Dynamical Task Switching in Cellular Computers
Abstract
1. Introduction
2. Background
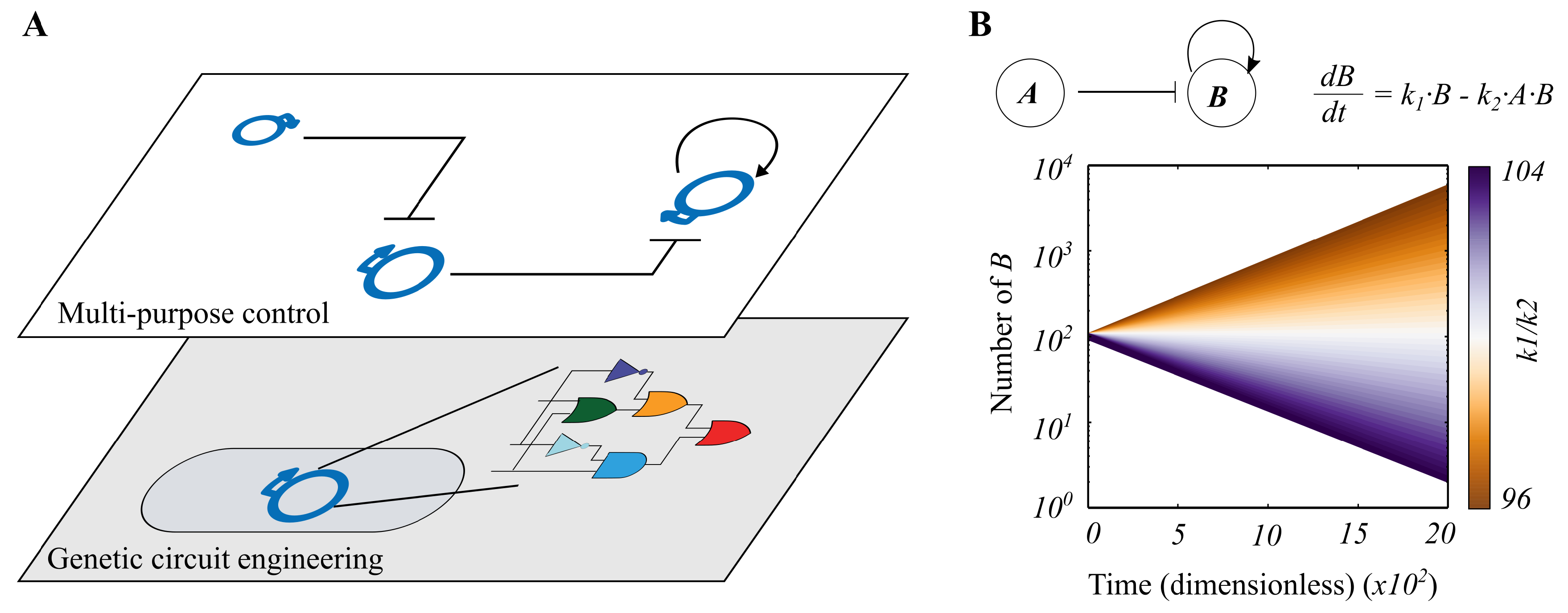
Our Task Switching Model
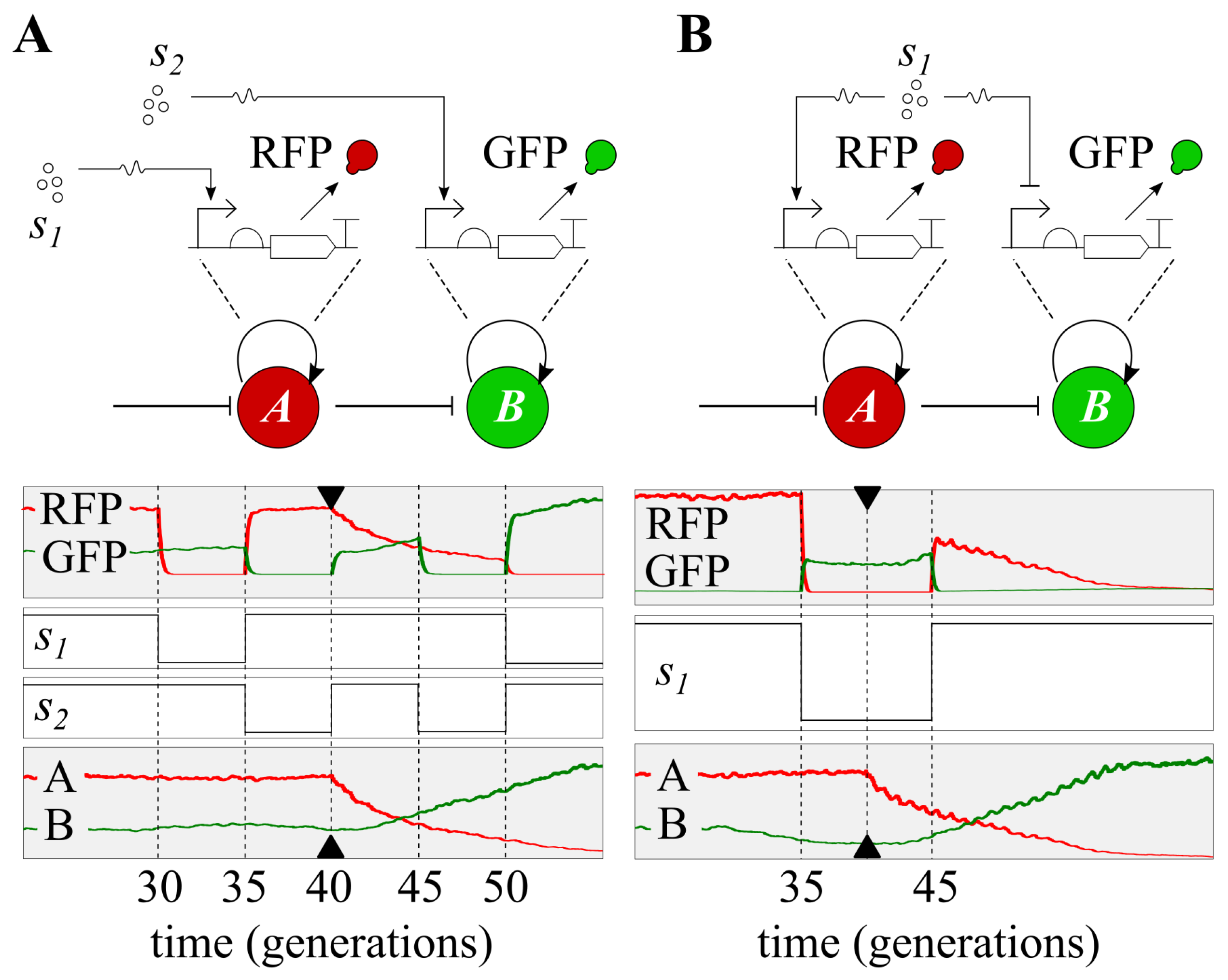
3. Results
3.1. Continuous Modelling
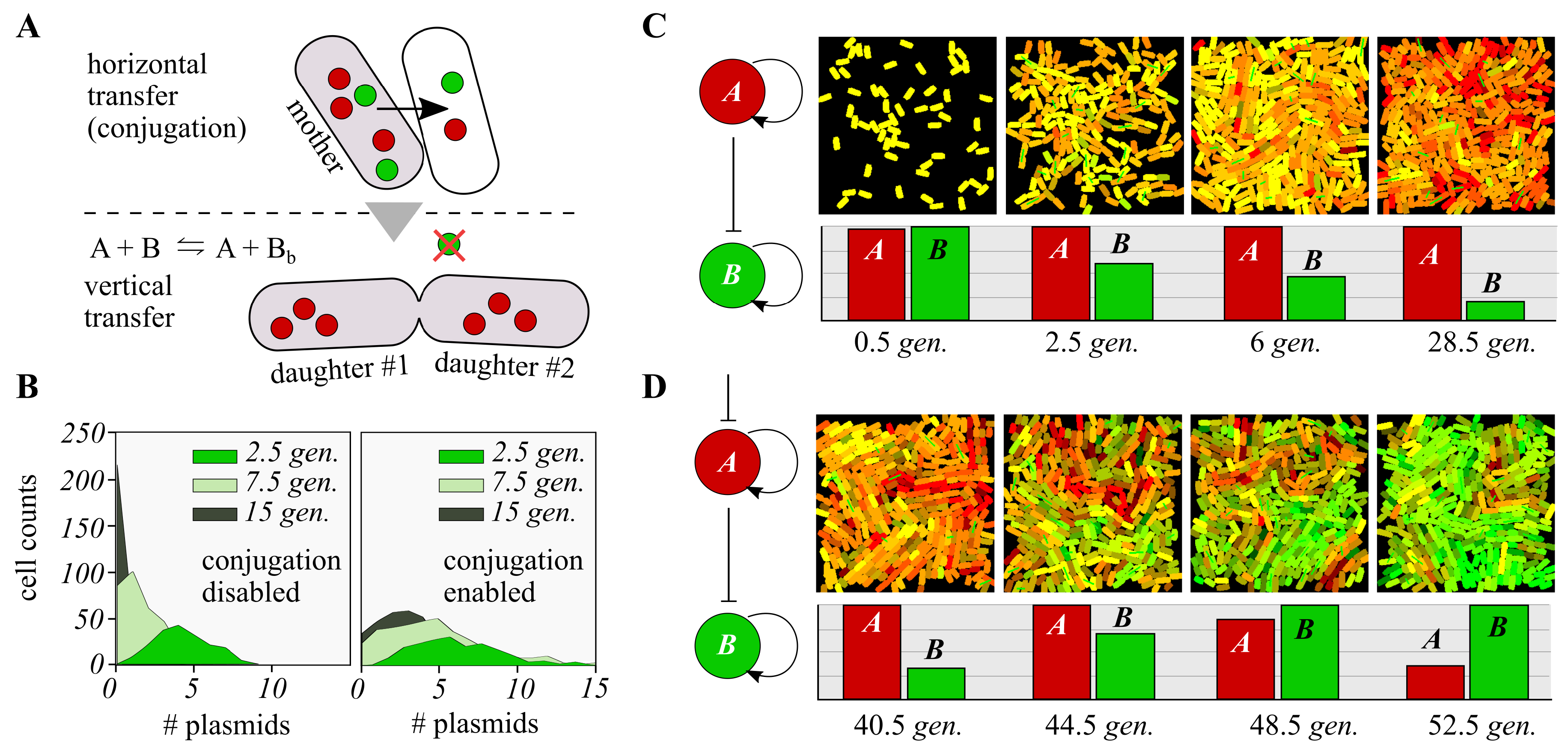
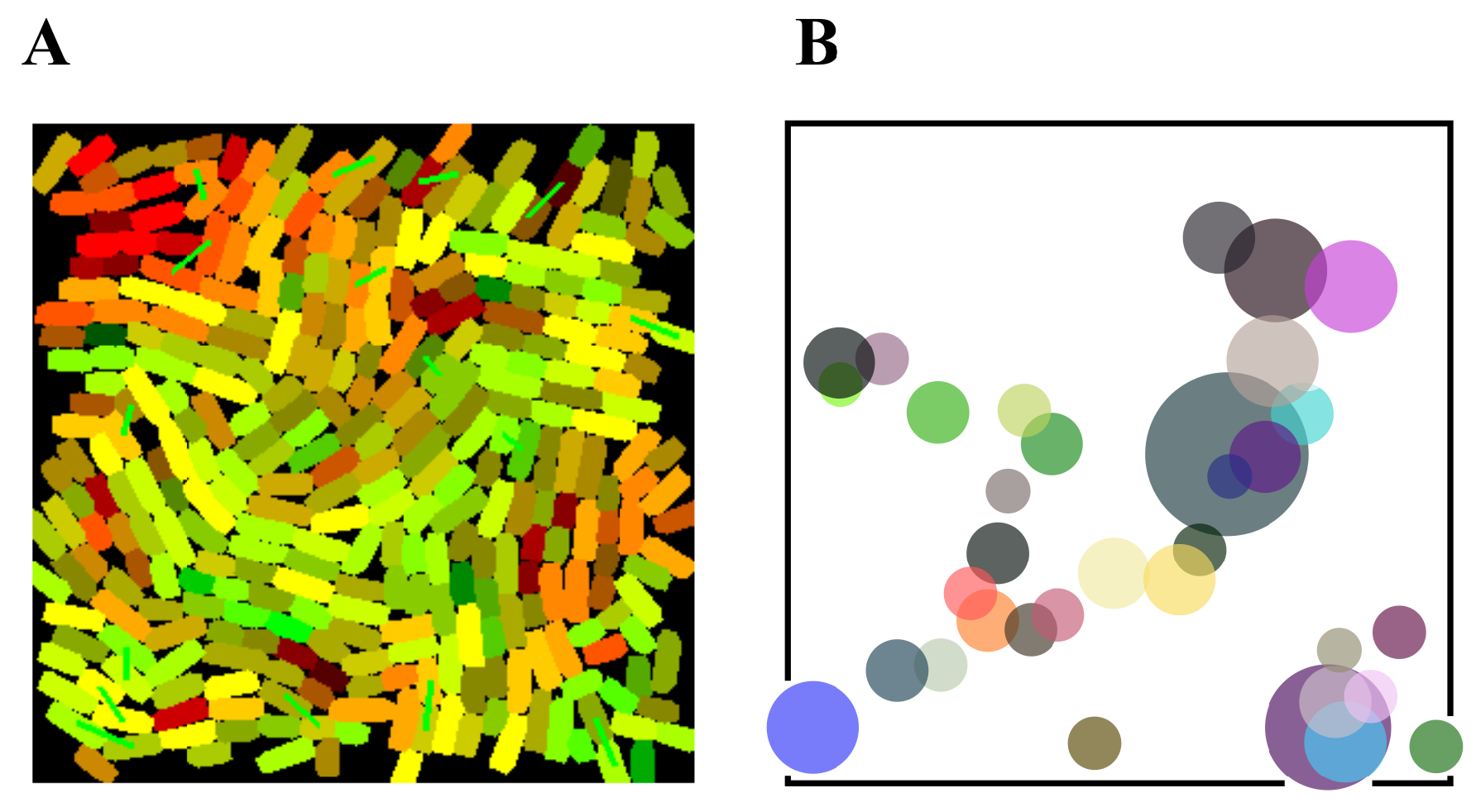
3.2. Discrete Simulation
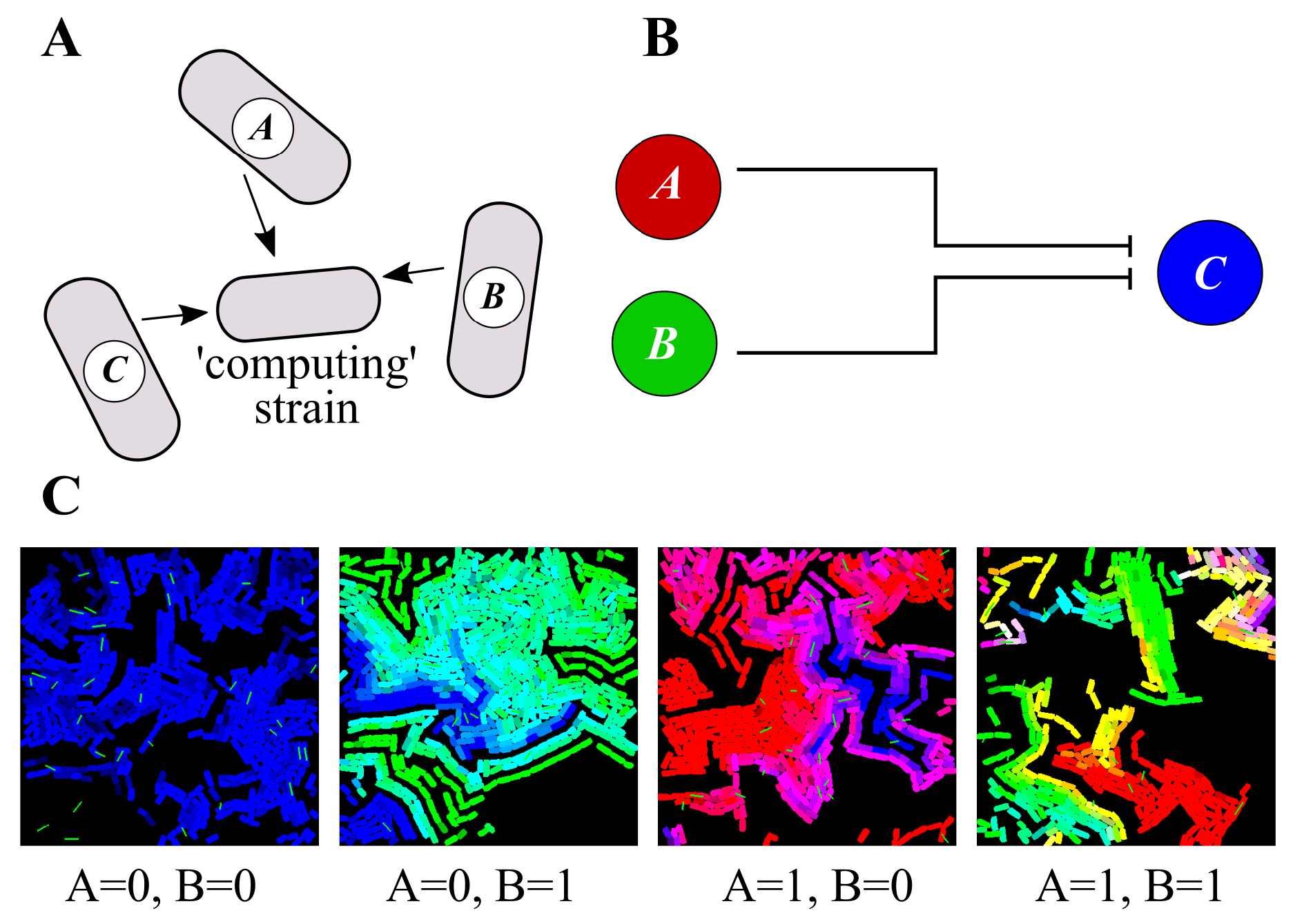
3.3. Distributed Computations Using Cell Consortia
3.4. Use-Casing the Potential of Task Switching
4. Materials and Methods
4.1. Differential models
4.2. Stochastic Models
4.3. Agent-Based Spatial Simulations
5. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Amos, M.; Goni-Moreno, A. Cellular computing and synthetic biology. In Computational Matter; Stepney, S., Rasmussen, S., Amos, M., Eds.; Springer: Berlin, Germany, 2018; pp. 93–110. [Google Scholar]
- Church, G.M.; Elowitz, M.B.; Smolke, C.D.; Voigt, C.A.; Weiss, R. Realizing the potential of synthetic biology. Nat. Rev. Mol. Cell Biol. 2014, 15, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Ro, D.K.; Paradise, E.M.; Ouellet, M.; Fisher, K.J.; Newman, K.L.; Ndungu, J.M.; Ho, K.A.; Eachus, R.A.; Ham, T.S.; Kirby, J.; et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 2006, 440, 940–943. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Jeong, H.; Lee, S.J. Synthetic biology for microbial heavy metal biosensors. Anal. Bioanal. Chem. 2018, 410, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Selvester, R.H.; Collier, C.R.; Pearson, R.B. Analog computer model of the vectorcardiogram. Circulation 1965, 31, 45–53. [Google Scholar] [CrossRef]
- Heinmets, F. Analog computer analysis of a model-system for the induced enzyme synthesis. J. Theor. Biol. 1964, 6, 60–75. [Google Scholar] [CrossRef]
- Nielsen, A.A.; Der, B.S.; Shin, J.; Vaidyanathan, P.; Paralanov, V.; Strychalski, E.A.; Ross, D.; Densmore, D.; Voigt, C.A. Genetic circuit design automation. Science 2016, 352, aac7341. [Google Scholar] [CrossRef] [PubMed]
- Wintle, B.C.; Boehm, C.R.; Rhodes, C.; Molloy, J.C.; Millett, P.; Adam, L.; Breitling, R.; Carlson, R.; Casagrande, R.; Dando, M.; et al. Point of View: A transatlantic perspective on 20 emerging issues in biological engineering. Elife 2017, 6, e30247. [Google Scholar] [CrossRef]
- Amos, M. Cellular Computing; Oxford University Press: Oxford, UK, 2004. [Google Scholar]
- Manzoni, R.; Urrios, A.; Velazquez-Garcia, S.; de Nadal, E.; Posas, F. Synthetic biology: insights into biological computation. Integr. Biol. 2016, 8, 518–532. [Google Scholar] [CrossRef]
- Benenson, Y. Biomolecular computing systems: principles, progress and potential. Nat. Rev. Genet. 2012, 13, 455–468. [Google Scholar] [CrossRef]
- Bonnet, J.; Yin, P.; Ortiz, M.E.; Subsoontorn, P.; Endy, D. Amplifying genetic logic gates. Science 2013, 340, 599–603. [Google Scholar] [CrossRef]
- Lou, C.; Liu, X.; Ni, M.; Huang, Y.; Huang, Q.; Huang, L.; Jiang, L.; Lu, D.; Wang, M.; Liu, C.; et al. Synthesizing a novel genetic sequential logic circuit: A push-on push-off switch. Mol. Syst. Biol. 2010, 6, 350. [Google Scholar] [CrossRef] [PubMed]
- Purcell, O.; Savery, N.J.; Grierson, C.S.; di Bernardo, M. A comparative analysis of synthetic genetic oscillators. J. R. Soc. Interface 2010, 7, 1503–1524. [Google Scholar] [CrossRef] [PubMed]
- Friedland, A.E.; Lu, T.K.; Wang, X.; Shi, D.; Church, G.; Collins, J.J. Synthetic gene networks that count. Science 2009, 324, 1199–1202. [Google Scholar] [CrossRef] [PubMed]
- Funnell, B.E.; Phillips, G.J. Plasmid Biology; ASM Press: Sterling, VA, USA, 2004. [Google Scholar]
- Smillie, C.; Garcillán-Barcia, M.P.; Francia, M.V.; Rocha, E.P.; de la Cruz, F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, E.; Aparicio, T.; Goñi-Moreno, A.; Fraile, S.; de Lorenzo, V. SEVA 2.0: An update of the Standard European Vector Architecture for de-/re-construction of bacterial functionalities. Nucleic Acids Res. 2014, 43, D1183–D1189. [Google Scholar] [CrossRef]
- Llosa, M.; Gomis-Rüth, F.X.; Coll, M.; Cruz, F.d.L. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 2002, 45, 1–8. [Google Scholar] [CrossRef]
- Tatum, E.; Lederberg, J. Gene recombination in the bacterium Escherichia coli. J. Bacteriol. 1947, 53, 673. [Google Scholar]
- Goñi-Moreno, A.; Amos, M.; de la Cruz, F. Multicellular computing using conjugation for wiring. PLoS ONE 2013, 8, e65986. [Google Scholar] [CrossRef]
- Goñi-Moreno, A.; Amos, M. A reconfigurable NAND/NOR genetic logic gate. BMC Syst. Biol. 2012, 6, 126. [Google Scholar] [CrossRef]
- Gil, D.; Bouché, J.P. ColE1-type vectors with fully repressible replication. Gene 1991, 105, 17–22. [Google Scholar] [CrossRef]
- Friehs, K. Plasmid copy number and plasmid stability. In New Trends and Developments in Biochemical Engineering; Scheper, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 47–82. [Google Scholar]
- Núñez, B.; Avila, P.; De La Cruz, F. Genes involved in conjugative DNA processing of plasmid R6K. Mol. Microbiol. 1997, 24, 1157–1168. [Google Scholar] [CrossRef] [PubMed]
- Goni-Moreno, A.; Amos, M. DiSCUS: A simulation platform for conjugation computing. In International Conference on Unconventional Computation and Natural Computation; Springer: Cham, Switzerland, 2015; pp. 181–191. [Google Scholar]
- García-Betancur, J.C.; Goñi-Moreno, A.; Horger, T.; Schott, M.; Sharan, M.; Eikmeier, J.; Wohlmuth, B.; Zernecke, A.; Ohlsen, K.; Kuttler, C.; et al. Cell differentiation defines acute and chronic infection cell types in Staphylococcus aureus. Elife 2017, 6, e28023. [Google Scholar] [CrossRef] [PubMed]
- Nikel, P.I.; Romero-Campero, F.J.; Zeidman, J.A.; Goñi-Moreno, Á.; de Lorenzo, V. The glycerol-dependent metabolic persistence of Pseudomonas putida KT2440 reflects the regulatory logic of the GlpR repressor. MBio 2015, 6, e00340-15. [Google Scholar] [CrossRef] [PubMed]
- Goñi-Moreno, A.; Redondo-Nieto, M.; Arroyo, F.; Castellanos, J. Biocircuit design through engineering bacterial logic gates. Nat. Comput. 2011, 10, 119–127. [Google Scholar] [CrossRef]
- Regot, S.; Macia, J.; Conde, N.; Furukawa, K.; Kjellén, J.; Peeters, T.; Hohmann, S.; de Nadal, E.; Posas, F.; Solé, R. Distributed biological computation with multicellular engineered networks. Nature 2011, 469, 207. [Google Scholar] [CrossRef] [PubMed]
- Macía, J.; Posas, F.; Solé, R.V. Distributed computation: the new wave of synthetic biology devices. Trends Biotechnol. 2012, 30, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Getino, M.; Palencia-Gándara, C.; Garcillán-Barcia, M.P.; de la Cruz, F. PifC and Osa, Plasmid Weapons against Rival Conjugative Coupling Proteins. Front. Microbiol. 2017, 8, 2260. [Google Scholar] [CrossRef]
- Miró-Bueno, J.M.; Rodríguez-Patón, A. A simple negative interaction in the positive transcriptional feedback of a single gene is sufficient to produce reliable oscillations. PloS ONE 2011, 6, e27414. [Google Scholar] [CrossRef]
- Goni-Moreno, A.; Amos, M. Model for a population-based microbial oscillator. BioSystems 2011, 105, 286–294. [Google Scholar] [CrossRef]
- Gillespie, D.T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977, 81, 2340–2361. [Google Scholar] [CrossRef]
- Espeso, D.R.; Martínez-García, E.; de Lorenzo, V.; Goñi-Moreno, Á. Physical forces shape group identity of swimming Pseudomonas putida cells. Front. Microbiol. 2016, 7, 1437. [Google Scholar] [CrossRef] [PubMed]
- Goñi-Moreno, A.; Carcajona, M.; Kim, J.; Martínez-García, E.; Amos, M.; de Lorenzo, V. An implementation-focused bio/algorithmic workflow for synthetic biology. ACS Synth. Biol. 2016, 5, 1127–1135. [Google Scholar] [CrossRef] [PubMed]
- Gama, J.A.; Zilhão, R.; Dionisio, F. Co-resident plasmids travel together. Plasmid 2017, 93, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Gama, J.A.; Zilhão, R.; Dionisio, F. Multiple plasmid interference–Pledging allegiance to my enemy’s enemy. Plasmid 2017, 93, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Del Campo, I.; Ruiz, R.; Cuevas, A.; Revilla, C.; Vielva, L.; de la Cruz, F. Determination of conjugation rates on solid surfaces. Plasmid 2012, 67, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Seoane, J.; Yankelevich, T.; Dechesne, A.; Merkey, B.; Sternberg, C.; Smets, B.F. An individual-based approach to explain plasmid invasion in bacterial populations. FEMS Microbiol. Ecol. 2011, 75, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Goñi-Moreno, A.; Benedetti, I.; Kim, J.; de Lorenzo, V. Deconvolution of gene expression noise into spatial dynamics of transcription factor–promoter interplay. ACS Synth. Biol. 2017, 6, 1359–1369. [Google Scholar] [CrossRef]
- Zatyka, M.; Thomas, C.M. Control of genes for conjugative transfer of plasmids and other mobile elements. FEMS Microbiol. Rev. 1998, 21, 291–319. [Google Scholar] [CrossRef]
- Daniel, R.; Rubens, J.R.; Sarpeshkar, R.; Lu, T.K. Synthetic analog computation in living cells. Nature 2013, 497, 619. [Google Scholar] [CrossRef]
- Farzadfard, F.; Lu, T.K. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 2014, 346, 1256272. [Google Scholar] [CrossRef]
- Tang, W.; Liu, D.R. Rewritable multi-event analog recording in bacterial and mammalian cells. Science 2018, 360, eaap8992. [Google Scholar] [CrossRef] [PubMed]
- Van de Guchte, M. Horizontal gene transfer and ecosystem function dynamics. Trends Microbiol. 2017, 25, 699–700. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goñi-Moreno, A.; de la Cruz, F.; Rodríguez-Patón, A.; Amos, M. Dynamical Task Switching in Cellular Computers. Life 2019, 9, 14. https://doi.org/10.3390/life9010014
Goñi-Moreno A, de la Cruz F, Rodríguez-Patón A, Amos M. Dynamical Task Switching in Cellular Computers. Life. 2019; 9(1):14. https://doi.org/10.3390/life9010014
Chicago/Turabian StyleGoñi-Moreno, Angel, Fernando de la Cruz, Alfonso Rodríguez-Patón, and Martyn Amos. 2019. "Dynamical Task Switching in Cellular Computers" Life 9, no. 1: 14. https://doi.org/10.3390/life9010014
APA StyleGoñi-Moreno, A., de la Cruz, F., Rodríguez-Patón, A., & Amos, M. (2019). Dynamical Task Switching in Cellular Computers. Life, 9(1), 14. https://doi.org/10.3390/life9010014




