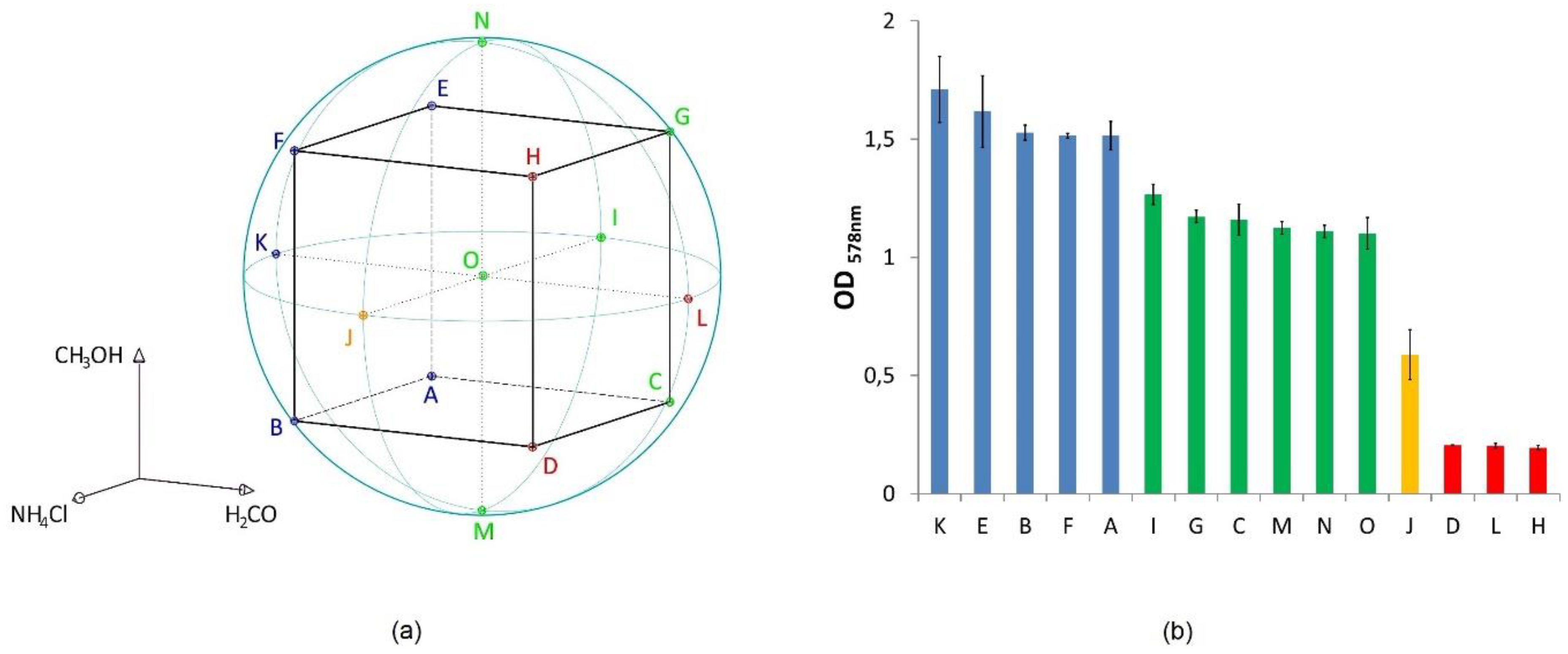
| Minimum Value | Mean Value | Maximum Value | |
|---|---|---|---|
| ODmax | 0.20 ± 0.01 (H) | 1.07 ± 0.05 | 1.71 ± 0.14 (K) |
| Turnover rate max [h−1] | 0.028 ± 0.003 (L) | 0.082 ± 0.002 | 0.098 ± 0.000 (G) |
| Diethers [ng mg−1 dw] | 198.41 ± 58.42 (L) | 583.96 ± 68.73 | 883.79 ± 197.54 (K) |
| Tetraethers [ng mg−1 dw] | 16.89 ± 15.16 (L) | 42.50 ± 15.63 | 68.36 ± 7.04 (A) |
| Diethers/tetraethers | 11.53 ± 2.14 (H) | 15.31 ± 5.10 | 19.43 ± 7.00 (G) |
| Total lipids [ng mg−1 dw] | 215.31 ± 73.42 (L) | 626.46 ± 78.95 | 938.11 ± 228.19 (K) |
| Total amino acids [µmol L−1] | 62.08 ± 9.82 (H) | 228.00 ± 34.27 | 663.58 ± 129.24 (I) |
| Minimum Value | Mean Value | Maximum Value | |
|---|---|---|---|
| ODmax | 0.22 ± 0.10 (Fo) | 0.73 ± 0.43 | 1.10 ± 0.05 (Min) |
| Turnover rate max [h−1] | 0.051 ± 0.021 (Fo) | 0.079 ± 0.019 | 0.094 ± 0.005 (Min) |
| Archaeol/macrocyclic archaeol | 0.76 ± 0.20 (Me) | 2.23 ± 2.76 | 6.38 ± 3.96 (Fo) |
| Diethers/tetraethers | 3.68 ± 1.51 (Min) | 5.48 ± 2.41 | 8.83 ± 2.34 (Fo) |
| GTGT/GDGT | 0.03 ± 0.01 (Min) | 0.07 ± 0.06 | 0.16 ± 0.05 (Fo) |
| GDGT/GMGT | 0.83 ± 0.12 (Me) | 1.42 ± 0.83 | 2.67 ± 0.60 (Fo) |
| GTGT/GMGT | 0.02 ± 0.01 (Me) | 0.14 ± 0.20 | 0.44 ±0.17 (Fo) |
| GMGT-0/0’ | 1.96 ± 0.09 (Me) | 2.18 ± 0.40 | 2.78 ± 0.58 (Fo) |
| Total amino acids [µmol L−1] | 47.22 ± 7.93 (Am) | 180.70 ± 94.02 | 268.08 ± 101.83 (Me) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taubner, R.-S.; Baumann, L.M.F.; Bauersachs, T.; Clifford, E.L.; Mähnert, B.; Reischl, B.; Seifert, R.; Peckmann, J.; Rittmann, S.K.-M.R.; Birgel, D. Membrane Lipid Composition and Amino Acid Excretion Patterns of Methanothermococcus okinawensis Grown in the Presence of Inhibitors Detected in the Enceladian Plume. Life 2019, 9, 85. https://doi.org/10.3390/life9040085
Taubner R-S, Baumann LMF, Bauersachs T, Clifford EL, Mähnert B, Reischl B, Seifert R, Peckmann J, Rittmann SK-MR, Birgel D. Membrane Lipid Composition and Amino Acid Excretion Patterns of Methanothermococcus okinawensis Grown in the Presence of Inhibitors Detected in the Enceladian Plume. Life. 2019; 9(4):85. https://doi.org/10.3390/life9040085
Chicago/Turabian StyleTaubner, Ruth-Sophie, Lydia M. F. Baumann, Thorsten Bauersachs, Elisabeth L. Clifford, Barbara Mähnert, Barbara Reischl, Richard Seifert, Jörn Peckmann, Simon K.-M. R. Rittmann, and Daniel Birgel. 2019. "Membrane Lipid Composition and Amino Acid Excretion Patterns of Methanothermococcus okinawensis Grown in the Presence of Inhibitors Detected in the Enceladian Plume" Life 9, no. 4: 85. https://doi.org/10.3390/life9040085
APA StyleTaubner, R.-S., Baumann, L. M. F., Bauersachs, T., Clifford, E. L., Mähnert, B., Reischl, B., Seifert, R., Peckmann, J., Rittmann, S. K.-M. R., & Birgel, D. (2019). Membrane Lipid Composition and Amino Acid Excretion Patterns of Methanothermococcus okinawensis Grown in the Presence of Inhibitors Detected in the Enceladian Plume. Life, 9(4), 85. https://doi.org/10.3390/life9040085





