Prebiotic Soup Components Trapped in Montmorillonite Nanoclay Form New Molecules: Car-Parrinello Ab Initio Simulations
Abstract
1. Introduction
1.1. Possible Origins of Life Scenarios—A Short Review
1.2. Catalytic Surfaces, Nanoconfinement, and Biogenesis
1.3. Theoretical Chemistry in Origins of Life Research
1.4. Our Aim
2. Methods
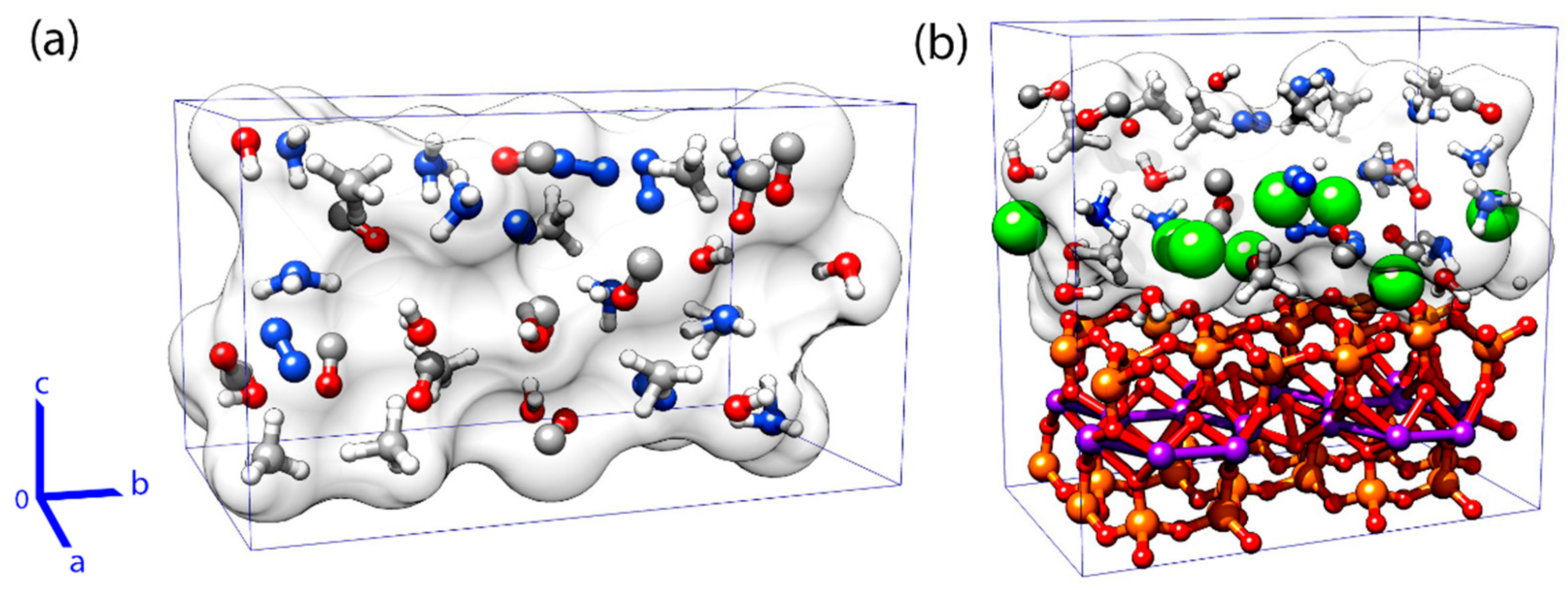
2.1. Systems
2.2. CPMD Simulations
3. Results and Discussion
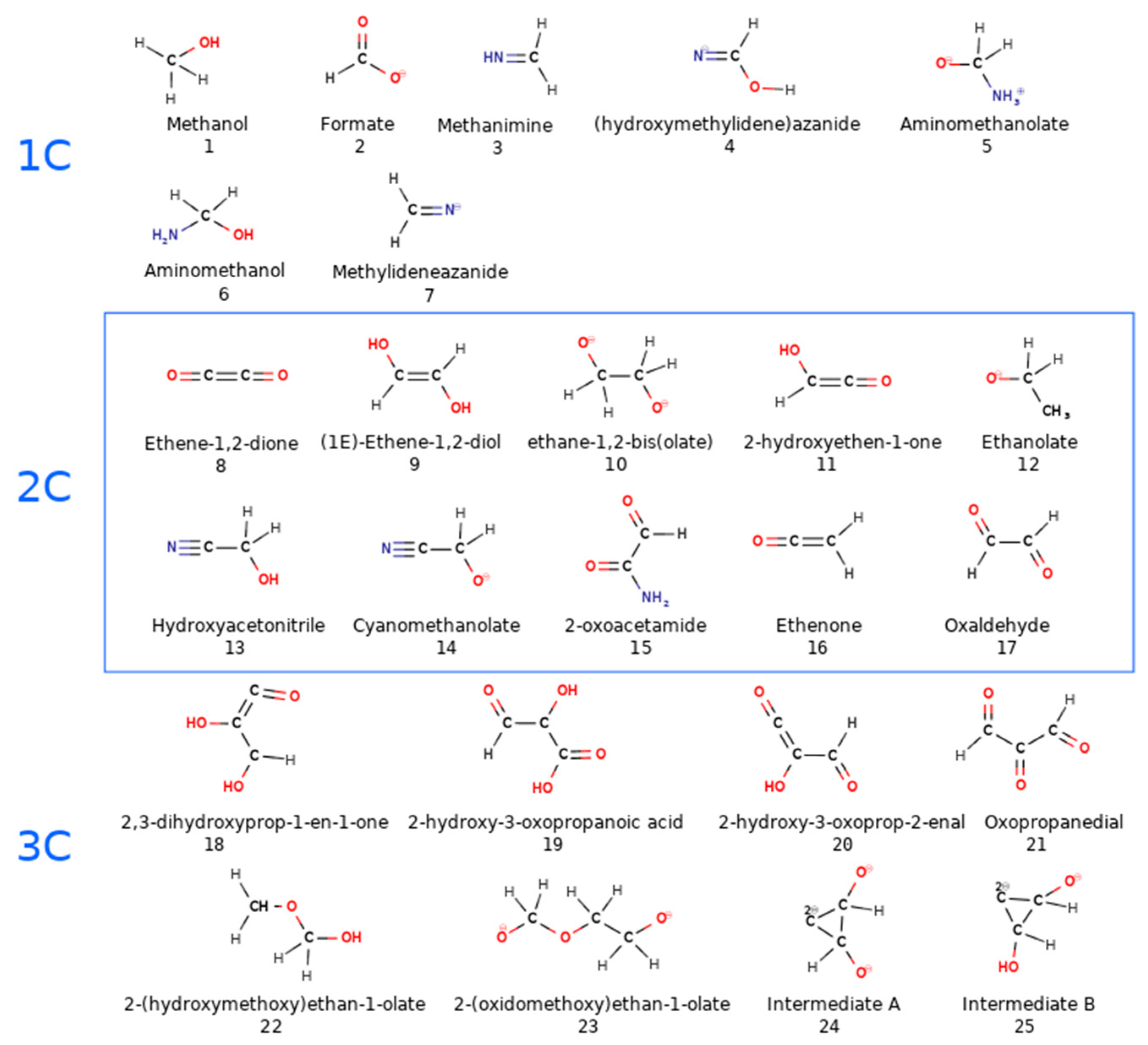
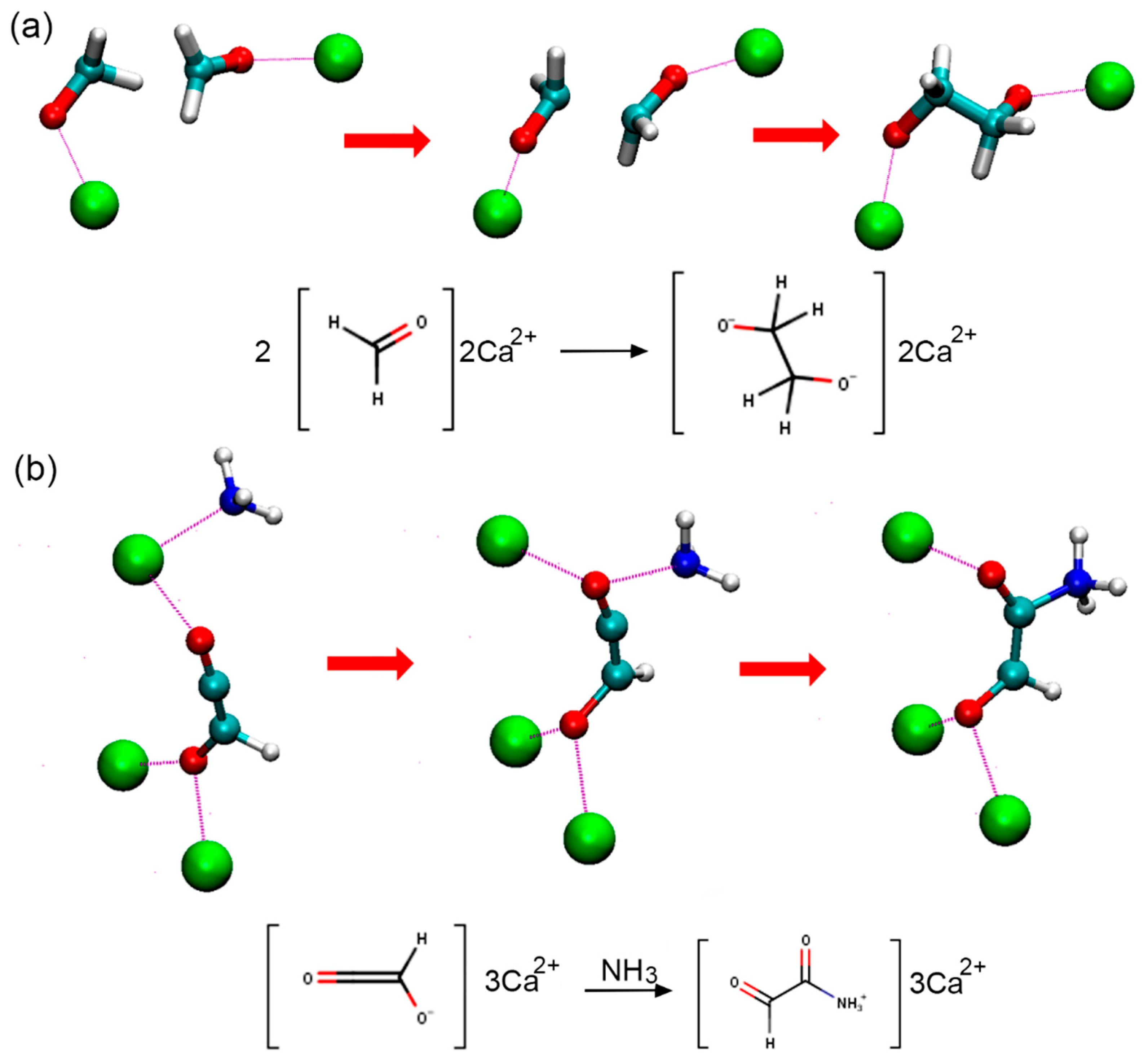
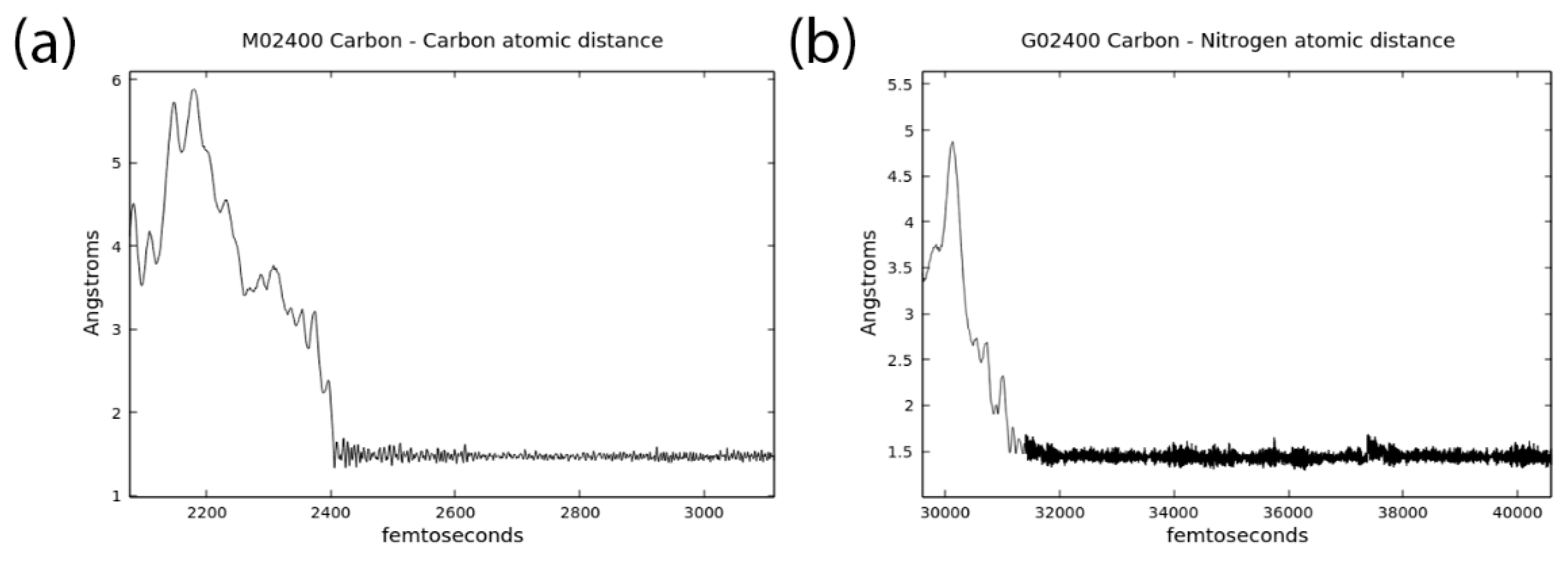
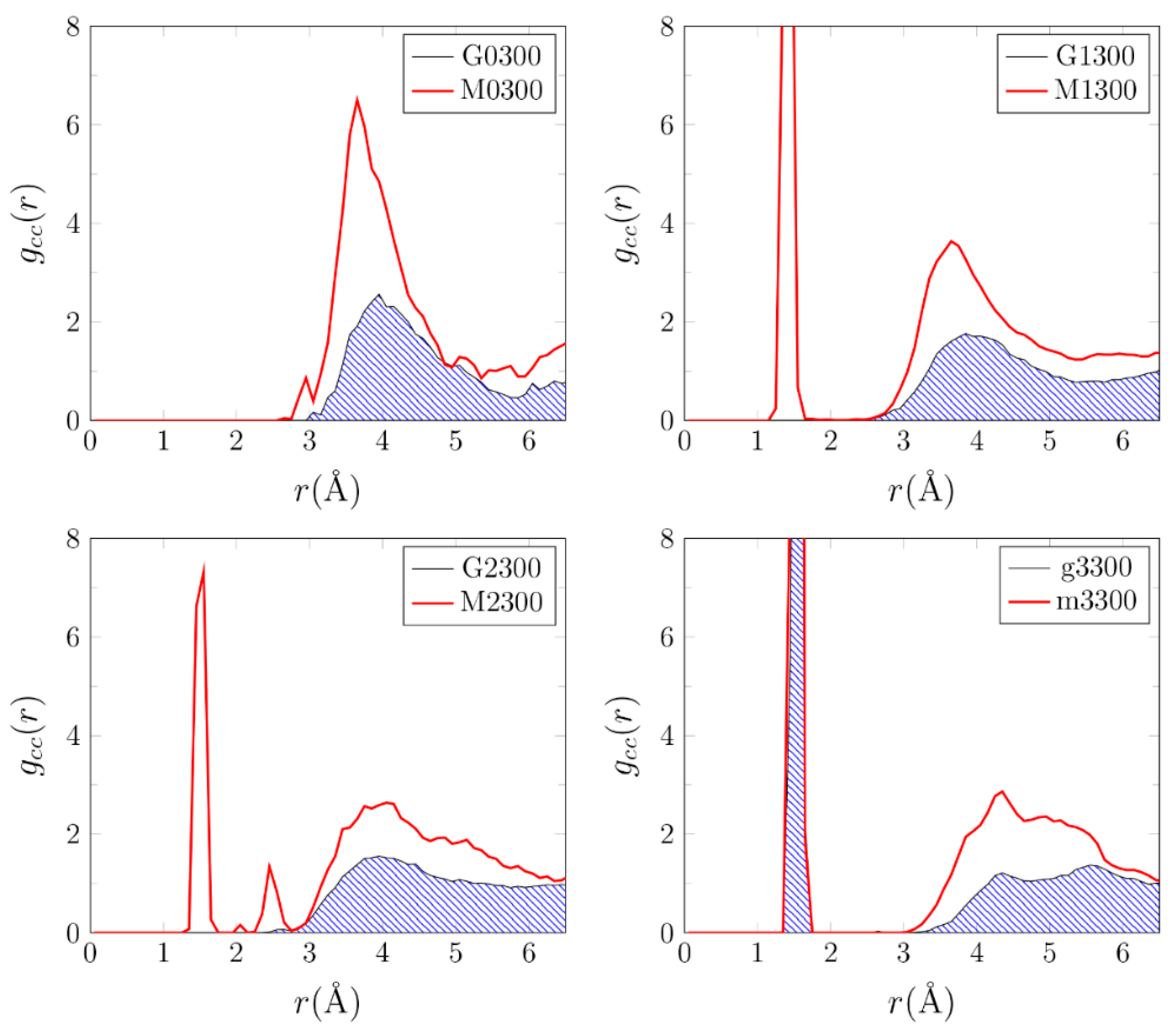
3.1. Effects of MMT on Chemical Reactivity
3.2. Primordial Soup Ingredient Dynamics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| a.u. | atomic units |
| Å | Angstroms |
| AIMD | ab initio molecular dynamics |
| Approx. | approximately |
| atm | atmospheres |
| CCL2 | CC chemokine ligand 2 |
| CCR2 | CC chemokine receptor 2 |
| CCR5 | CC chemokine receptor 5 |
| CPMD | Car-Parrinello molecular dynamics |
| DFT | density functional theory |
| Dimension c | refers to the size of the vector c |
| fs | femtoseconds |
| g | gases only box |
| g/mL | grams per milliliter |
| refers to the atom-atom radial distribution function | |
| HCN | hydrogen cyanide |
| i.a. | Latin meaning “among other things” |
| i.e., | Latin meaning “in essence/in other words” |
| K | Kelvin |
| kcal/mol | kilocalories per mol |
| m | montmorillonite and gases box |
| MD | molecular dynamics |
| MMT | montmorillonite |
| nm | nanometer |
| NVT | refers to the canonical enssemble at |
| PBC | periodic boundary conditions |
| pH | potential hydrogen |
| ps | picoseconds |
| RMSD | root mean square deviation |
| RNA | ribonucleic acid |
| Ry | Rydberg atomic units |
| THz | terahertz |
| TLC | thin layer chromatography |
| UV | ultraviolet |
| UV–Vis | ultraviolet-visible |
References
- Bhushan, B.; Nayak, A.; Kamaluddin. Catalytic Role of Manganese Oxides in Prebiotic Nucleobases Synthesis from Formamide. Orig. Life Evol. Biosph. 2016, 46, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Pollet, R.; Boehme, C.; Marx, D. Ab Initio Simulations of Desorption and Reactivity of Glycine at a Water-Pyrite Interface at “Iron-Sulfur World” Prebiotic Conditions. Orig. Life Evol. Biosph. 2006, 36, 363–379. [Google Scholar] [CrossRef] [PubMed]
- Ritson, D.J.; Battilocchio, C.; Ley, S.V.; Sutherland, J.D. Mimicking the Surface and Prebiotic Chemistry of Early Earth Using Flow Chemistry. Nat. Commun. 2018, 9, 1821. [Google Scholar] [CrossRef] [PubMed]
- Szostak, N.; Synak, J.; Borowski, M.; Wasik, S.; Blazewicz, J. Simulating the Origins of Life: The Dual Role of RNA Replicases as an Obstacle to Evolution. PLoS ONE 2017, 12, e0180827. [Google Scholar] [CrossRef] [PubMed]
- Szostak, J.W. The Origin of Life on Earth and the Design of Alternative Life Forms. Mol. Front. J. 2017, 01, 121–131. [Google Scholar] [CrossRef]
- Kitadai, N.; Maruyama, S. Origins of Building Blocks of Life: A Review. Geosci. Front. 2018, 9, 1117–1153. [Google Scholar] [CrossRef]
- Ranjan, S.; Sasselov, D.D. Influence of the UV Environment on the Synthesis of Prebiotic Molecules. Astrobiology 2016, 16, 68–88. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.-F. Adsorption and Polymerization of Amino Acids on Mineral Surfaces: A Review. Orig. Life Evol. Biosph. 2008, 38, 211–242. [Google Scholar] [CrossRef]
- Ruiz-Mirazo, K.; Briones, C.; de la Escosura, A. Chemical Roots of Biological Evolution: The Origins of Life as a Process of Development of Autonomous Functional Systems. Open Biol. 2017, 7, 170050. [Google Scholar] [CrossRef]
- Patel, B.H.; Percivalle, C.; Ritson, D.J.; Duffy, C.D.; Sutherland, J.D. Common Origins of RNA, Protein and Lipid Precursors in a Cyanosulfidic Protometabolism. Nat. Chem. 2015, 7, 301–307. [Google Scholar] [CrossRef]
- Saladino, R.; Crestini, C.; Pino, S.; Costanzo, G.; Di Mauro, E. Formamide and the Origin of Life. Phys. Life Rev. 2012, 9, 84–104. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J.D. The Origin of Life-Out of the Blue. Angew. Chem. Int. Ed. 2016, 55, 104–121. [Google Scholar] [CrossRef] [PubMed]
- Trevors, J.T. Hypothesized Origin of Microbial Life in a Prebiotic Gel and the Transition to a Living Biofilm and Microbial Mats. C. R. Biol. 2011, 334, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Wächtershäuser, G. Groundworks for an Evolutionary Biochemistry: The Iron-Sulphur World. Prog. Biophys. Mol. Biol. 1992, 58, 85–201. [Google Scholar] [CrossRef]
- Miller, S.L. A Production of Amino Acids Under Possible Primitive Earth Conditions. Science 1953, 117, 528–529. [Google Scholar] [CrossRef] [PubMed]
- Bartley, B.A.; Kim, K.; Medley, J.K.; Sauro, H.M. Synthetic Biology: Engineering Living Systems from Biophysical Principles. Biophys. J. 2017, 112, 1050–1058. [Google Scholar] [CrossRef]
- Ferus, M.; Pietrucci, F.; Saitta, A.M.; Knížek, A.; Kubelík, P.; Ivanek, O.; Shestivska, V.; Civiš, S. Formation of Nucleobases in a Miller-Urey Reducing Atmosphere. Proc. Natl. Acad. Sci. USA 2017, 114, 4306–4311. [Google Scholar] [CrossRef]
- Leyton, P. The Role of Minerals on Prebiotic Synthesis: Comment on “Formamide and the Origin of Life” by R. Saladino et al. Phys. Life Rev. 2012, 9, 116–117. [Google Scholar] [CrossRef]
- Huber, C.; Eisenreich, W.; Hecht, S.; Wächtershäuser, G. A Possible Primordial Peptide Cycle. Science 2003, 301, 938–940. [Google Scholar] [CrossRef]
- Huber, C.; Wächtershäuser, G. Peptides by Activation of Amino Acids with CO on (Ni,Fe)S Surfaces: Implications for the Origin of Life. Science 1998, 281, 670–672. [Google Scholar] [CrossRef]
- Chandru, K.; Gilbert, A.; Butch, C.; Aono, M.; Cleaves, H.J. The Abiotic Chemistry of Thiolated Acetate Derivatives and the Origin of Life. Sci. Rep. 2016, 6, 29883. [Google Scholar] [CrossRef] [PubMed]
- Powner, M.W.; Gerland, B.; Sutherland, J.D. Synthesis of Activated Pyrimidine Ribonucleotides in Prebiotically Plausible Conditions. Nature 2009, 459, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Stairs, S.; Nikmal, A.; Bučar, D.-K.; Zheng, S.-L.; Szostak, J.W.; Powner, M.W. Divergent Prebiotic Synthesis of Pyrimidine and 8-Oxo-Purine Ribonucleotides. Nat. Commun. 2017, 8, 15270. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J.D. Opinion: Studies on the Origin of Life—The End of the Beginning. Nat. Rev. Chem. 2017, 1, 0012. [Google Scholar] [CrossRef]
- Xu, J.; Tsanakopoulou, M.; Magnani, C.J.; Szabla, R.; Šponer, J.E.; Šponer, J.; Góra, R.W.; Sutherland, J.D. A Prebiotically Plausible Synthesis of Pyrimidine β-Ribonucleosides and Their Phosphate Derivatives Involving Photoanomerization. Nat. Chem. 2017, 9, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Carota, E.; Botta, G.; Rotelli, L.; Di Mauro, E.; Saladino, R. Current Advances in Prebiotic Chemistry Under Space Conditions. Curr. Org. Chem. 2015, 19, 1963–1979. [Google Scholar] [CrossRef]
- Costanzo, G.; Saladino, R.; Crestini, C.; Ciciriello, F.; Di Mauro, E. Formamide as the Main Building Block in the Origin of Nucleic Acids. BMC Evol. Biol. 2007, 7 (Suppl. 2), S1. [Google Scholar] [CrossRef]
- Costanzo, G.; Giorgi, A.; Scipioni, A.; Timperio, A.M.; Mancone, C.; Tripodi, M.; Kapralov, M.; Krasavin, E.; Kruse, H.; Sponer, J.E.J.; et al. Non-Enzymatic Oligomerization of 3’,5’ Cyclic CMP Induced by Proton- and UV-Irradiation Hints at a Non-Fastidious Origin of RNA. ChemBioChem 2017. [Google Scholar] [CrossRef]
- Ferus, M.; Knizek, A.; Sponer, J.; Sponer, J.E.; Civis, S. RADICAL SYNTHESIS OF NUCLEIC ACID BASES FROM FORMAMIDE IN IMPACT PLASMA. Chem. List. 2015, 109, 406–414. [Google Scholar]
- Ferus, M.; Nesvorný, D.; Šponer, J.; Kubelík, P.; Michalčíková, R.; Shestivská, V.; Šponer, J.E.; Civiš, S. High-Energy Chemistry of Formamide: A Unified Mechanism of Nucleobase Formation. Proc. Natl. Acad. Sci. USA 2015, 112, 657–662. [Google Scholar] [CrossRef]
- Jeilani, Y.A.; Nguyen, H.T.; Newallo, D.; Dimandja, J.-M.D.; Nguyen, M.T. Free Radical Routes for Prebiotic Formation of DNA Nucleobases from Formamide. Phys. Chem. Chem. Phys. 2013, 15, 21084. [Google Scholar] [CrossRef] [PubMed]
- Saladino, R.; Botta, G.; Pino, S.; Costanzo, G.; Di Mauro, E. Materials for the Onset. A Story of Necessity and Chance. Front. Biosci. (Landmark Ed.) 2013, 18, 1275–1289. [Google Scholar] [PubMed]
- Nguyen, H.T.; Jeilani, Y.A.; Hung, H.M.; Nguyen, M.T. Radical Pathways for the Prebiotic Formation of Pyrimidine Bases from Formamide. J. Phys. Chem. A 2015, 119, 8871–8883. [Google Scholar] [CrossRef] [PubMed]
- Cossetti, C.; Crestini, C.; Saladino, R.; di Mauro, E. Borate Minerals and RNA Stability. Polymers 2010, 2, 211–228. [Google Scholar] [CrossRef]
- Saladino, R.; Neri, V.; Crestini, C.; Costanzo, G.; Graciotti, M.; Di Mauro, E. Synthesis and Degradation of Nucleic Acid Components by Formamide and Iron Sulfur Minerals. J. Am. Chem. Soc. 2008, 130, 15512–15518. [Google Scholar] [CrossRef] [PubMed]
- Martins, Z.; Botta, O.; Fogel, M.L.; Sephton, M.A.; Glavin, D.P.; Watson, J.S.; Dworkin, J.P.; Schwartz, A.W.; Ehrenfreund, P. Extraterrestrial Nucleobases in the Murchison Meteorite. Earth Planet. Sci. Lett. 2008, 270, 130–136. [Google Scholar] [CrossRef]
- Saladino, R.; Botta, G.; Delfino, M.; Di Mauro, E. Meteorites as Catalysts for Prebiotic Chemistry. Chem. Eur. J. 2013, 19, 16916–16922. [Google Scholar] [CrossRef] [PubMed]
- Botta, L.; Saladino, R.; Bizzarri, B.M.; Cobucci-Ponzano, B.; Iacono, R.; Avino, R.; Caliro, S.; Carandente, A.; Lorenzini, F.; Tortora, A.; et al. Formamide-Based Prebiotic Chemistry in the Phlegrean Fields. Adv. Sp. Res. 2018, 62, 2372–2379. [Google Scholar] [CrossRef]
- Saitta, A.M.; Saija, F. Miller Experiments in Atomistic Computer Simulations. Proc. Natl. Acad. Sci. USA 2014, 111, 13768–13773. [Google Scholar] [CrossRef] [PubMed]
- Cafferty, B.J.; Fialho, D.M.; Khanam, J.; Krishnamurthy, R.; Hud, N.V. Spontaneous Formation and Base Pairing of Plausible Prebiotic Nucleotides in Water. Nat. Commun. 2016, 7, 11328. [Google Scholar] [CrossRef] [PubMed]
- Attwater, J.; Holliger, P. Origins of Life: The Cooperative Gene. Nature 2012, 491, 48–49. [Google Scholar] [CrossRef] [PubMed]
- Dass, A.; Jaber, M.; Brack, A.; Foucher, F.; Kee, T.; Georgelin, T.; Westall, F. Potential Role of Inorganic Confined Environments in Prebiotic Phosphorylation. Life 2018, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Hanczyc, M.M.; Fujikawa, S.M.; Szostak, J.W. Experimental Models of Primitive Cellular Compartments: Encapsulation, Growth, and Division. Science 2003, 302, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.P. Catalysis and Prebiotic Synthesis. Rev. Mineral. Geochem. 2005, 59, 187–210. [Google Scholar] [CrossRef]
- Joshi, P.C.; Pitsch, S.; Ferris, J.P. Selectivity of Montmorillonite Catalyzed Prebiotic Reactions of D, L-Nucleotides. Orig. Life Evol. Biosph. 2007, 37, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Jheeta, S.; Joshi, P. Prebiotic RNA Synthesis by Montmorillonite Catalysis. Life 2014, 4, 318–330. [Google Scholar] [CrossRef] [PubMed]
- Cygan, R.T.; Romanov, V.N.; Myshakin, E.M. Molecular Simulation of Carbon Dioxide Capture by Montmorillonite Using an Accurate and Flexible Force Field. J. Phys. Chem. C 2012, 116, 13079–13091. [Google Scholar] [CrossRef]
- Michalkova, A.; Robinson, T.L.; Leszczynski, J. Adsorption of Thymine and Uracil on 1:1 Clay Mineral Surfaces: Comprehensive Ab Initio Study on Influence of Sodium Cation and Water. Phys. Chem. Chem. Phys. 2011, 13, 7862–7881. [Google Scholar] [CrossRef] [PubMed]
- Dawley, M.M.; Scott, A.M.; Hill, F.C.; Leszczynski, J.; Orlando, T.M. Adsorption of Formamide on Kaolinite Surfaces: A Combined Infrared Experimental and Theoretical Study. J. Phys. Chem. C 2012, 116, 23981–23991. [Google Scholar] [CrossRef]
- Michalkova Scott, A.; Dawley, M.M.; Orlando, T.M.; Hill, F.C.; Leszczynski, J. Theoretical Study of the Roles of Na+ and Water on the Adsorption of Formamide on Kaolinite Surfaces. J. Phys. Chem. C 2012, 116, 23992–24005. [Google Scholar] [CrossRef]
- Saladino, R.; Ciambecchini, U.; Crestini, C.; Costanzo, G.; Negri, R.; Di Mauro, E. One-Pot TiO2-Catalyzed Synthesis of Nucleic Bases and Acyclonucleosides from Formamide: Implications for the Origin of Life. Chembiochem 2003, 4, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Senanayake, S.D.; Idriss, H. Photocatalysis and the Origin of Life: Synthesis of Nucleoside Bases from Formamide on TiO2(001) Single Surfaces. Proc. Natl. Acad. Sci. USA 2006, 103, 1194–1198. [Google Scholar] [CrossRef] [PubMed]
- Goldman, N.; Reed, E.J.; Fried, L.E.; William Kuo, I.-F.; Maiti, A. Synthesis of Glycine-Containing Complexes in Impacts of Comets on Early Earth. Nat. Chem. 2010, 2, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Martins, Z.; Price, M.C.; Goldman, N.; Sephton, M.A.; Burchell, M.J. Shock Synthesis of Amino Acids from Impacting Cometary and Icy Planet Surface Analogues. Nat. Geosci. 2013, 6, 1045–1049. [Google Scholar] [CrossRef]
- Schreiner, E.; Nair, N.N.; Wittekindt, C.; Marx, D. Peptide Synthesis in Aqueous Environments: The Role of Extreme Conditions and Pyrite Mineral Surfaces on Formation and Hydrolysis of Peptides. J. Am. Chem. Soc. 2011. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Santiburcio, D.; Marx, D. Chemistry in Nanoconfined Water. Chem. Sci. 2017, 8, 3444–3452. [Google Scholar] [CrossRef] [PubMed]
- Car, R.; Parrinello, M. Unified Approach for Molecular Dynamics and Density-Functional Theory. Phys. Rev. Lett. 1985, 55, 2471–2474. [Google Scholar] [CrossRef] [PubMed]
- Stirling, A.; Rozgonyi, T.; Krack, M.; Bernasconi, M. Prebiotic NH3 Formation: Insights from Simulations. Inorg. Chem. 2016, 55, 1934–1939. [Google Scholar] [CrossRef]
- Pietrucci, F.; Saitta, A.M. Formamide Reaction Network in Gas Phase and Solution via a Unified Theoretical Approach: Toward a Reconciliation of Different Prebiotic Scenarios. Proc. Natl. Acad. Sci. USA 2015, 112, 15030–15035. [Google Scholar] [CrossRef]
- Pérez-Villa, A.; Saitta, A.M.; Georgelin, T.; Lambert, J.-F.; Guyot, F.; Maurel, M.-C.; Pietrucci, F. Synthesis of RNA Nucleotides in Plausible Prebiotic Conditions from Ab Initio Computer Simulations. J. Phys. Chem. Lett. 2018, 9, 4981–4987. [Google Scholar] [CrossRef]
- Sena, M.M.; Morrow, C.P.; Kirkpatrick, R.J.; Krishnan, M. Supercritical Carbon Dioxide at Smectite Mineral–Water Interfaces: Molecular Dynamics and Adaptive Biasing Force Investigation of CO2/H2O Mixtures Nanoconfined in Na-Montmorillonite. Chem. Mater. 2015, 27, 6946–6959. [Google Scholar] [CrossRef]
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M.; Martinez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A Package for Building Initial Configurations for Molecular Dynamics Simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.J.; Holian, B.L. The Nose–Hoover Thermostat. J. Chem. Phys. 1985, 83, 4069–4074. [Google Scholar] [CrossRef]
- Giannozzi, P.; Baroni, S.; Bonini, N.; Calandra, M.; Car, R.; Cavazzoni, C.; Ceresoli, D.; Chiarotti, G.L.; Cococcioni, M.; Dabo, I.; et al. QUANTUM ESPRESSO: A Modular and Open-Source Software Project for Quantum Simulations of Materials. J. Phys. Condens. Matter 2009, 21, 395502. [Google Scholar] [CrossRef] [PubMed]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef]
- Ahmed Adllan, A.; Dal Corso, A. Ultrasoft Pseudopotentials and Projector Augmented-Wave Data Sets: Application to Diatomic Molecules. J. Phys. Condens. Matter 2011, 23, 425501. [Google Scholar] [CrossRef] [PubMed]
- Verlet, L. Computer “Experiments” on Classical Fluids. I. Thermodynamical Properties of Lennard-Jones Molecules. Phys. Rev. 1967, 159, 98–103. [Google Scholar] [CrossRef]
- Tribello, G.A.; Bonomi, M.; Branduardi, D.; Camilloni, C.; Bussi, G. PLUMED 2: New Feathers for an Old Bird. Comput. Phys. Commun. 2014, 185, 604–613. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Laporte, S.; Finocchi, F.; Paulatto, L.; Blanchard, M.; Balan, E.; Guyot, F.; Saitta, A.M. Strong Electric Fields at a Prototypical Oxide/Water Interface Probed by Ab Initio Molecular Dynamics: MgO(001). Phys. Chem. Chem. Phys. 2015, 17, 20382–20390. [Google Scholar] [CrossRef]
- Ranjan, S.; Todd, Z.R.; Sutherland, J.D.; Sasselov, D.D. Sulfidic Anion Concentrations on Early Earth for Surficial Origins-of-Life Chemistry. Astrobiology 2018, 18, 1023–1040. [Google Scholar] [CrossRef] [PubMed]
- Rydzewski, J.; Nowak, W. Ligand Diffusion in Proteins via Enhanced Sampling in Molecular Dynamics. Phys. Life Rev. 2017, 22–23, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Powner, M.W. Prebiotic Systems Chemistry: Complexity Overcoming Clutter. Chem 2017, 2, 470–501. [Google Scholar] [CrossRef]
| Box | Temperature | No. Atoms | Chemical Species | Dimension c [Å] |
|---|---|---|---|---|
| g0300 | 300 | 176 | 32 H2O; 8 NH3; 8 CH4; 4 H2 | 10.43 |
| g0400 | 400 | |||
| g0600 | 600 | |||
| g1300 | 300 | 126 | 8 H2O; 8 NH3; 8 CH4; 5 N2; 10 CO | 7.41 |
| g1400 | 400 | |||
| g1600 | 600 | |||
| g2300 | 300 | 126 | 9 H2O; 9 NH3; 9 Formaldehyde; 9 HCN | 7.41 |
| g2400 | 400 | |||
| g2600 | 600 | |||
| g3300 | 300 | 126 | 9 NH3; 9 Glycine | 7.41 |
| g3400 | 400 | |||
| g3600 | 600 | |||
| m0300 | 300 | 328 | 4 MMT; 32 H2O; 8 NH3; 8 CH4; 4 H2 | 18.07 |
| m0400 | 400 | |||
| m0600 | 600 | |||
| m1300 | 300 | 278 | 4 MMT; 8 H2O; 8 NH3; 8 CH4; 5 N2; 10 CO | 17.90 |
| m1400 | 400 | |||
| m1600 | 600 | |||
| m2300 | 300 | 278 | 4 MMT; 9 H2O; 9 NH3; 9 Formaldehyde; 9 HCN | 17.90 |
| m2400 | 400 | |||
| m2600 | 600 | |||
| m3300 | 300 | 278 | 4 MMT; 9 NH3; 9 Glycine | 17.90 |
| m3400 | 400 | |||
| m3600 | 600 |
| Box | C C | C N | C O | Products |
|---|---|---|---|---|
| g0300 | 0 | 0 | 0 | 0 |
| g0400 | 0 | 0 | 0 | 0 |
| g0600 | 0 | 0 | 0 | 0 |
| g1300 | 0 | 0 | 0 | 0 |
| g1400 | 0 | 0 | 0 | 0 |
| g1600 | 0 | 0 | 0 | 0 |
| g2300 | 0 | 15 | 0 | 6 |
| g2400 | 0 | 1 | 43 | 6 |
| g2600 | 6 | 0 | 0 | 14 |
| g3300 | 0 | 0 | 0 | 0 |
| g3400 | 0 | 0 | 0 | 0 |
| g3600 | 0 | 0 | 0 | 0 |
| m0300 | 0 | 0 | 0 | 0 |
| m0400 | 0 | 0 | 0 | 0 |
| m0600 | 0 | 0 | 0 | 0 |
| m1300 | 30 | 0 | 5 | 24, 25 |
| m1400 | 24 | 0 | 12 | 8, 9, 11, 18, 19, 20, 21 |
| m1600 | 4 | 0 | 3 | 9, 11, 12, 15, 18, 21 |
| m2300 | 9 | 7 | 2 | 1, 3, 4, 5, 13, 22 |
| m2400 | 17 | 2 | 9 | 1, 5, 6, 7, 10, 14, 22, 23 |
| m2600 | 17 | 1 | 6 | 1, 2, 3, 6, 7, 10, 12, 13 |
| m3300 | 0 | 7 | 16 | 0 |
| m3400 | 0 | 20 | 15 | 0 |
| m3600 | 0 | 7 | 4 | 0 |
| Box | C | H | N | O |
|---|---|---|---|---|
| m0300 | 0 | 126 | 30 | 905 |
| m0400 | 0 | 108 | 30 | 964 |
| m0600 | 0 | 140 | 32 | 1083 |
| m1300 | 10 | 41 | 37 | 649 |
| m1400 | 10 | 19 | 56 | 666 |
| m1600 | 8 | 47 | 50 | 882 |
| m2300 | 3 | 13 | 70 | 518 |
| m2400 | 4 | 28 | 90 | 603 |
| m2600 | 9 | 8 | 181 | 568 |
| m3300 | 0 | 1 | 9 | 825 |
| m3400 | 0 | 3 | 30 | 915 |
| m3600 | 0 | 4 | 20 | 985 |
| Box | Al | Si | Ca | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Avg. | Min. | Max. | Avg. | Min. | Max. | Avg. | Min. | Max. | |
| m0300 | 0.17 | 0.03 | 0.24 | 0.18 | 0.04 | 0.24 | 0.63 | 0.03 | 0.86 |
| m0400 | 0.20 | 0.04 | 0.27 | 0.21 | 0.08 | 0.24 | 0.43 | 0.03 | 0.75 |
| m0600 | 0.28 | 0.06 | 0.39 | 0.29 | 0.06 | 0.39 | 0.62 | 0.05 | 1.00 |
| m1300 | 0.38 | 0.01 | 0.58 | 0.36 | 0.01 | 0.51 | 0.66 | 0.05 | 1.00 |
| m1400 | 0.21 | 0.07 | 0.34 | 0.25 | 0.04 | 0.46 | 0.51 | 0.04 | 0.82 |
| m1600 | 0.27 | 0.11 | 0.39 | 0.54 | 0.06 | 1.00 | 0.58 | 0.04 | 0.99 |
| m2300 | 0.16 | 0.04 | 0.38 | 0.46 | 0.03 | 0.55 | 0.66 | 0.05 | 1.00 |
| m2400 | 0.21 | 0.04 | 0.34 | 0.43 | 0.03 | 0.59 | 0.72 | 0.03 | 1.00 |
| m2600 | 0.28 | 0.04 | 0.45 | 0.57 | 0.04 | 0.73 | 0.71 | 0.04 | 0.99 |
| m3300 | 0.25 | 0.04 | 0.45 | 0.22 | 0.04 | 0.30 | 0.28 | 0.03 | 1.00 |
| m3400 | 0.18 | 0.05 | 0.50 | 0.25 | 0.05 | 0.42 | 0.36 | 0.03 | 0.52 |
| m3600 | 0.29 | 0.05 | 0.28 | 0.37 | 0.06 | 0.48 | 0.56 | 0.05 | 1.00 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrascoza Mayén, J.F.; Rydzewski, J.; Szostak, N.; Blazewicz, J.; Nowak, W. Prebiotic Soup Components Trapped in Montmorillonite Nanoclay Form New Molecules: Car-Parrinello Ab Initio Simulations. Life 2019, 9, 46. https://doi.org/10.3390/life9020046
Carrascoza Mayén JF, Rydzewski J, Szostak N, Blazewicz J, Nowak W. Prebiotic Soup Components Trapped in Montmorillonite Nanoclay Form New Molecules: Car-Parrinello Ab Initio Simulations. Life. 2019; 9(2):46. https://doi.org/10.3390/life9020046
Chicago/Turabian StyleCarrascoza Mayén, Juan Francisco, Jakub Rydzewski, Natalia Szostak, Jacek Blazewicz, and Wieslaw Nowak. 2019. "Prebiotic Soup Components Trapped in Montmorillonite Nanoclay Form New Molecules: Car-Parrinello Ab Initio Simulations" Life 9, no. 2: 46. https://doi.org/10.3390/life9020046
APA StyleCarrascoza Mayén, J. F., Rydzewski, J., Szostak, N., Blazewicz, J., & Nowak, W. (2019). Prebiotic Soup Components Trapped in Montmorillonite Nanoclay Form New Molecules: Car-Parrinello Ab Initio Simulations. Life, 9(2), 46. https://doi.org/10.3390/life9020046





