The Anabaena sp. PCC 7120 Exoproteome: Taking a Peek outside the Box
Abstract
:1. Introduction
2. Experimental Section
3. Results and Discussion
3.1. The Exoproteome of Anabaena sp. PCC 7120
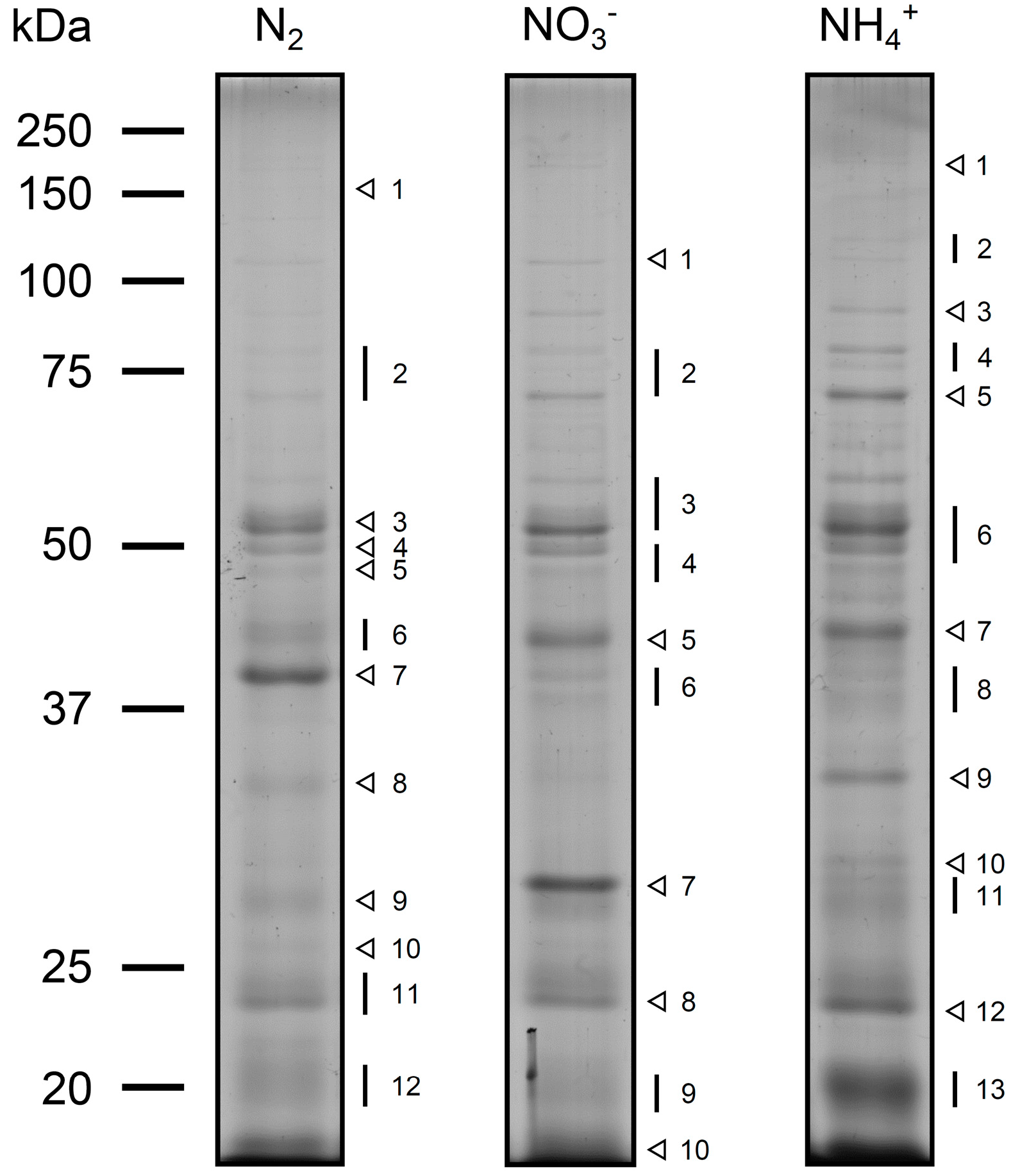
| Cyanobase a | Annotation b | Growth Condition c | Functional Category d | Previously Identified in e |
|---|---|---|---|---|
| All0004 | ATP synthase gamma chain | NH4+ | Photosynthesis and respiration | - |
| All0005 | ATP synthase subunit alpha | N2, NH4+ | Photosynthesis and respiration | Anabaena |
| All0167 | Maltooligosyltrehalose synthase | NH4+ | Other categories | - |
| All0168 | Alpha-amylase | NO3− | Other categories | - |
| All0207 | All0207 protein | NO3− | Conserved hypothetical protein | - |
| All0259 | Cytochrome c-550 | NH4+ | Photosynthesis and respiration | - |
| All0268 | All0268 protein | NH4+ | Conserved hypothetical protein | - |
| All0275 | Glycerophosphoryl diester phosphodiesterase | NO3−, NH4+ | Other categories | Anabaena |
| All0458 | Uncharacterized low temperature-induced protein all0458 | NH4+ | Conserved hypothetical protein | Anabaena |
| All0875 | All0875 protein | N2, NH4+ | Other categories | Anabaena |
| All1220 | All1220 protein | NH4+ | Conserved hypothetical protein | - |
| All1342 | All1342 protein | NO3− | No similarity | - |
| All1380 | All1380 protein | NO3−, NH4+ | No similarity | - |
| All1683 | Phosphoserine aminotransferase | N2 | Amino acid biosynthesis | - |
| All1750 | All1750 protein | NO3− | No similarity | - |
| All1951 | Substrate-binding protein of ABC transporter | N2, NO3−, NH4+ | Transport and binding proteins | - |
| All2105 | FMN-dependent NADH-azoreductase | NH4+ | Fatty acid, phospholipid and sterol metabolism | Anabaena |
| All2108 | All2108 protein | NO3− | Conserved hypothetical protein | - |
| All2315 | Ketol-acid reductoisomerase | NO3−, NH4+ | Amino acid biosynthesis | - |
| All2316 | Aldo/keto reductase | NH4+ | Other categories | - |
| All2375 | All2375 protein | NH4+ | Transport and binding proteins | - |
| All2425 | All2425 protein | NH4+ | No similarity | - |
| All2453 | Cytochrome b6-f complex iron-sulphur subunit 1 | NH4+ | Photosynthesis and respiration | - |
| All2498 | N-acetyl-gamma-glutamyl-phosphate reductase 2 | NH4+ | Transport and binding proteins | - |
| All2533 | Prolyl endopeptidase | N2, NH4+ | Translation | - |
| All2563 | Transaldolase | N2, NO3−, NH4+ | Energy metabolism | Anabaena |
| All2567 | Probable phosphoketolase 2 | NH4+ | Conserved hypothetical protein | - |
| All2655 | All2655 protein | NH4+ | No similarity | - |
| All2843 | Alkaline phosphatase | NO3−, NH4+ | Other categories | N. commune |
| All3149 | All3149 protein | NO3−, NH4+ | Conserved hypothetical protein | - |
| All3325 | All3325 protein | NH4+ | Conserved hypothetical protein | - |
| All3538 | Enolase | N2, NH4+ | Energy metabolism | N. punctiforme, Anabaena |
| All3556 | Succinate-semialdehyde dehydrogenase | N2, NO3− | Energy metabolism | - |
| All3643 | All3643 protein | NH4+ | No similarity | - |
| All3653 | Allophycocyanin subunit alpha-B | NH4+ | Photosynthesis and respiration | - |
| All3791 | Ribonuclease D | NH4+ | Transcription | - |
| All3797 | Beta-Ig-H3/fasciclin (Fragment) | N2, NH4+ | Conserved hypothetical protein | Anabaena |
| All3909 | Uroporphyrinogen decarboxylase | NO3−, NH4+ | Biosynthesis of cofactors, prosthetic groups, and carriers | - |
| All3964 | Phosphoglucomutase/phosphomannomutase | N2, NO3−, NH4+ | Central intermediary metabolism | Anabaena |
| All3984 | All3984 protein | N2 | Conserved hypothetical protein | N. punctiforme |
| All4038 | All4038 protein | NH4+ | No similarity | - |
| All4050 | All4050 protein | NH4+ | Conserved hypothetical protein | Anabaena |
| All4121 | Ferredoxin--NADP reductase | N2, NO3−, NH4+ | Photosynthesis and respiration | Anabaena |
| All4131 | Phosphoglycerate kinase | N2, NO3−, NH4+ | Regulatory functions | N. commune, N. punctiforme, Anabaena |
| All4145 | All4145 protein | NH4+ | Other categories | - |
| All4191 | DNA-directed RNA polymerase subunit alpha | NH4+ | Transcription | - |
| All4214 | 50S ribosomal protein L4 | NH4+ | Translation | - |
| All4287 | Peptidyl-prolyl cis-trans isomerase B | NH4+ | Translation | N. punctiforme |
| All4388 | All4388 protein | N2 | Conserved hypothetical protein | - |
| All4464 | Phosphoadenosine phosphosulfate reductase | NO3−, NH4+ | Amino acid biosynthesis | - |
| All4499 | All4499 protein | N2, NO3− | Conserved hypothetical protein | - |
| All4539 | l-sorbosone dehydrogenase | N2, NO3−, NH4+ | Other categories | N. punctiforme |
| All4563 | Fructose-bisphosphate aldolase | NH4+ | Other categories | - |
| All4575 | Phosphate-binding periplasmic protein of phosphate ABC transporter | N2, NO3−, NH4+ | Transport and binding proteins | - |
| All4749 | All4749 protein | NO3−, NH4+ | Conserved hypothetical protein | - |
| All4906 | Phosphoglycerate mutase | N2 | Energy metabolism | - |
| All4968 | Glutathione reductase | N2, NO3− | Biosynthesis of cofactors, prosthetic groups, and carriers | Anabaena |
| All4985 | Sucrose synthase | NH4+ | Energy metabolism | - |
| All5039 | ATP synthase subunit beta | N2, NO3−, NH4+ | Photosynthesis and respiration | Anabaena |
| All5062 | Glyceraldehyde-3-phosphate dehydrogenase 2 | N2, NH4+ | Photosynthesis and respiration | - |
| All7614 | All7614 protein | N2, NO3− | Conserved hypothetical protein | - |
| All7633 | All7633 protein | NO3−, NH4+ | Conserved hypothetical protein | - |
| Alr0020 | Phycobiliprotein ApcE | NH4+ | Photosynthesis and respiration | - |
| Alr0021 | Allophycocyanin subunit alpha 1 | N2, NO3−, NH4+ | Photosynthesis and respiration | N. punctiforme, Anabaena |
| Alr0022 | Allophycocyanin subunit beta | N2, NO3−, NH4+ | Photosynthesis and respiration | Anabaena |
| Alr0051 | IMP dehydrogenase | NO3− | Purines, pyrimidines, nucleosides, and nucleotides | - |
| Alr0069 | Ribonuclease PH | NH4+ | Transcription | - |
| Alr0132 | Alr0132 protein | NO3−, NH4+ | Conserved hypothetical protein | Anabaena |
| Alr0140 | Periplasmic oligopeptide-binding protein of oligopeptide ABC transporter | N2, NO3−, NH4+ | Transport and binding proteins | - |
| Alr0169 | Cyclomaltodextrin glucanotransferase | NH4+ | Other categories | - |
| Alr0237 | Probable cytosol aminopeptidase | N2, NH4+ | Translation | - |
| Alr0267 | Alr0267 protein | N2, NO3− | No similarity | N. punctiforme, Anabaena |
| Alr0474 | Alr0474 protein | NO3−, NH4+ | No similarity | - |
| Alr0523 | Phycoerythrocyanin subunit beta | N2, NO3−, NH4+ | Photosynthesis and respiration | N. commune |
| Alr0525 | Phycobilisome 34.5 kDa linker polypeptide, phycoerythrocyanin-associated, rod | NO3− | Photosynthesis and respiration | N. commune |
| Alr0528 | C-phycocyanin subunit beta | N2, NO3−, NH4+ | Photosynthesis and respiration | N. punctiforme, Anabaena |
| Alr0529 | C-phycocyanin alpha chain | N2, NO3−, NH4+ | Photosynthesis and respiration | N. punctiforme, Anabaena |
| Alr0530 | Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod | N2, NO3−, NH4+ | Photosynthesis and respiration | N. commune |
| Alr0534 | Phycobilisome rod-core linker polypeptide CpcG1 | NO3−, NH4+ | Photosynthesis and respiration | N. commune |
| Alr0608 | Nitrate transport protein NrtA | N2, NO3−, NH4+ | Amino acid biosynthesis | Anabaena |
| Alr0782 | Ribulose-phosphate 3-epimerase | NH4+ | Central intermediary metabolism | N. commune, Anabaena |
| Alr0834 | Porin major outer membrane protein | N2, NO3− | Cell envelope | - |
| Alr0880 | Oligopeptidase A | NH4+ | Translation | - |
| Alr0996 | Protease | N2 | Translation | - |
| Alr1004 | Alanine--glyoxylate aminotransferase | N2 | Amino acid biosynthesis | - |
| Alr1050 | Glucose-6-phosphate isomerase | N2, NO3−, NH4+ | Energy metabolism | Anabaena |
| Alr1080 | Acetylornithine aminotransferase | N2 | Amino acid biosynthesis | - |
| Alr1299 | Phosphoribosylglycinamide formyltransferase 2 | N2 | Purines, pyrimidines, nucleosides, and nucleotides | - |
| Alr1310 | Alr1310 protein | NH4+ | Conserved hypothetical protein | - |
| Alr1313 | 3-isopropylmalate dehydrogenase | NH4+ | Amino acid biosynthesis | - |
| Alr1329 | Alr1329 protein | N2 | No similarity | - |
| Alr1348 | Ferredoxin-sulphite reductase | NO3− | Other categories | - |
| Alr1362 | Alr1362 protein | NO3− | Other categories | - |
| Alr1364 | Alr1364 protein | NH4+ | Conserved hypothetical protein | - |
| Alr1381 | Calcium-dependent protease | NO3− | Translation | - |
| Alr1520 | Alr1520 protein | NO3− | Conserved hypothetical protein | - |
| Alr1524 | Ribulose bisphosphate carboxylase large chain | N2, NO3− | Photosynthesis and respiration | N. punctiforme |
| Alr1548 | Alr1548 protein | NO3−, NH4+ | Conserved hypothetical protein | - |
| Alr1742 | Chaperone protein DnaK2 | N2, NH4+ | Cellular processes | - |
| Alr1834 | Alr1834 protein | N2, NO3−, NH4+ | Transport and binding proteins | - |
| Alr1965 | ATP phosphoribosyltransferase | NO3−, NH4+ | Other categories | - |
| Alr2190 | Alpha-amylase | N2, NO3− | Other categories | - |
| Alr2313 | Alr2313 protein | NO3−, NH4+ | No similarity | - |
| Alr2328 | Glutamine synthetase | N2, NO3−, NH4+ | Amino acid biosynthesis | N. punctiforme, Anabaena |
| Alr2535 | Branched-chain amino-acid ABC transport system periplasmic binding protein | N2 | Transport and binding proteins | - |
| Alr2709 | Alr2709 protein | N2 | Conserved hypothetical protein | - |
| Alr2771 | Dihydroxy-acid dehydratase | NO3− | Amino acid biosynthesis | Anabaena |
| Alr2877 | Bicarbonate transport bicarbonate-binding protein | N2, NO3−, NH4+ | Transport and binding proteins | - |
| Alr2887 | Alr2887 protein | N2, NO3− | Conserved hypothetical protein | - |
| Alr2938 | Superoxide dismutase | N2, NO3−, NH4+ | Cellular processes | N. commune, N. punctiforme, Anabaena |
| Alr2948 | Alr2948 protein | NO3−, NH4+ | Other categories | - |
| Alr2973 | Glucokinase | NH4+ | Energy metabolism | - |
| Alr3090 | Alr3090 protein | NH4+ | Conserved hypothetical protein | Anabaena |
| Alr3344 | Transketolase | N2, NH4+ | Other categories | - |
| Alr3402 | Nucleoside diphosphate kinase | NH4+ | Purines, pyrimidines, nucleosides, and nucleotides | - |
| Alr3539 | Alr3539 protein | N2, NO3−, NH4+ | No similarity | Anabaena |
| Alr3588 | Alr3588 protein | NO3−, NH4+ | No similarity | Anabaena |
| Alr3607 | Alr3607 protein | NO3−, NH4+ | No similarity | - |
| Alr3608 | Alr3608 protein | N2 | Other categories | - |
| Alr3659 | Alr3659 protein | N2, NH4+ | Energy metabolism | N. punctiforme |
| Alr3808 | Nutrient stress-induced DNA-binding protein | N2, NH4+ | Other categories | N. commune, Anabaena |
| Alr4072 | Alr4072 protein | N2, NO3−, NH4+ | Other categories | - |
| Alr4123 | Phosphoribulokinase | N2, NH4+ | Photosynthesis and respiration | - |
| Alr4238 | Alr4238 protein | NH4+ | Other categories | Anabaena |
| Alr4385 | Triosephosphate isomerase | NH4+ | Energy metabolism | - |
| Alr4448 | Endo-1,4-beta-xylanase | N2 | Other categories | - |
| Alr4550 | Uncharacterized protein alr4550 | N2, NO3−, NH4+ | Conserved hypothetical protein | N. punctiforme, Anabaena |
| Alr4641 | Peroxiredoxin | NO3−, NH4+ | Cellular processes | Anabaena |
| Alr4745 | Dihydrolipoyl dehydrogenase | N2, NO3−, NH4+ | Energy metabolism | - |
| Alr4794 | Alr4794 protein | NO3−, NH4+ | Conserved hypothetical protein | Anabaena |
| Alr4853 | Aspartate aminotransferase | N2 | Amino acid biosynthesis | - |
| Alr4907 | Ornithine carbamoyltransferase | N2, NO3−, NH4+ | Amino acid biosynthesis | - |
| Alr4976 | Phosphodiesterase/alkaline phosphatase D | NO3− | Other categories | N. commune |
| Alr4979 | Alr4979 protein | NH4+ | Conserved hypothetical protein | Anabaena |
| Alr5103 | ll-diaminopimelate aminotransferase 1 | N2, NO3− | Other categories | Anabaena |
| Alr5182 | Oxidoreductase | NH4+ | Other categories | - |
| Alr7261 | Alr7261 protein | N2, NO3−, NH4+ | Other categories | - |
| Alr7346 | Alr7346 protein | N2, NH4+ | No similarity | - |
| Alr7524 | Alr7524 protein | N2, NO3−, NH4+ | Conserved hypothetical protein | - |
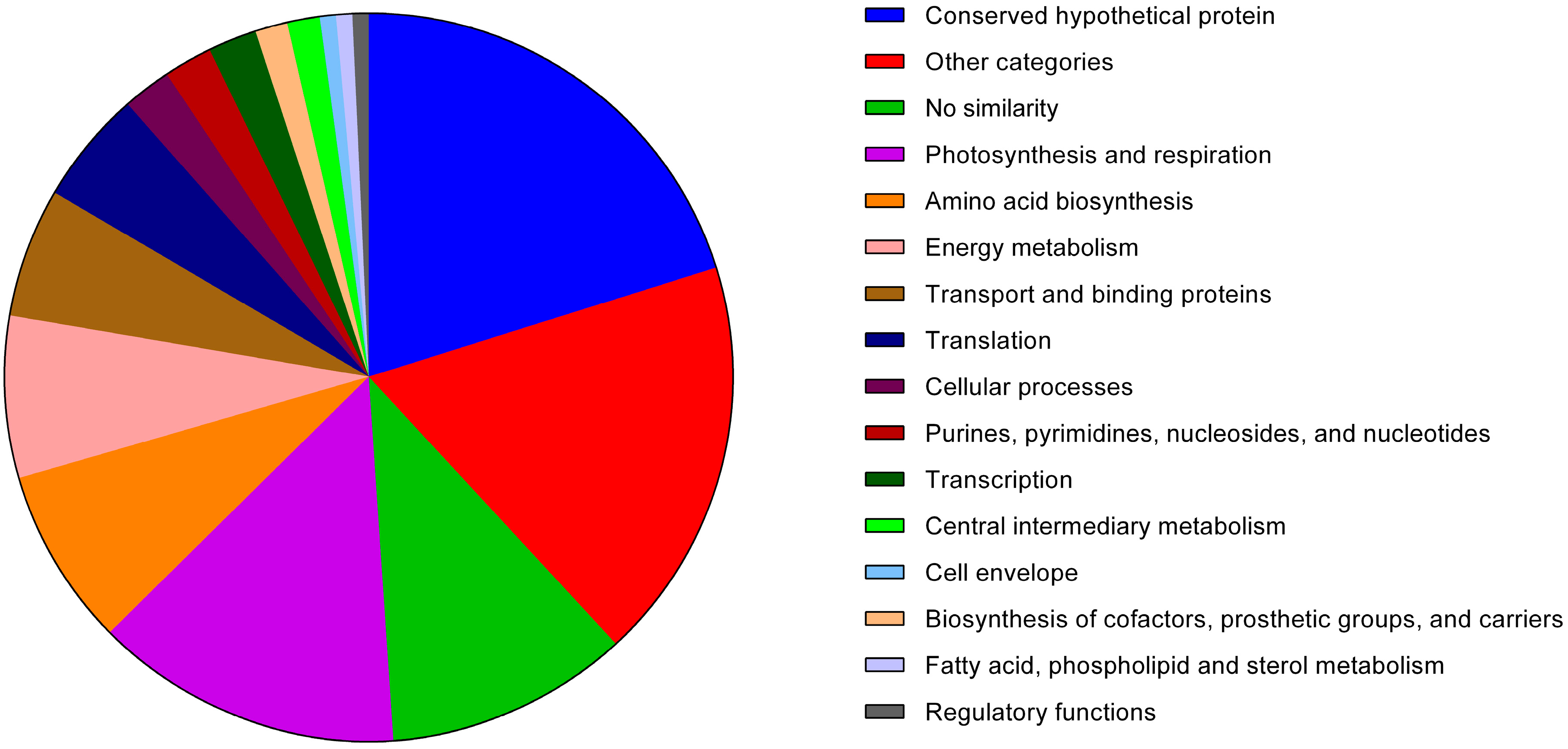
3.1.1. Analysis of the Anabaena sp. PCC 7120 Identified Exoproteins
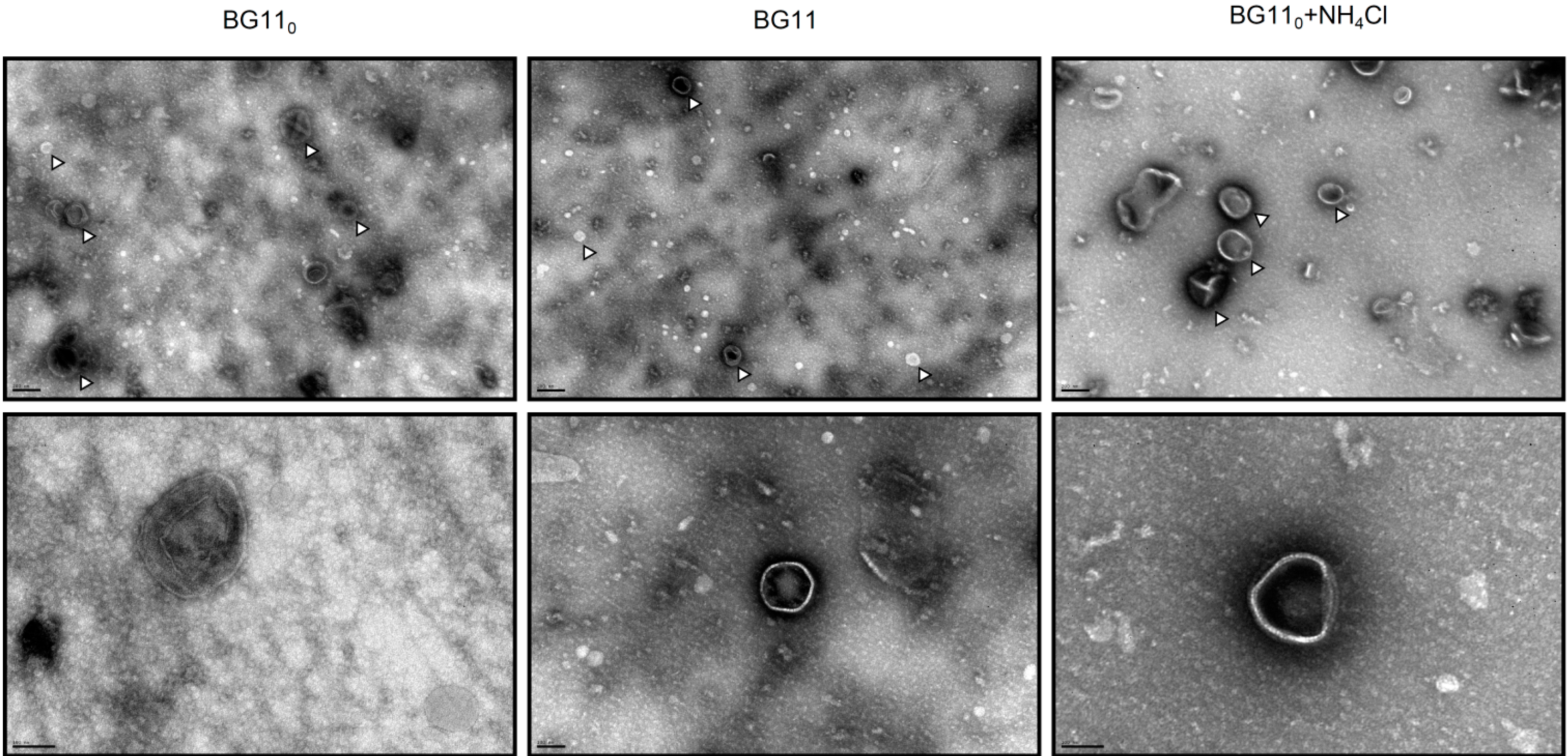
3.1.2. Membrane Vesicle Formation
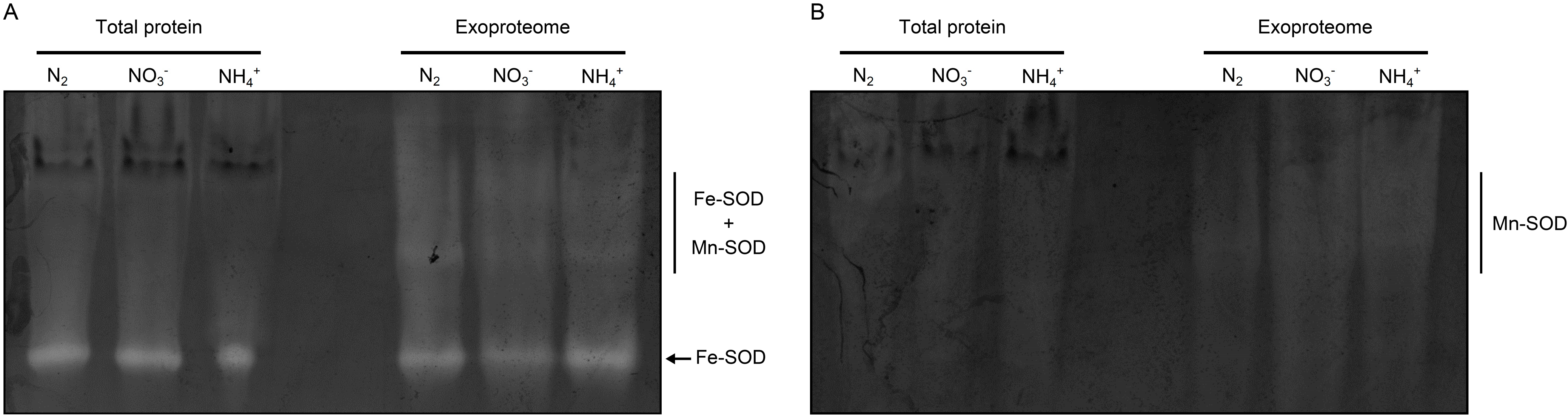
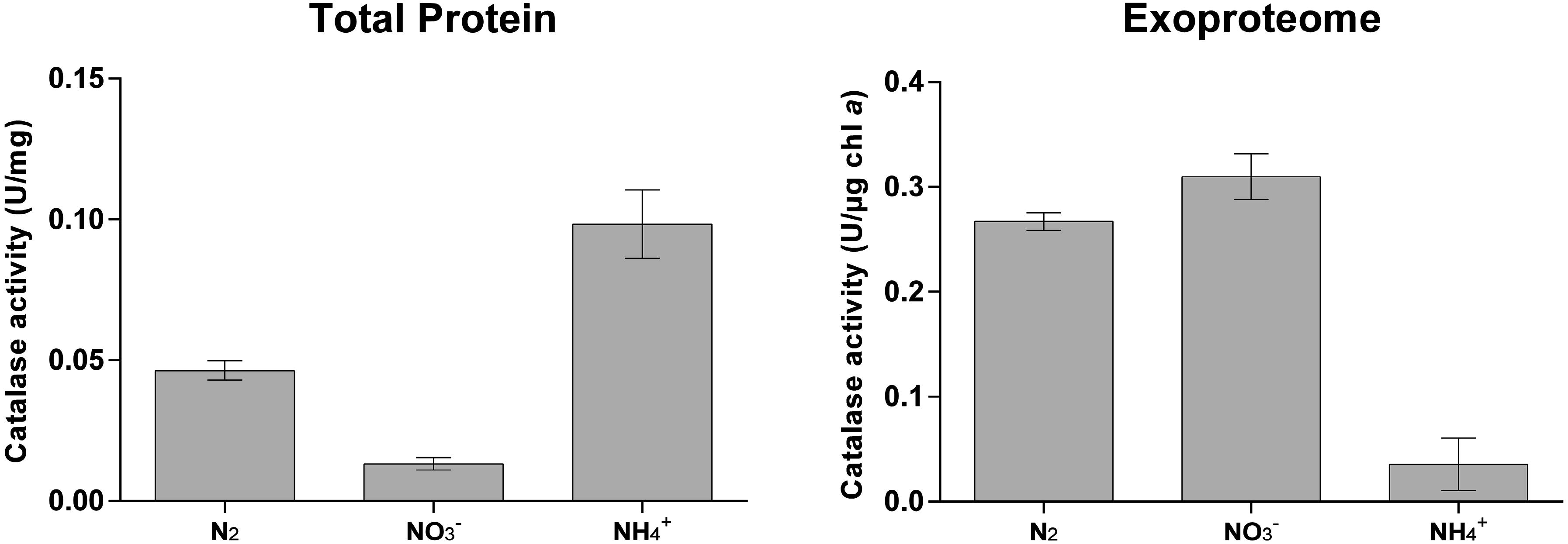
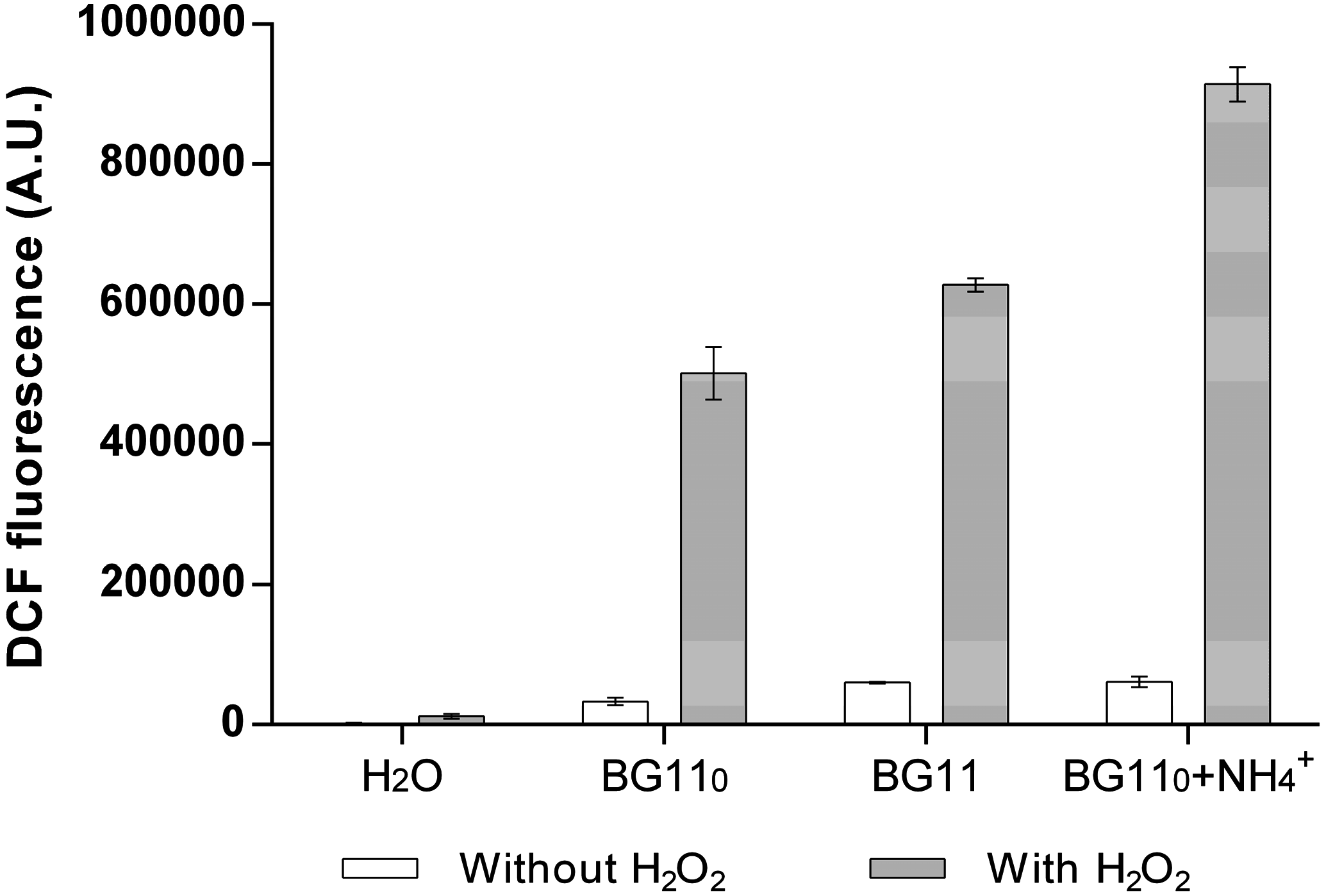
3.2. Cyanobacterial Exoproteins Involved in Redox Homeostasis
3.3. Modulating Cyanobacterial Protein Secretion: Future Perspectives
4. Conclusions
Acknowledgments
Author Contributions
Supplementary Materials
Conflicts of Interest
References
- Varki, A. Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2009. [Google Scholar]
- Sazuka, T.; Ohara, O. Towards a proteome project of cyanobacterium Synechocystis sp. strain PCC6803: Linking 130 protein spots with their respective genes. Electrophoresis 1997, 18, 1252–1258. [Google Scholar] [CrossRef] [PubMed]
- Sazuka, T.; Yamaguchi, M.; Ohara, O. Cyano2Dbase updated: Linkage of 234 protein spots to corresponding genes through N-terminal microsequencing. Electrophoresis 1999, 20, 2160–2171. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Kim, D.S.; Lee, J.; Kim, S.J.; Kim, S.I.; Kim, Y.H.; Hong, J.; Yoo, J.S.; Suh, K.H.; Park, Y.M. Proteome analysis of light-induced proteins in Synechocystis sp. PCC 6803: Identification of proteins separated by 2D-PAGE using N-terminal sequencing and MALDI-TOF MS. Mol. Cells 2000, 10, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Wegener, K.M.; Singh, A.K.; Jacobs, J.M.; Elvitigala, T.; Welsh, E.A.; Keren, N.; Gritsenko, M.A.; Ghosh, B.K.; Camp, D.G., II; Smith, R.D.; et al. Global proteomics reveal an atypical strategy for carbon/nitrogen assimilation by a cyanobacterium under diverse environmental perturbations. Mol. Cell. Proteomics 2010, 9, 2678–2689. [Google Scholar] [CrossRef] [PubMed]
- Pandhal, J.; Ow, S.Y.; Wright, P.C.; Biggs, C.A. Comparative proteomics study of salt tolerance between a nonsequenced extremely halotolerant cyanobacterium and its mildly halotolerant relative using in vivo metabolic labeling and in vitro isobaric labeling. J. Proteome Res. 2009, 8, 818–828. [Google Scholar] [CrossRef] [PubMed]
- Pandhal, J.; Wright, P.C.; Biggs, C.A. Proteomics with a pinch of salt: A cyanobacterial perspective. Saline Syst. 2008, 4. [Google Scholar] [CrossRef]
- Rowland, J.G.; Simon, W.J.; Prakash, J.S.; Slabas, A.R. Proteomics reveals a role for the RNA helicase CrhR in the modulation of multiple metabolic pathways during cold acclimation of Synechocystis sp. PCC6803. J. Proteome Res. 2011, 10, 3674–3689. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Selão, T.T.; Pisareva, T.; Qian, J.; Sze, S.K.; Carlberg, I.; Norling, B. Deletion of Synechocystis sp. PCC 6803 leader peptidase LepB1 affects photosynthetic complexes and respiration. Mol. Cell. Proteomics 2013, 12, 1192–1203. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, H.M.; Cui, S.X.; Suzuki, I.; Zhang, L.F.; Li, L.; Bo, T.T.; Wang, J.; Murata, N.; Huang, F. Proteomic study of the impact of hik33 mutation in Synechocystis sp. PCC 6803 under normal and salt stress conditions. J. Proteome Res. 2012, 11, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Barrios-Llerena, M.E.; Reardon, K.F.; Wright, P.C. 2-DE proteomic analysis of the model cyanobacterium Anabaena variabilis. Electrophoresis 2007, 28, 1624–1632. [Google Scholar] [CrossRef] [PubMed]
- Gan, C.S.; Reardon, K.F.; Wright, P.C. Comparison of protein and peptide prefractionation methods for the shotgun proteomic analysis of Synechocystis sp. PCC 6803. Proteomics 2005, 5, 2468–2478. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.B.; Ow, S.Y.; Barrios-Llerena, M.E.; Wright, P.C.; Moradas-Ferreira, P.; Tamagnini, P. iTRAQ-based quantitative proteomic analysis of Gloeothece sp. PCC 6909: Comparison with its sheathless mutant and adaptations to nitrate deficiency and sulfur limitation. J. Proteomics 2011, 75, 270–283. [Google Scholar] [CrossRef] [PubMed]
- Ow, S.Y.; Noirel, J.; Cardona, T.; Taton, A.; Lindblad, P.; Stensjo, K.; Wright, P.C. Quantitative overview of N2 fixation in Nostoc punctiforme ATCC 29133 through cellular enrichments and iTRAQ shotgun proteomics. J. Proteome Res. 2009, 8, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Ow, S.Y.; Cardona, T.; Taton, A.; Magnuson, A.; Lindblad, P.; Stensjo, K.; Wright, P.C. Quantitative shotgun proteomics of enriched heterocysts from Nostoc sp. PCC 7120 using 8-plex isobaric peptide tags. J. Proteome Res. 2008, 7, 1615–1628. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.C.; Campbell, E.L.; Meeks, J.C. A soluble 3D LC/MS/MS proteome of the filamentous cyanobacterium Nostoc punctiforme. J. Proteome Res. 2006, 5, 3096–3104. [Google Scholar] [CrossRef] [PubMed]
- Aryal, U.K.; Callister, S.J.; McMahon, B.H.; McCue, L.A.; Brown, J.; Stockel, J.; Liberton, M.; Mishra, S.; Zhang, X.; Nicora, C.D.; et al. Proteomic profiles of five strains of oxygenic photosynthetic cyanobacteria of the genus Cyanothece. J. Proteome Res. 2014, 13, 3262–3276. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Hedman, E.; Funk, C.; Kieselbach, T.; Schroder, W.P.; Norling, B. Isolation of outer membrane of Synechocystis sp. PCC 6803 and its proteomic characterization. Mol. Cell. Proteomics 2004, 3, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Moslavac, S.; Bredemeier, R.; Mirus, O.; Granvogl, B.; Eichacker, L.A.; Schleiff, E. Proteomic analysis of the outer membrane of Anabaena sp. strain PCC 7120. J. Proteome Res. 2005, 4, 1330–1338. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S.; Huang, F.; Nilsson, F.; Hagemann, M.; Norling, B. Proteomics of Synechocystis sp. strain PCC 6803. Identification of periplasmic proteins in cells grown at low and high salt concentrations. Eur. J. Biochem. 2000, 267, 5900–5907. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.F.; Yang, H.M.; Cui, S.X.; Hu, J.; Wang, J.; Kuang, T.Y.; Norling, B.; Huang, F. Proteomic analysis of plasma membranes of cyanobacterium Synechocystis sp. strain PCC 6803 in response to high pH stress. J. Proteome Res. 2009, 8, 2892–2902. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Parmryd, I.; Nilsson, F.; Persson, A.L.; Pakrasi, H.B.; Andersson, B.; Norling, B. Proteomics of Synechocystis sp. strain PCC 6803: Identification of plasma membrane proteins. Mol. Cell. Proteomics 2002, 1, 956–966. [Google Scholar] [CrossRef] [PubMed]
- Simon, W.J.; Hall, J.J.; Suzuki, I.; Murata, N.; Slabas, A.R. Proteomic study of the soluble proteins from the unicellular cyanobacterium Synechocystis sp. PCC6803 using automated matrix-assisted laser desorption/ionization-time of flight peptide mass fingerprinting. Proteomics 2002, 2, 1735–1742. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, R.; Matros, A.; Melzer, M.; Mock, H.P.; Sainis, J.K. Heterogeneity in thylakoid membrane proteome of Synechocystis 6803. J. Proteomics 2010, 73, 976–991. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Pisareva, T.; Norling, B. Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 2005, 5, 4905–4916. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Stevanovic, M.; Brouwer, E.; Bublak, D.; Tripp, J.; Schorge, T.; Karas, M.; Schleiff, E. Secretome analysis of Anabaena sp. PCC 7120 and the involvement of the TolC-homologue HgdD in protein secretion. Environ. Microbiol. 2014. [Google Scholar] [CrossRef]
- Helm, R.D.; Potts, M. Extracellular matrix (ECM). In Ecology of Cyanobacteria II: Their Diversity in Space and Time; Whitton, B.A., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 461–480. [Google Scholar]
- Kehr, J.C.; Zilliges, Y.; Springer, A.; Disney, M.D.; Ratner, D.D.; Bouchier, C.; Seeberger, P.H.; de Marsac, N.T.; Dittmann, E. A mannan binding lectin is involved in cell-cell attachment in a toxic strain of Microcystis aeruginosa. Mol. Microbiol. 2006, 59, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Sergeyenko, T.V.; Los, D.A. Identification of secreted proteins of the cyanobacterium Synechocystis sp. strain PCC 6803. FEMS Microbiol. Lett. 2000, 193, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Vilhauer, L.; Jervis, J.; Ray, W.K.; Helm, R.F. The exo-proteome and exo-metabolome of Nostoc punctiforme (cyanobacteria) in the presence and absence of nitrate. Arch. Microbiol. 2014, 196, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Zilliges, Y.; Kehr, J.C.; Mikkat, S.; Bouchier, C.; de Marsac, N.T.; Borner, T.; Dittmann, E. An extracellular glycoprotein is implicated in cell-cell contacts in the toxic cyanobacterium Microcystis aeruginosa PCC 7806. J. Bacteriol. 2008, 190, 2871–2879. [Google Scholar] [CrossRef] [PubMed]
- Desvaux, M.; Hebraud, M.; Talon, R.; Henderson, I.R. Secretion and subcellular localizations of bacterial proteins: A semantic awareness issue. Trends Microbiol. 2009, 17, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
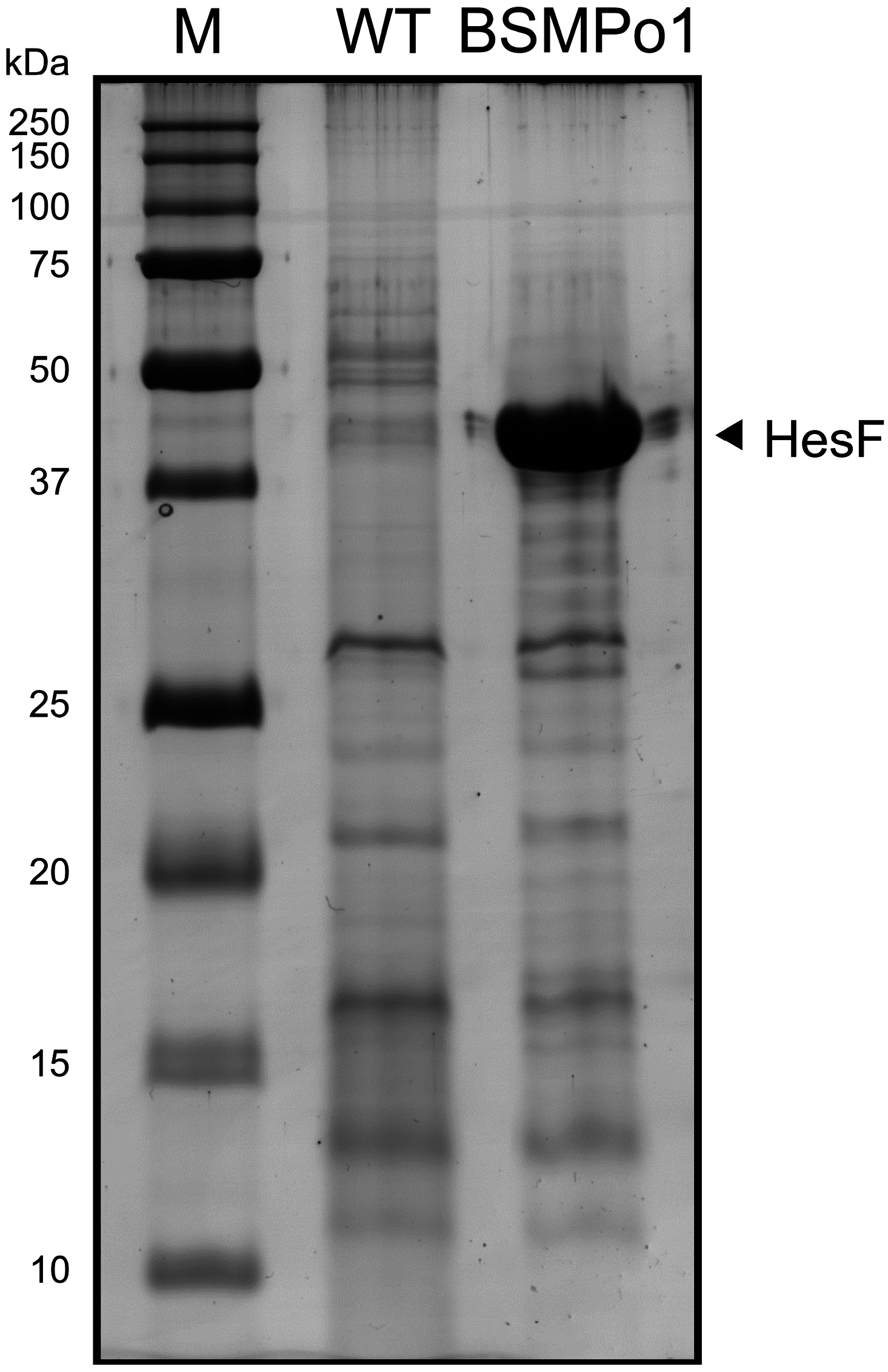
- Oliveira, P.; Pinto, F.; Pacheco, C.C.; Mota, R.; Tamagnini, P. HesF, an exoprotein required for filament adhesion and aggregation in Anabaena sp. PCC 7120. Environ. Microbiol. 2014. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Gluck, F.; Hoogland, C.; Antinori, P.; Robin, X.; Nikitin, F.; Zufferey, A.; Pasquarello, C.; Fetaud, V.; Dayon, L.; Muller, M.; et al. EasyProt—An easy-to-use graphical platform for proteomics data analysis. J. Proteomics 2013, 79, 146–160. [Google Scholar] [CrossRef] [PubMed]
- Elias, J.E.; Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Lopes Pinto, F.; Erasmie, S.; Blikstad, C.; Lindblad, P.; Oliveira, P. FtsZ degradation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Plant Physiol. 2011, 168, 1934–1942. [Google Scholar] [CrossRef] [PubMed]
- Pitcher, L.H.; Brennan, E.; Zilinskas, B.A. The antiozonant ethylenediurea does not act via superoxide dismutase induction in bean. Plant Physiol. 1992, 99, 1388–1392. [Google Scholar] [CrossRef] [PubMed]
- Hempel, S.L.; Buettner, G.R.; O’Malley, Y.Q.; Wessels, D.A.; Flaherty, D.M. Dihydrofluorescein diacetate is superior for detecting intracellular oxidants: Comparison with 2’,7’-dichlorodihydrofluorescein diacetate, 5(and 6)-carboxy-2’,7’-dichlorodihydrofluorescein diacetate, and dihydrorhodamine 123. Free Radic. Biol. Med. 1999, 27, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Zille, A.; Micheletti, E.; Moradas-Ferreira, P.; De Philippis, R.; Tamagnini, P. Complexity of cyanobacterial exopolysaccharides: Composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol. Rev. 2009, 33, 917–941. [Google Scholar] [CrossRef] [PubMed]
- Heidorn, T.; Camsund, D.; Huang, H.H.; Lindberg, P.; Oliveira, P.; Stensjo, K.; Lindblad, P. Synthetic biology in cyanobacteria engineering and analyzing novel functions. Methods Enzymol. 2011, 497, 539–579. [Google Scholar] [PubMed]
- Nanni, P.; Levander, F.; Roda, G.; Caponi, A.; James, P.; Roda, A. A label-free nano-liquid chromatography-mass spectrometry approach for quantitative serum peptidomics in Crohn’s disease patients. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2009, 877, 3127–3136. [Google Scholar] [CrossRef]
- Koropatkin, N.M.; Pakrasi, H.B.; Smith, T.J. Atomic structure of a nitrate-binding protein crucial for photosynthetic productivity. Proc. Natl. Acad. Sci. USA 2006, 103, 9820–9825. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Igual, R.; Picossi, S.; Lopez-Garrido, J.; Flores, E.; Herrero, A. N and C control of ABC-type bicarbonate transporter Cmp and its LysR-type transcriptional regulator CmpR in a heterocyst-forming cyanobacterium, Anabaena sp. Environ. Microbiol. 2012, 14, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Omata, T. Substrate-binding lipoprotein of the cyanobacterium Synechococcus sp. strain PCC 7942 involved in the transport of nitrate and nitrite. J. Biol. Chem. 1997, 272, 3036–3041. [Google Scholar] [CrossRef] [PubMed]
- Kellett, L.E.; Poole, D.M.; Ferreira, L.M.; Durrant, A.J.; Hazlewood, G.P.; Gilbert, H.J. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem. J. 1990, 272, 369–376. [Google Scholar]
- Meiss, G.; Gimadutdinow, O.; Haberland, B.; Pingoud, A. Mechanism of DNA cleavage by the DNA/RNA-non-specific Anabaena sp. PCC 7120 endonuclease NucA and its inhibition by NuiA. J. Mol. Biol. 2000, 297, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Muro-Pastor, A.M.; Flores, E.; Herrero, A.; Wolk, C.P. Identification, genetic analysis and characterization of a sugar-non-specific nuclease from the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 1992, 6, 3021–3030. [Google Scholar] [CrossRef] [PubMed]
- Ekman, M.; Picossi, S.; Campbell, E.L.; Meeks, J.C.; Flores, E. A Nostoc punctiforme sugar transporter necessary to establish a cyanobacterium-plant symbiosis. Plant Physiol. 2013, 161, 1984–1992. [Google Scholar] [CrossRef] [PubMed]
- Mastronunzio, J.E.; Huang, Y.; Benson, D.R. Diminished exoproteome of Frankia spp. in culture and symbiosis. Appl. Environ. Microbiol. 2009, 75, 6721–6728. [Google Scholar] [CrossRef] [PubMed]
- Osbourne, D.; Aruni, A.W.; Dou, Y.; Perry, C.; Boskovic, D.S.; Roy, F.; Fletcher, H.M. VimA-dependent modulation of the secretome in Porphyromonas gingivalis. Mol. Oral Microbiol. 2012, 27, 420–435. [Google Scholar] [CrossRef] [PubMed]
- Tjalsma, H. Feature-based reappraisal of the Bacillus subtilis exoproteome. Proteomics 2007, 7, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Tjalsma, H.; Antelmann, H.; Jongbloed, J.D.; Braun, P.G.; Darmon, E.; Dorenbos, R.; Dubois, J.Y.; Westers, H.; Zanen, G.; Quax, W.J.; et al. Proteomics of protein secretion by Bacillus subtilis: Separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 2004, 68, 207–233. [Google Scholar] [CrossRef] [PubMed]
- Ziebandt, A.K.; Kusch, H.; Degner, M.; Jaglitz, S.; Sibbald, M.J.; Arends, J.P.; Chlebowicz, M.A.; Albrecht, D.; Pantucek, R.; Doskar, J.; et al. Proteomics uncovers extreme heterogeneity in the Staphylococcus aureus exoproteome due to genomic plasticity and variant gene regulation. Proteomics 2010, 10, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Silva, W.M.; Seyffert, N.; Santos, A.V.; Castro, T.L.; Pacheco, L.G.; Santos, A.R.; Ciprandi, A.; Dorella, F.A.; Andrade, H.M.; Barh, D.; et al. Identification of 11 new exoproteins in Corynebacterium pseudotuberculosis by comparative analysis of the exoproteome. Microb. Pathog. 2013, 61–62, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Mashburn-Warren, L.M.; Whiteley, M. Special delivery: Vesicle trafficking in prokaryotes. Mol. Microbiol. 2006, 61, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Kulp, A.; Kuehn, M.J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010, 64, 163–184. [Google Scholar] [CrossRef] [PubMed]
- Biller, S.J.; Schubotz, F.; Roggensack, S.E.; Thompson, A.W.; Summons, R.E.; Chisholm, S.W. Bacterial vesicles in marine ecosystems. Science 2014, 343, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Walda, C.; Stucken, K.; Dagan, T. Characterization of outer membrane vesicles in cyanobacteria. In Proceedings of the 9th European Workshop on the Molecular Biology of Cyanobacteria, Texel, The Netherlands, 7–11 September 2014; p. 91.
- Raynaud, C.; Etienne, G.; Peyron, P.; Laneelle, M.A.; Daffe, M. Extracellular enzyme activities potentially involved in the pathogenicity of Mycobacterium tuberculosis. Microbiology 1998, 144, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Smith, I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 2003, 16, 463–496. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, D.; Reichardt, W.; Vettermann, S. Extracellular superoxide dismutase from Streptococcus pyogenes type 12 strain is manganese-dependent. FEMS Microbiol. Lett. 1998, 160, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.O.; Man, S.M.; Lamb, S.; Raftery, M.J.; Wilkins, M.R.; Kovach, Z.; Mitchell, H. The secretome of Campylobacter concisus. FEBS J. 2010, 277, 1606–1617. [Google Scholar] [CrossRef] [PubMed]
- Shirkey, B.; Kovarcik, D.P.; Wright, D.J.; Wilmoth, G.; Prickett, T.F.; Helm, R.F.; Gregory, E.M.; Potts, M. Active Fe-containing superoxide dismutase and abundant sodF mRNA in Nostoc commune (cyanobacteria) after years of desiccation. J. Bacteriol. 2000, 182, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Regelsberger, G.; Obinger, C.; Zoder, R.; Altmann, F.; Peschek, G.A. Purification and characterization of a hydroperoxidase from the cyanobacterium Synechocystis PCC 6803: Identification of its gene by peptide mass mapping using matrix assisted laser desorption ionization time-of-flight mass spectrometry. FEMS Microbiol. Lett. 1999, 170, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tichy, M.; Vermaas, W. In vivo role of catalase-peroxidase in Synechocystis sp. strain PCC 6803. J Bacteriol. 1999, 181, 1875–1882. [Google Scholar] [PubMed]
- Pascual, M.B.; Mata-Cabana, A.; Florencio, F.J.; Lindahl, M.; Cejudo, F.J. Overoxidation of 2-Cys peroxiredoxin in prokaryotes: Cyanobacterial 2-Cys peroxiredoxins sensitive to oxidative stress. J. Biol. Chem. 2010, 285, 34485–34492. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Are polyphenols antioxidants or pro-oxidants? What do we learn from cell culture and in vivo studies? Arch. Biochem. Biophys. 2008, 476, 107–112. [Google Scholar] [CrossRef] [PubMed]
- López, D.; Vlamakis, H.; Kolter, R. Biofilms. Cold Spring Harb. Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef]
- Theoret, J.R.; Cooper, K.K.; Zekarias, B.; Roland, K.L.; Law, B.F.; Curtiss, R., III; Joens, L.A. The Campylobacter jejuni Dps homologue is important for in vitro biofilm formation and cecal colonization of poultry and may serve as a protective antigen for vaccination. Clin. Vaccine Immunol. 2012, 19, 1426–1431. [Google Scholar] [CrossRef] [PubMed]
- Enany, S.; Yoshida, Y.; Magdeldin, S.; Bo, X.; Zhang, Y.; Enany, M.; Yamamoto, T. Two dimensional electrophoresis of the exo-proteome produced from community acquired methicillin resistant Staphylococcus aureus belonging to clonal complex 80. Microbiol. Res. 2013, 168, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Park, M.K.; Kim, S.H.; Oh, K.G.; Jung, K.H.; Hong, C.H.; Yoon, J.W.; Chai, Y.G. Identification of stringent response-related and potential serological proteins released from Bacillus anthracis overexpressing the RelA/SpoT homolog, RshBant. Curr. Microbiol. 2014, 69, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Avila-Calderon, E.D.; Lopez-Merino, A.; Jain, N.; Peralta, H.; Lopez-Villegas, E.O.; Sriranganathan, N.; Boyle, S.M.; Witonsky, S.; Contreras-Rodriguez, A. Characterization of outer membrane vesicles from Brucella melitensis and protection induced in mice. Clin. Dev. Immunol. 2012, 2012. [Google Scholar] [CrossRef]
- Christie-Oleza, J.A.; Pina-Villalonga, J.M.; Bosch, R.; Nogales, B.; Armengaud, J. Comparative proteogenomics of twelve Roseobacter exoproteomes reveals different adaptive strategies among these marine bacteria. Mol. Cell. Proteomics 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Pittman, M.S.; Robinson, H.C.; Poole, R.K. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J. Biol. Chem. 2005, 280, 32254–32261. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.O.; Gho, Y.S.; Lee, J.C.; Kim, S.I. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol. Lett. 2009, 297, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Reed, B.; Chen, R. Biotechnological applications of bacterial protein secretion: From therapeutics to biofuel production. Res. Microbiol. 2013, 164, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Lindblad, P.; Lindberg, P.; Oliveira, P.; Stensjo, K.; Heidorn, T. Design, engineering, and construction of photosynthetic microbial cell factories for renewable solar fuel production. Ambio 2012, 41, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, S.; Aslim, B.; Suludere, Z.; Tan, S. Metal removal of cyanobacterial exopolysaccharides by uronic acid content and monosaccharide composition. Carbohydr. Polym. 2014, 101, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Haselkorn, R. Heterocyst differentiation and nitrogen fixation in cyanobacteria. In Associative and Endophytic Nitrogen-Fixing Bacteria and Cyanobacterial Associations; Elmrich, C., Newton, W.E., Eds.; Springer: Dordrecht, The Netherlands, 2007; Volume 5, pp. 233–255. [Google Scholar]
- Burnat, M.; Herrero, A.; Flores, E. Compartmentalized cyanophycin metabolism in the diazotrophic filaments of a heterocyst-forming cyanobacterium. Proc. Natl. Acad. Sci. USA 2014, 111, 3823–3828. [Google Scholar] [CrossRef] [PubMed]
- Tamagnini, P.; Leitão, E.; Oliveira, P.; Ferreira, D.; Pinto, F.; Harris, D.J.; Heidorn, T.; Lindblad, P. Cyanobacterial hydrogenases: Diversity, regulation and applications. FEMS Microbiol. Rev. 2007, 31, 692–720. [Google Scholar] [CrossRef] [PubMed]
- Camsund, D.; Heidorn, T.; Lindblad, P. Design and analysis of LacI-repressed promoters and DNA-looping in a cyanobacterium. J. Biol. Eng. 2014, 8. [Google Scholar] [CrossRef]
- Huang, H.H.; Lindblad, P. Wide-dynamic-range promoters engineered for cyanobacteria. J. Biol. Eng. 2013, 7. [Google Scholar] [CrossRef]
- Chen, D.J.; Osterrieder, N.; Metzger, S.M.; Buckles, E.; Doody, A.M.; DeLisa, M.P.; Putnam, D. Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc. Natl. Acad. Sci. USA 2010, 107, 3099–3104. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, P.; Martins, N.M.; Santos, M.; Couto, N.A.S.; Wright, P.C.; Tamagnini, P. The Anabaena sp. PCC 7120 Exoproteome: Taking a Peek outside the Box. Life 2015, 5, 130-163. https://doi.org/10.3390/life5010130
Oliveira P, Martins NM, Santos M, Couto NAS, Wright PC, Tamagnini P. The Anabaena sp. PCC 7120 Exoproteome: Taking a Peek outside the Box. Life. 2015; 5(1):130-163. https://doi.org/10.3390/life5010130
Chicago/Turabian StyleOliveira, Paulo, Nuno M. Martins, Marina Santos, Narciso A. S. Couto, Phillip C. Wright, and Paula Tamagnini. 2015. "The Anabaena sp. PCC 7120 Exoproteome: Taking a Peek outside the Box" Life 5, no. 1: 130-163. https://doi.org/10.3390/life5010130
APA StyleOliveira, P., Martins, N. M., Santos, M., Couto, N. A. S., Wright, P. C., & Tamagnini, P. (2015). The Anabaena sp. PCC 7120 Exoproteome: Taking a Peek outside the Box. Life, 5(1), 130-163. https://doi.org/10.3390/life5010130






