The Heptameric SmAP1 and SmAP2 Proteins of the Crenarchaeon Sulfolobus Solfataricus Bind to Common and Distinct RNA Targets
Abstract
:1. Introduction
2. Materials and Methods
2.1. Construction of Plasmids pMJ05-SmAP1-His and pMJO5-SmAP2-His and Transformation of Sso Cells
2.2. Synthesis of His-tagged Sso-SmAP Proteins and Purification
2.3. Deep Sequencing Analysis
2.4. Bioinformatic Analyses
2.5. Expression of Sso ORF 5410 in E. coli and Purification of Sso-SmAP2
2.6. Crystallization, Data Collection and Structure Determination
| Sso-SmAP2 | |
|---|---|
| Data collection | |
| Beamline | ESRF ID23-1 |
| Wavelength (Å) | 0.9762 |
| Unit cell | a = 197.21 Å, b = 197.21 Å, c = 38.88 Å, α = 90.0°, β = 90.0°, γ = 120.0° |
| Space group | R3: H |
| Resolution range (Å) * | 49.3-2.6 (2.74-2.6) |
| Completeness (%) | 100 (100) |
| Multiplicity | 5.3 (5.1) |
| Rsym | 0.127 (0.990) |
| Rpim | 0.061 (0.49) |
| Mean I/σI | 29.2 (2.1) |
| Unique reflections | 17319 (2519) |
| Refinement | |
| R/Rfree Twin_fraction | 0.140/0.177 0.36 |
| r.m.s.-deviations | |
| bond length (Å) | 0.006 |
| bond angle (°) | 1.09 |
| Number of atoms | |
| protein | 44384 |
| water | 550 |
| B-factors (Å2) | |
| protein | 55.10 |
| water | 54.81 |
3. Results and Discussion
3.1. 3D-Structure and Surface Charge of Sso-SmAP2 in Comparison with Other SmAPs
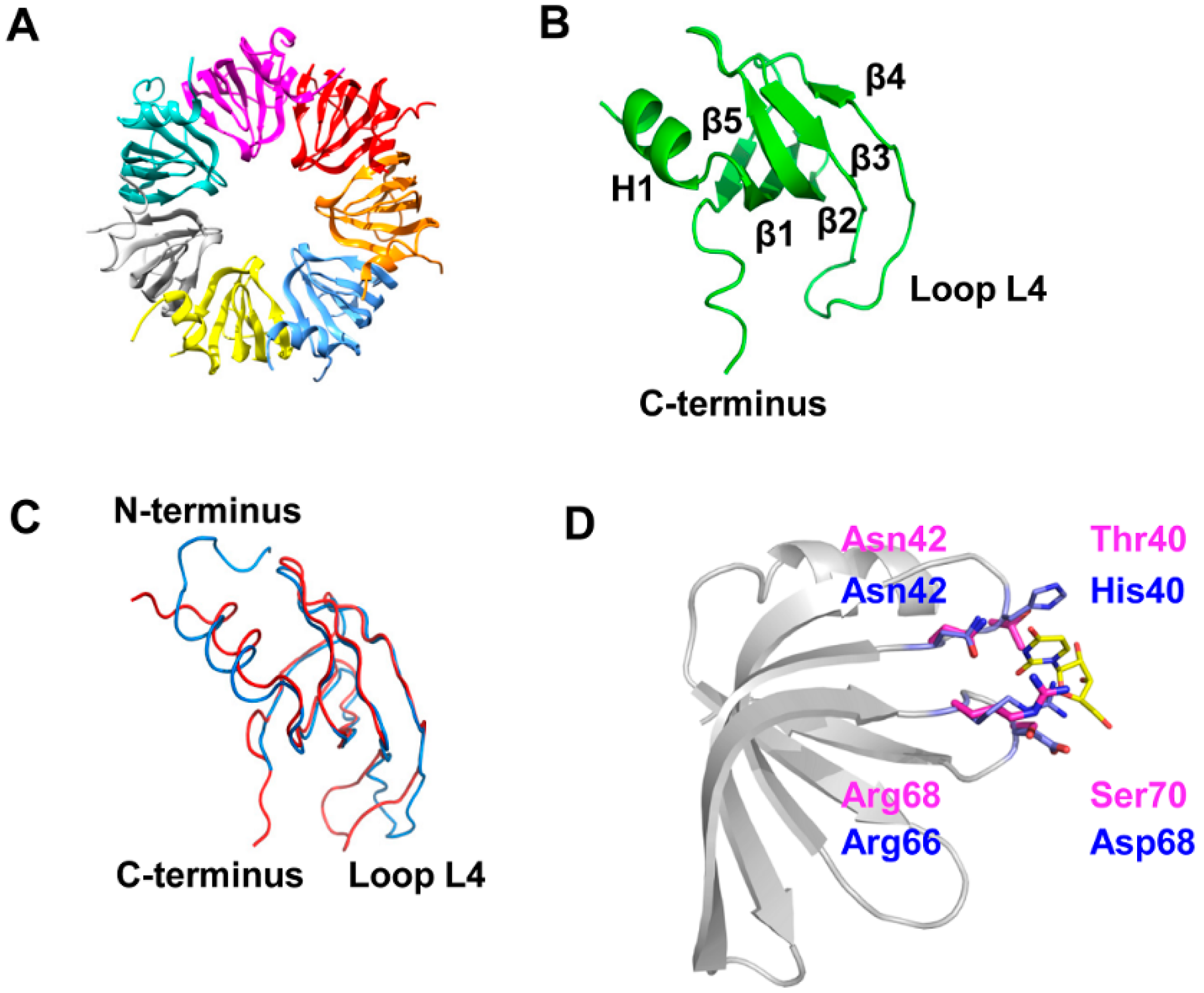
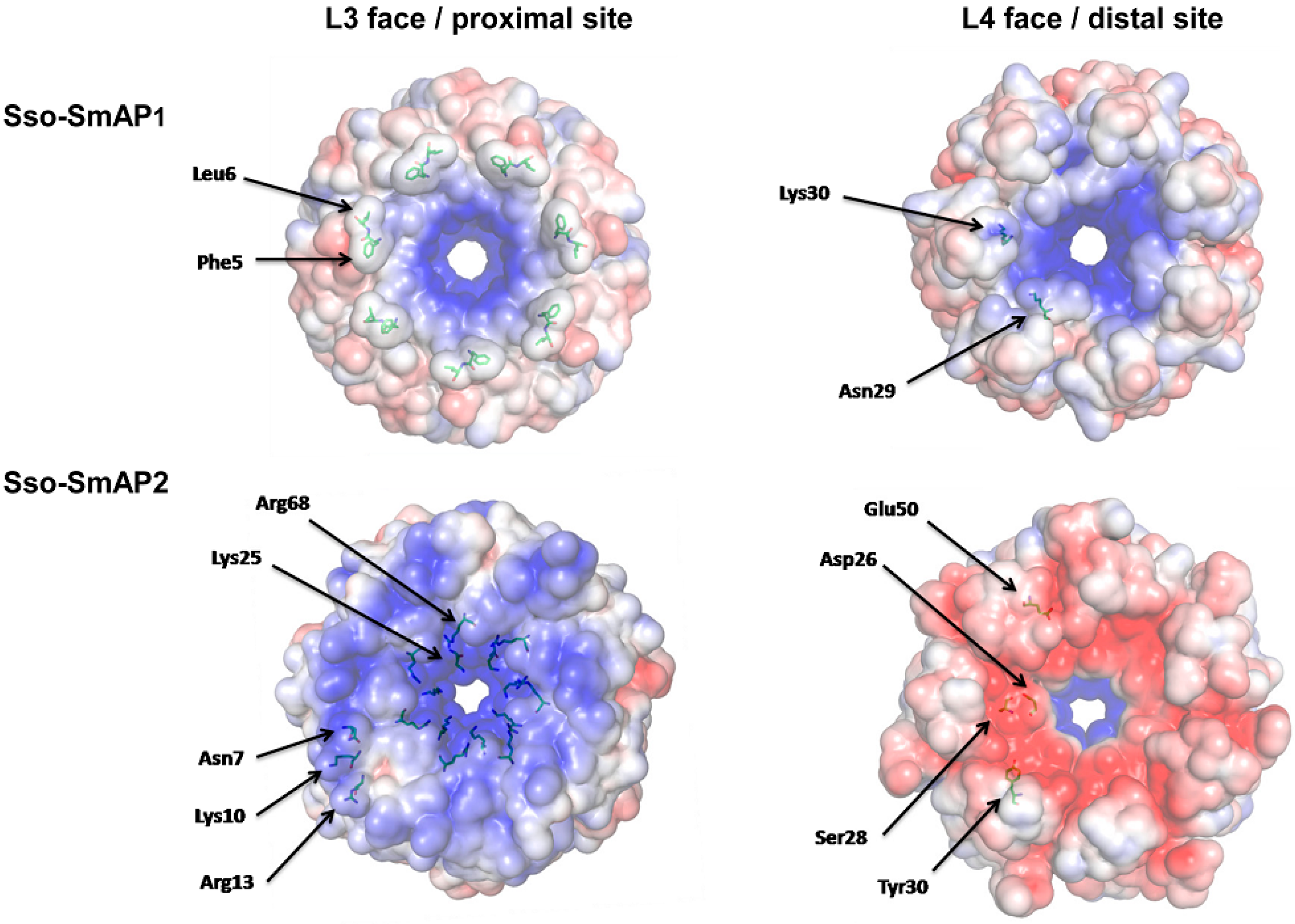
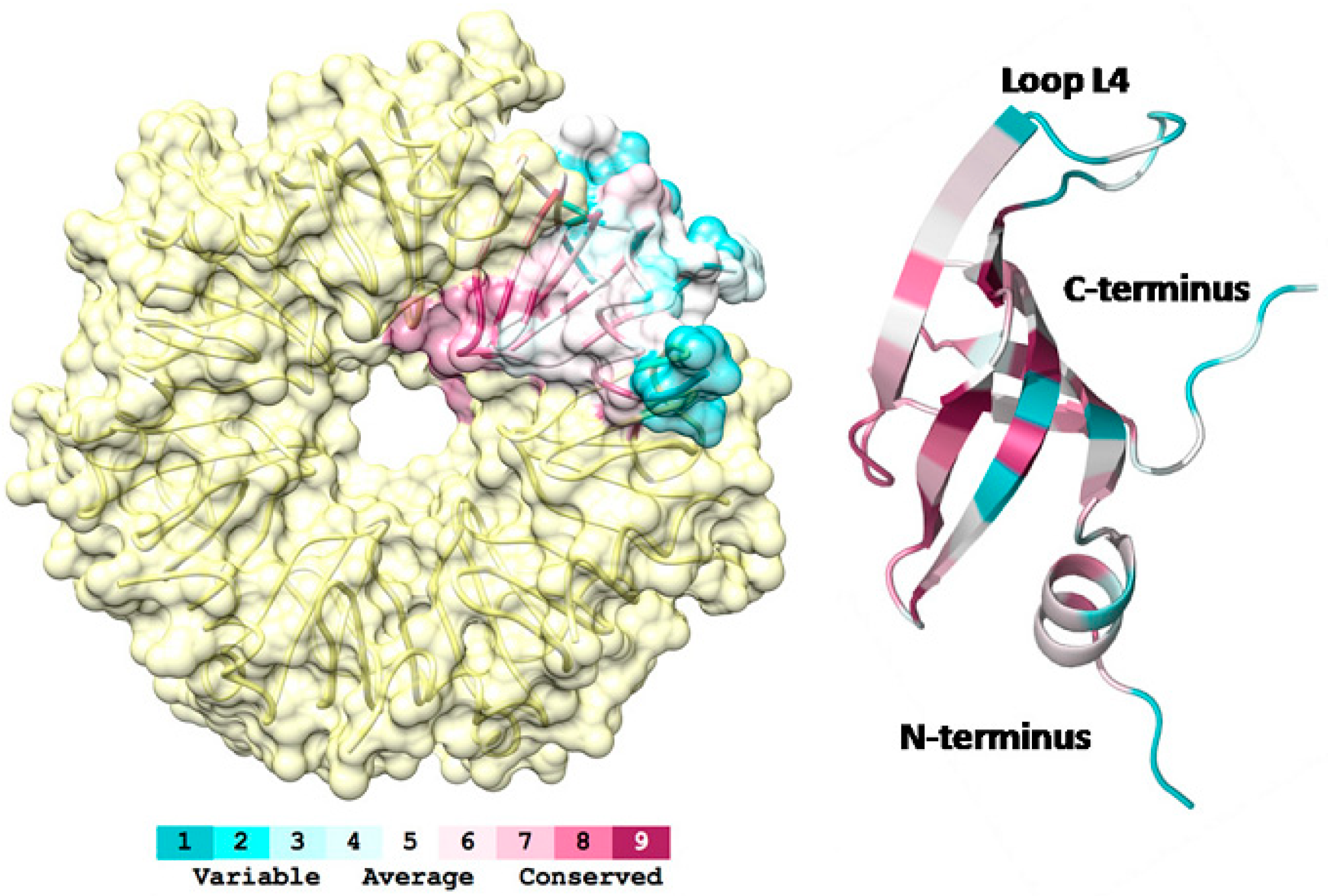
3.2. Sso-SmAP1 and SsoAP2 Bind to Common and Distinct RNA Substrates
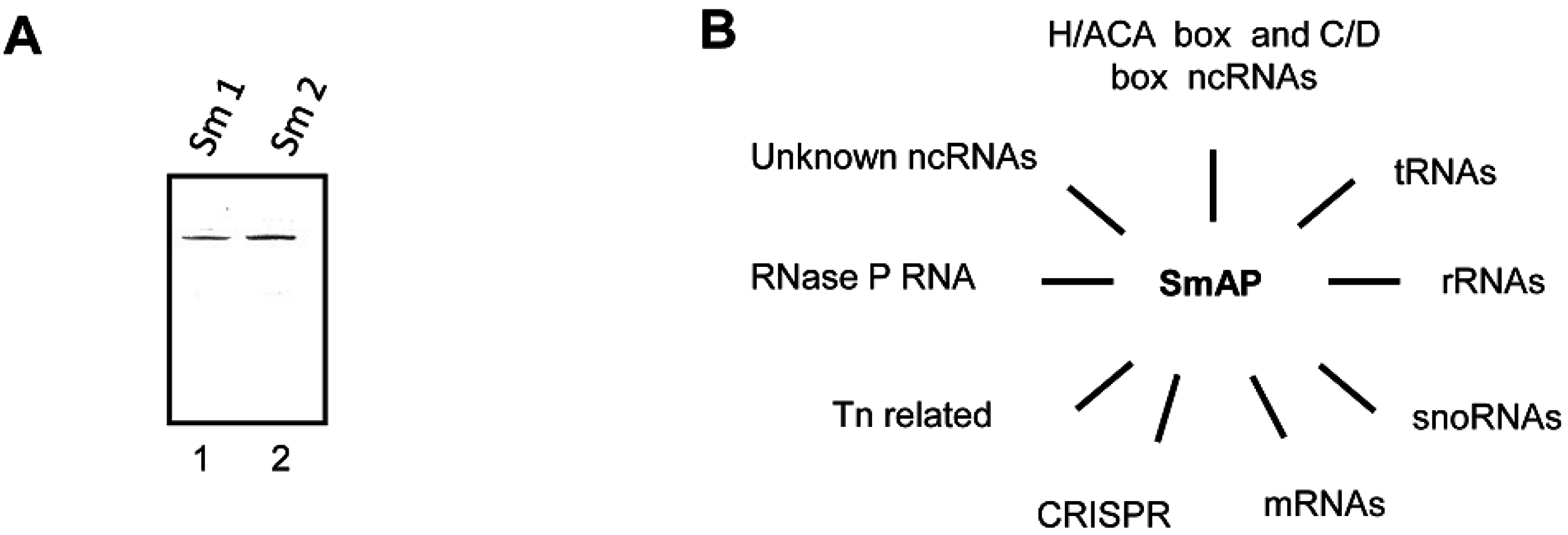
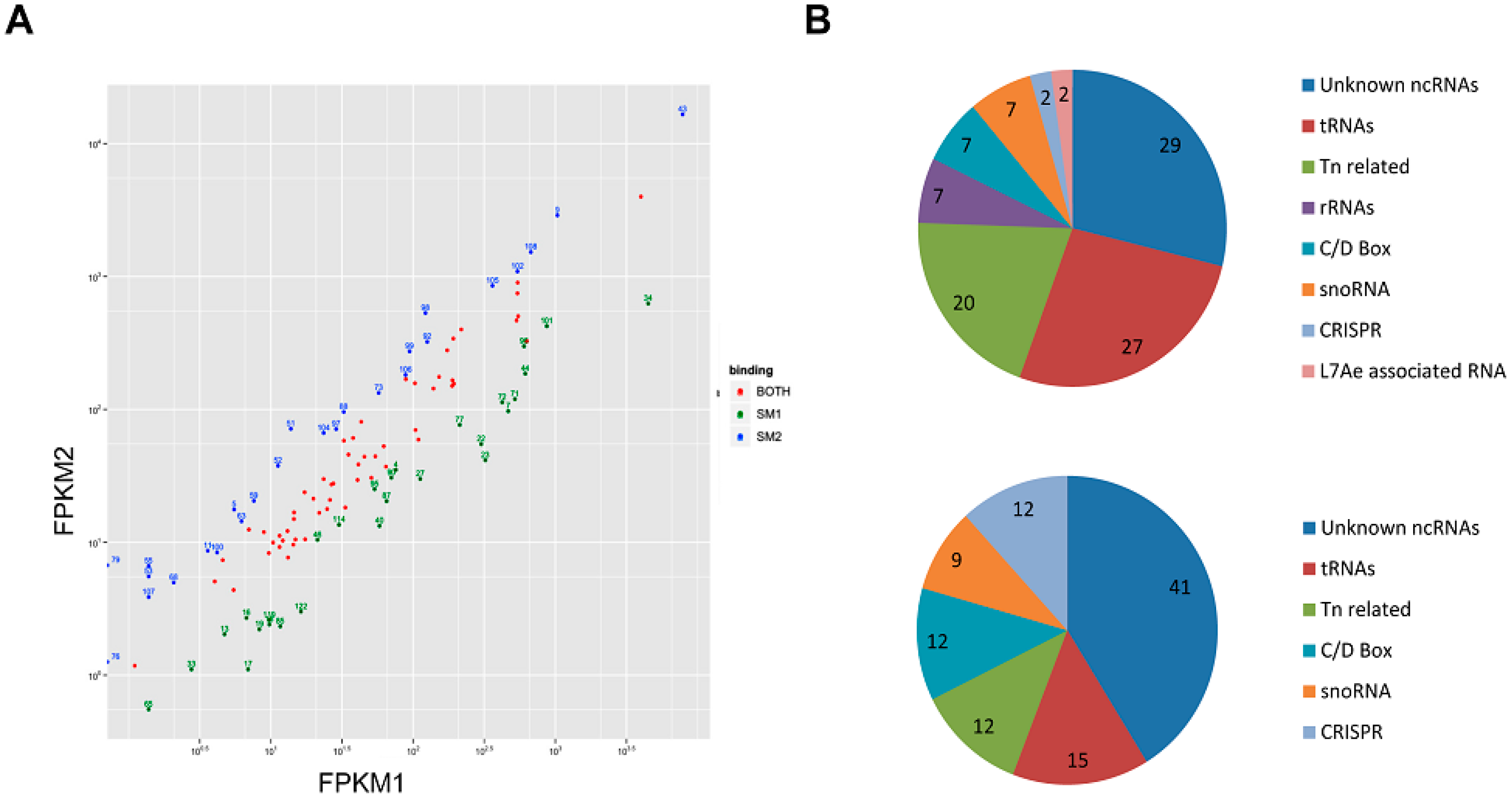

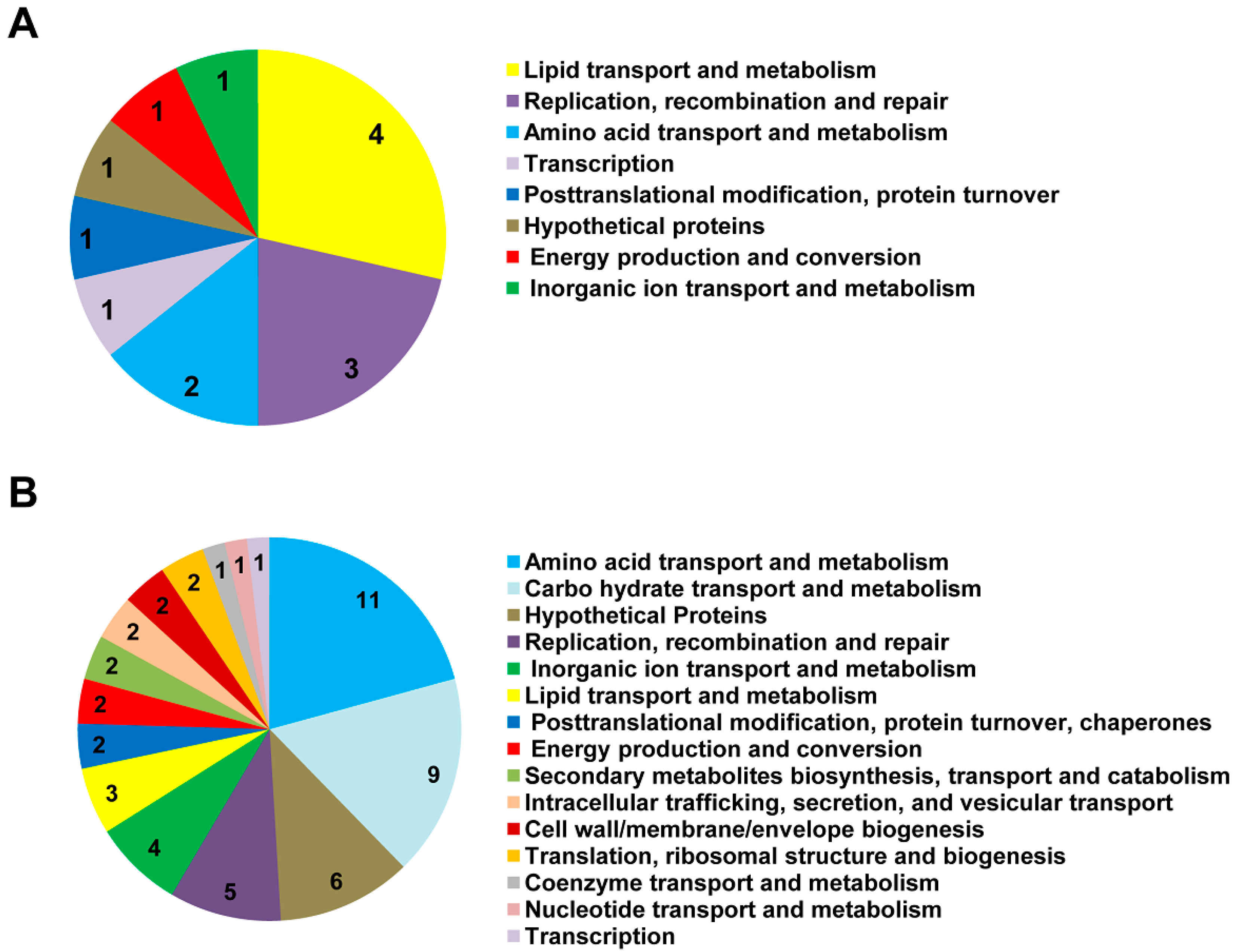
4. Outlook
Acknowledgements
Author Contributions
Supplementary Materials
Conflicts of Interest
References
- Wilusz, C.J.; Wilusz, J. Eukaryotic Lsm proteins: Lessons from bacteria. Nat. Struct. Mol. Biol. 2005, 12, 1031–1036. [Google Scholar] [CrossRef] [PubMed]
- Weichenrieder, O. RNA binding by Hfq and ring-forming (L)Sm proteins: A trade-off between optimal sequence readout and RNA backbone conformation. RNA Biol. 2014, 11, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Mura, C.; Randolph, P.S.; Patterson, J.; Cozen, A.E. Archaeal and eukaryotic homologs of Hfq: A structural and evolutionary perspective on Sm function. RNA Biol. 2013, 10, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Tomasevic, N.; Peculis, B.A. Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol. Cell. Biol. 2002, 22, 4101–4112. [Google Scholar] [CrossRef] [PubMed]
- Kufel, J.; Allmang, C.; Verdone, L.; Beggs, J.D.; Tollervey, D. Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p. Mol. Cell. Biol. 2002, 22, 5248–5256. [Google Scholar] [CrossRef] [PubMed]
- Schümperli, D.; Pillai, R.S. The special Sm core structure of the U7 snRNP: Far-reaching significance of a small nuclear ribonucleoprotein. Cell.Mol. Life Sci. 2004, 61, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Dominski, Z.; Marzluff, W.F. Formation of the 3' end of histone mRNA: Getting closer to the end. Gene 2007, 396, 373–390. [Google Scholar] [CrossRef] [PubMed]
- Tharun, S.; He, W.; Mayes, A.E.; Lennertz, P.; Beggs, J.D.; Parker, R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 2000, 404, 515–518. [Google Scholar] [CrossRef]
- Seto, A.G.; Zaug, A.J.; Sobel, S.G.; Wolin, S.L.; Cech, T.R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 1999, 401, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Sauer, E.; Schmidt, S.; Weichenrieder, O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. USA 2012, 109, 9396–9401. [Google Scholar] [CrossRef] [PubMed]
- Panja, S.; Schu, D.J.; Woodson, S.A. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 2013, 15, 7536–7546. [Google Scholar] [CrossRef]
- Ribeiro, E.A., Jr.; Beich-Frandsen, M.; Konarev, P.V.; Shang, W.; Vecerek, B.; Kontaxis, G.; Hämmerle, H.; Peterlik, H.; Svergun, D.I.; Bläsi, U.; et al. Structural flexibility of RNA as molecular basis for Hfq chaperone function. Nucleic Acids Res. 2012, 40, 8072–8084. [Google Scholar] [CrossRef] [PubMed]
- Weber, G.; Trowitzsch, S.; Kastner, B.; Lührmann, R.; Wahl, M.C. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 2010, 29, 4172–4184. [Google Scholar] [CrossRef] [PubMed]
- Mura, C.; Phillips, M.; Kozhukhovsky, A.; Eisenberg, D. Structure and assembly of an augmented Sm-like archaeal protein 14-mer. Proc. Natl. Acad. Sci. USA 2003, 100, 4539–4544. [Google Scholar] [CrossRef] [PubMed]
- Khusial, P.; Plaag, R.; Zieve, G.W. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem. Sci. 2005, 30, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Benz, J.; Späth, B.; Maier, L.K.; Straub, J.; Granzow, M.; Raabe, M.; Urlaub, H.; Hoffmann, J.; Brutschy, B.; et al. The archaeal Lsm protein binds to small RNAs. J. Biol. Chem. 2010, 285, 34429–34438. [Google Scholar] [CrossRef] [PubMed]
- Kilic, T.; Thore, S.; Suck, D. Crystal structure of an archaeal Sm protein from Sulfolobus solfataricus. Proteins 2005, 61, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Törö, I.; Basquin, J.; Teo-Dreher, H.; Suck, D. Archaeal Sm proteins form heptameric and hexameric complexes: crystal structures of the Sm1 and Sm2 proteins from the hyperthermophile Archaeoglobus fulgidus. J. Mol. Biol. 2002, 320, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Törö, I.; Thore, S.; Mayer, C.; Basquin, J.; Seraphin, B.; Suck, D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001, 20, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- Thore, S.; Mayer, C.; Sauter, C.; Weeks, S.; Suck, D. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya. J. Biol. Chem. 2003, 278, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Albers, S.V.; Jonuscheit, M.; Dinkelaker, S.; Urich, T.; Kletzin, A.; Tampe, R.; Driessen, A.J.; Schleper, C. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl. Environ. Microbiol. 2006, 72, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D 2006, 62, 72–82. [Google Scholar] [CrossRef]
- Collaborative Computational Project. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. Sect. D 1994, 50, 760–763. [Google Scholar]
- Evans, P.R. An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. Sect. D 2011, 67, 282–292. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Kleywegt, G.J.; Brunger, A.T. Checking your imagination: applications of the free R value. Structure 1996, 4, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Armon, A.; Graur, D.; Ben-Tal, N. ConSurf: An algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J. Mol. Biol. 2001, 307, 447–463. [Google Scholar] [CrossRef] [PubMed]
- Beich-Frandsen, M.; Vecerek, B.; Konarev, P.V.; Sjoblom, B.; Kloiber, K.; Hämmerle, H.; Rajkowitsch, L.; Miles, A.J.; Kontaxis, G.; Wallace, B.A.; et al. Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res. 2011, 39, 4900–4915. [Google Scholar] [CrossRef] [PubMed]
- Vecerek, B.; Rajkowitsch, L.; Sonnleitner, E.; Schröder, R.; Bläsi, U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008, 36, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Wurtzel, O.; Sapra, R.; Chen, F.; Zhu, Y.; Simmons, B.A.; Sorek, R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010, 20, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Zago, M.A.; Dennis, P.P.; Omer, A.D. The expanding world of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005, 55, 1812–1828. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.H.; Polacek, N.; Zywicki, M.; Huber, H.; Brugger, K.; Garrett, R.; Bachellerie, J.P.; Hüttenhofer, A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005, 55, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, J.; Aagaard, C.; Semionenkov, M.; Garrett, R.A. Archaeal introns: Splicing, intercellular mobility and evolution. Trends Biochem. Sci. 1997, 22, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Murina, V.N.; Nikulin, A.D. RNA-binding Sm-like proteins of bacteria and archaea. Similarity and difference in structure and function. Biochem. (Mosc.) 2012, 76, 1434–1449. [Google Scholar] [CrossRef]
- Omer, A.D.; Lowe, T.M.; Russell, A.G.; Ebhardt, H.; Eddy, S.R.; Dennis, P.P. Homologs of small nucleolar RNAs in Archaea. Science 2000, 288, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.M.; Lai, L.B.; Susanti, D.; Mukhopadhyay, B.; Gopalan, V. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc. Natl. Acad. Sci. USA 2010, 107, 14573–14578. [Google Scholar] [CrossRef] [PubMed]
- Märtens, B.; Manoharadas, S.; Hasenöhrl, D.; Manica, A.; Bläsi, U. Antisense regulation by transposon-derived RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO Rep. 2013, 14, 527–533. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Märtens, B.; Bezerra, G.A.; Kreuter, M.J.; Grishkovskaya, I.; Manica, A.; Arkhipova, V.; Djinovic-Carugo, K.; Bläsi, U. The Heptameric SmAP1 and SmAP2 Proteins of the Crenarchaeon Sulfolobus Solfataricus Bind to Common and Distinct RNA Targets. Life 2015, 5, 1264-1281. https://doi.org/10.3390/life5021264
Märtens B, Bezerra GA, Kreuter MJ, Grishkovskaya I, Manica A, Arkhipova V, Djinovic-Carugo K, Bläsi U. The Heptameric SmAP1 and SmAP2 Proteins of the Crenarchaeon Sulfolobus Solfataricus Bind to Common and Distinct RNA Targets. Life. 2015; 5(2):1264-1281. https://doi.org/10.3390/life5021264
Chicago/Turabian StyleMärtens, Birgit, Gustavo Arruda Bezerra, Mathias Josef Kreuter, Irina Grishkovskaya, Andrea Manica, Valentina Arkhipova, Kristina Djinovic-Carugo, and Udo Bläsi. 2015. "The Heptameric SmAP1 and SmAP2 Proteins of the Crenarchaeon Sulfolobus Solfataricus Bind to Common and Distinct RNA Targets" Life 5, no. 2: 1264-1281. https://doi.org/10.3390/life5021264
APA StyleMärtens, B., Bezerra, G. A., Kreuter, M. J., Grishkovskaya, I., Manica, A., Arkhipova, V., Djinovic-Carugo, K., & Bläsi, U. (2015). The Heptameric SmAP1 and SmAP2 Proteins of the Crenarchaeon Sulfolobus Solfataricus Bind to Common and Distinct RNA Targets. Life, 5(2), 1264-1281. https://doi.org/10.3390/life5021264






