Author Contributions
Conceptualization, S.B. and M.A.A.K.; Methodology, S.B., M.A.A.K., M.H.A., J.R., S.P., M.I.A.K., S.K.C. and B.K.P.; Validation, S.B., M.A.A.K., M.H.A., J.R., S.P. and M.I.A.K.; Formal analysis, S.B., M.A.A.K., M.H.A. and J.R.; Investigation, S.B., M.A.A.K., M.H.A., J.R., S.P., M.I.A.K., S.K.C. and B.K.P.; Resources, S.B., M.A.A.K., M.H.A., J.R., S.P., M.I.A.K., S.K.C. and B.K.P.; Writing—original draft, S.B., M.A.A.K., M.H.A., J.R., S.P. and M.I.A.K.; Writing—review & editing, S.B., M.A.A.K., M.H.A., J.R., S.P. and M.I.A.K.; Visualization, S.B., M.A.A.K., M.H.A., J.R., S.P., M.I.A.K., S.K.C. and B.K.P.; Supervision, S.B.; Project administration, S.B.; Funding acquisition, S.B. All authors have read and agreed to the published version of the manuscript.
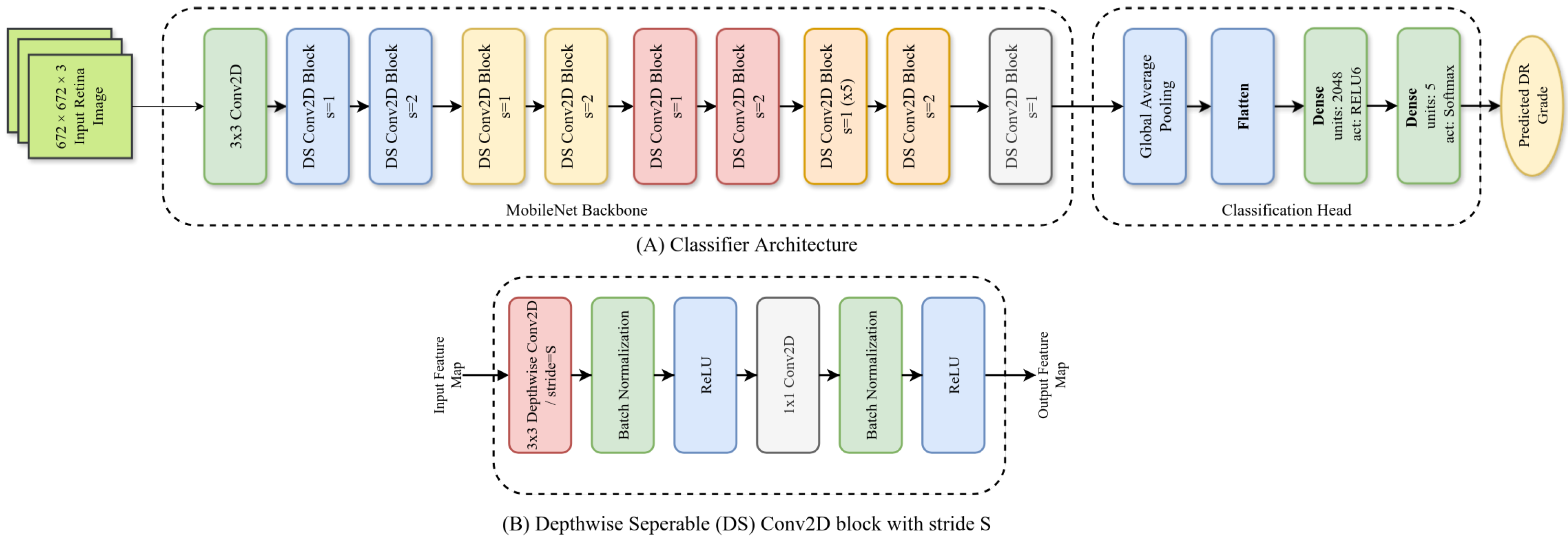
Figure 1.
Architecture of our ADRCs. Here, (A): overall architecture; (B): architecture of a single Depthwise Separable (DS) block.
Figure 1.
Architecture of our ADRCs. Here, (A): overall architecture; (B): architecture of a single Depthwise Separable (DS) block.
Figure 2.
Architecture of our U-net-based segmentation model with the MobileNet encoder and a customized decoder. Here, (A): overall segmentation model architecture; (B): a single upsampling block.
Figure 2.
Architecture of our U-net-based segmentation model with the MobileNet encoder and a customized decoder. Here, (A): overall segmentation model architecture; (B): a single upsampling block.
Figure 3.
Confusion matrices of our ADRCs evaluated on the Kaggle EyePACS, IDRiD, and Messidor-2 datasets. Each subfigure shows mean ± std over each class of the three datasets.
Figure 3.
Confusion matrices of our ADRCs evaluated on the Kaggle EyePACS, IDRiD, and Messidor-2 datasets. Each subfigure shows mean ± std over each class of the three datasets.
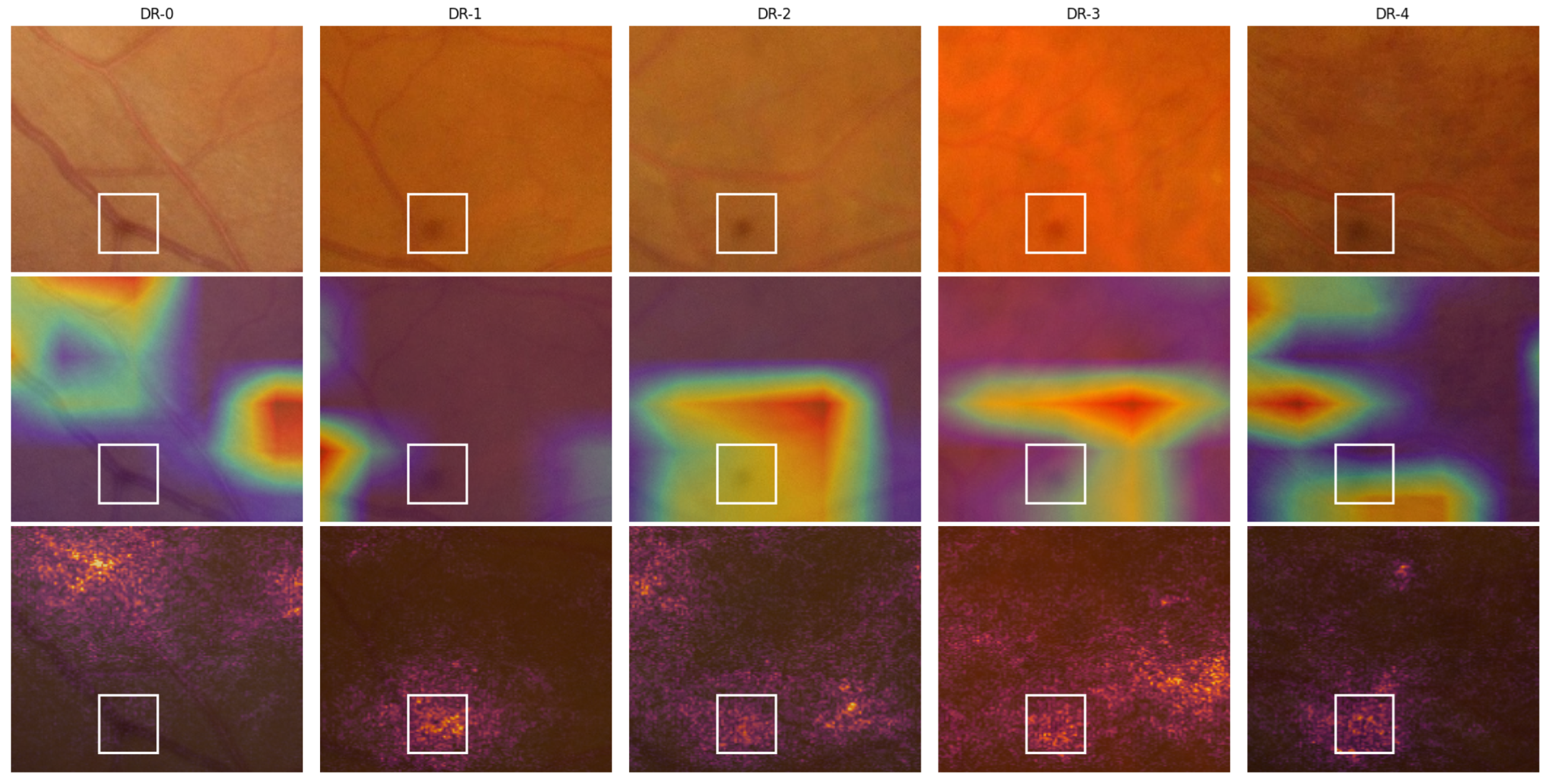
Figure 4.
Visualization of Grad-CAM and IG for sample images from the IDRiD-DR dataset. Each row shows the original image, the Grad-CAM heatmap and overlay, and the IG heatmap and overlay. The five columns correspond to images from DR-0 through DR-4 grades, where the ADRC made correct predictions in all cases except DR-1.
Figure 4.
Visualization of Grad-CAM and IG for sample images from the IDRiD-DR dataset. Each row shows the original image, the Grad-CAM heatmap and overlay, and the IG heatmap and overlay. The five columns correspond to images from DR-0 through DR-4 grades, where the ADRC made correct predictions in all cases except DR-1.
Figure 5.
A close-up view of the lower region of the fundus images from
Figure 4 showcasing a spot caused by an imaging equipment flaw. Here, row 1 contains the cropped position of the original images, and rows 2 and 3 contain the Grad-CAM and IG overlays, respectively. The white boxes highlight the identified spots.
Figure 5.
A close-up view of the lower region of the fundus images from
Figure 4 showcasing a spot caused by an imaging equipment flaw. Here, row 1 contains the cropped position of the original images, and rows 2 and 3 contain the Grad-CAM and IG overlays, respectively. The white boxes highlight the identified spots.
Figure 6.
Our procedure of automatically evaluating ADRC interpretability. We demonstrate the process of interpreting the Grad-CAM activation around the exudates and hemorrhages. Here: (a) original fundus image, (b) Grad-CAM heatmap (with color-map) highlighting the ADRC-activated regions of the fundus image, (c) binary thresholded heatmap showing the highest activations (90th percentile), (d,f) segmented lesion masks from our trained segmentation models (exudates and hemorrhages), and (e,g) intersection map from the logical AND operation between the thresholded heatmap and lesion masks. The white regions represent overlapping pixels, whereas red regions correspond to non-overlapping regions. The non-overlapping regions refer to regions with lesion masks but no heatmap activation.
Figure 6.
Our procedure of automatically evaluating ADRC interpretability. We demonstrate the process of interpreting the Grad-CAM activation around the exudates and hemorrhages. Here: (a) original fundus image, (b) Grad-CAM heatmap (with color-map) highlighting the ADRC-activated regions of the fundus image, (c) binary thresholded heatmap showing the highest activations (90th percentile), (d,f) segmented lesion masks from our trained segmentation models (exudates and hemorrhages), and (e,g) intersection map from the logical AND operation between the thresholded heatmap and lesion masks. The white regions represent overlapping pixels, whereas red regions correspond to non-overlapping regions. The non-overlapping regions refer to regions with lesion masks but no heatmap activation.
Table 1.
Datasets employed at each stage of our study.
Table 1.
Datasets employed at each stage of our study.
| Task | Datasets |
|---|
| Building ADRCs | Kaggle EyePACS [71,72], IDRiD-DR [73], Messidor-2 [74] |
| Building Segmentation Models | IDRiD [73], E-ophtha [75], PALM [76] |
| Analyzing ADRCs’ Decision | IDRiD-DR [73], Retinal-Lesions [77] |
Table 2.
Distribution of images in the DR severity classification datasets used for building ADRCs.
Table 2.
Distribution of images in the DR severity classification datasets used for building ADRCs.
| Dataset Name | Train | Test | Total |
|---|
| DR-0 | DR-1 | DR-2 | DR-3 | DR-4 | Subtotal | DR-0 | DR-1 | DR-2 | DR-3 | DR-4 | Subtotal |
| Kaggle EyePACS | 25,810 | 2443 | 5292 | 873 | 708 | 35,126 | 39,533 | 3762 | 7861 | 1214 | 1206 | 53,576 | 88,702 |
| IDRiD-DR | 134 | 20 | 136 | 74 | 49 | 413 | 34 | 5 | 32 | 19 | 13 | 103 | 516 |
| Messidor-2 | - | - | - | - | - | - | 1017 | 270 | 347 | 75 | 35 | 1744 | 1744 |
Table 3.
Distribution of images used for building segmentation models. Here, Type: type of segmentation area, #Patches: number of patches per image, #Train: number of training images, #Valid: number of validation images, #Test: number of test images, MA: microaneurysms, EX: hard exudates, HE: haemorrhage, SE: soft exudates, OD: optic disc, MC: macula.
Table 3.
Distribution of images used for building segmentation models. Here, Type: type of segmentation area, #Patches: number of patches per image, #Train: number of training images, #Valid: number of validation images, #Test: number of test images, MA: microaneurysms, EX: hard exudates, HE: haemorrhage, SE: soft exudates, OD: optic disc, MC: macula.
| Type | Datasets | #Patches | #Train | #Valid | #Test |
|---|
| MA | IDRiD + E-ophtha | 25 | 155 | 35 | 39 |
| EX | IDRiD + E-ophtha | 25 | 86 | 20 | 22 |
| HE | IDRiD | 25 | 53 | 13 | 14 |
| SE | IDRiD | 25 | 26 | 7 | 7 |
| OD | IDRiD + PALM | 1 | 289 | 64 | 73 |
| MC | PALM | 1 | 234 | 52 | 59 |
Table 4.
Distribution of images in our datasets used for ADRCs’ decision analysis.
Table 4.
Distribution of images in our datasets used for ADRCs’ decision analysis.
| Dataset | DR-0 | DR-1 | DR-2 | DR-3 | DR-4 | Total |
|---|
| IDRiD-DR | 34 | 5 | 32 | 19 | 13 | 103 |
| Retinal-Lesions (A) | 67 | 184 | 561 | 120 | 45 | 977 |
| Retinal-Lesions (B) | - | - | 415 | 136 | 65 | 616 |
Table 5.
Performance of our DNN-based ADRCs for DR severity classification.
Table 5.
Performance of our DNN-based ADRCs for DR severity classification.
| Train Dataset | Test Dataset | Accuracy | F1 Score | Specificity | AUC |
|---|
| Kaggle EyePACS | Kaggle EyePACS | 0.8287 | 0.8102 | 0.7246 | 0.8785 |
| IDRiD-DR | 0.4951 | 0.4765 | 0.8414 | 0.8352 |
| Messidor-2 | 0.7466 | 0.7120 | 0.7844 | 0.8674 |
| IDRiD-DR | Kaggle EyePACS | 0.7042 | 0.6507 | 0.4492 | 0.6589 |
| IDRiD-DR | 0.6214 | 0.6089 | 0.8655 | 0.8412 |
| Messidor-2 | 0.5826 | 0.5338 | 0.6575 | 0.7020 |
Table 6.
Statistical analysis of the probability scores for correct and incorrect predictions of our DNN-based ADRCs for DR severity classification.
Table 6.
Statistical analysis of the probability scores for correct and incorrect predictions of our DNN-based ADRCs for DR severity classification.
|
TrainDB |
TestDB |
Grade | Correct Prediction | Incorrect Prediction |
|---|
| Count | Min | Max | Mean | Median | Std | Count | Min | Max | Mean | Median | Std |
| Kaggle EyePACS |
Kaggle EyePACS | 0 | 38,199 | 0.2978 | 1.0000 | 0.9422 | 0.9800 | 0.0946 | 1334 | 0.2889 | 1.0000 | 0.6361 | 0.6107 | 0.1624 |
| 1 | 596 | 0.3153 | 0.9840 | 0.6800 | 0.6824 | 0.1526 | 3166 | 0.2838 | 1.0000 | 0.8523 | 0.9258 | 0.1613 |
| 2 | 4211 | 0.2988 | 0.9988 | 0.7832 | 0.8195 | 0.1606 | 3650 | 0.2977 | 1.0000 | 0.7616 | 0.7893 | 0.1841 |
| 3 | 617 | 0.3458 | 0.9986 | 0.7663 | 0.7892 | 0.1599 | 597 | 0.3392 | 1.0000 | 0.7304 | 0.7376 | 0.1685 |
| 4 | 773 | 0.3598 | 1.0000 | 0.9118 | 0.9957 | 0.1476 | 433 | 0.3192 | 0.9990 | 0.7212 | 0.7353 | 0.1738 |
|
IDRiD-DR | 0 | 22 | 0.5599 | 0.9990 | 0.9024 | 0.9313 | 0.1270 | 12 | 0.3918 | 0.6568 | 0.4961 | 0.4773 | 0.0895 |
| 1 | 0 | - | - | - | - | - | 5 | 0.3893 | 0.9932 | 0.6285 | 0.5188 | 0.2603 |
| 2 | 6 | 0.5877 | 0.8786 | 0.6890 | 0.6447 | 0.1233 | 26 | 0.4320 | 0.9823 | 0.7082 | 0.7212 | 0.1448 |
| 3 | 16 | 0.4993 | 0.9658 | 0.7695 | 0.8338 | 0.1617 | 3 | 0.5036 | 0.9936 | 0.7712 | 0.8163 | 0.2481 |
| 4 | 7 | 0.7129 | 0.9714 | 0.8496 | 0.8585 | 0.0810 | 6 | 0.5334 | 0.8767 | 0.6868 | 0.6334 | 0.1484 |
|
Messidor-2 | 0 | 954 | 0.3746 | 0.9997 | 0.9283 | 0.9749 | 0.1112 | 63 | 0.3703 | 0.9613 | 0.6420 | 0.6235 | 0.1516 |
| 1 | 38 | 0.3634 | 0.7954 | 0.5915 | 0.6150 | 0.1207 | 232 | 0.3661 | 0.9992 | 0.8076 | 0.8644 | 0.1775 |
| 2 | 225 | 0.3180 | 0.9857 | 0.7269 | 0.7619 | 0.1581 | 122 | 0.3216 | 0.9979 | 0.7043 | 0.6930 | 0.1786 |
| 3 | 63 | 0.5104 | 0.9971 | 0.8378 | 0.8931 | 0.1475 | 12 | 0.4974 | 0.9490 | 0.6705 | 0.7025 | 0.1558 |
| 4 | 22 | 0.4267 | 0.9999 | 0.8174 | 0.9691 | 0.2231 | 13 | 0.4046 | 0.9738 | 0.6919 | 0.6238 | 0.2094 |
|
IDRiD-DR |
Kaggle EyePACS | 0 | 36,222 | 0.2848 | 1.0000 | 0.9687 | 0.9988 | 0.0853 | 3311 | 0.2825 | 1.0000 | 0.7555 | 0.7810 | 0.1939 |
| 1 | 34 | 0.3771 | 0.9556 | 0.6723 | 0.6863 | 0.1775 | 3728 | 0.2962 | 1.0000 | 0.9564 | 0.9984 | 0.1070 |
| 2 | 631 | 0.3211 | 0.9993 | 0.7145 | 0.7135 | 0.1764 | 7230 | 0.2881 | 1.0000 | 0.9101 | 0.9891 | 0.1496 |
| 3 | 134 | 0.3480 | 0.9971 | 0.7244 | 0.7242 | 0.1766 | 1080 | 0.3007 | 1.0000 | 0.8238 | 0.8938 | 0.1846 |
| 4 | 708 | 0.2761 | 1.0000 | 0.8959 | 0.9723 | 0.1474 | 498 | 0.2869 | 1.0000 | 0.8021 | 0.8690 | 0.1927 |
|
IDRiD-DR | 0 | 28 | 0.4664 | 1.0000 | 0.9627 | 0.9962 | 0.1011 | 6 | 0.5833 | 0.9436 | 0.7346 | 0.6735 | 0.1662 |
| 1 | 1 | 0.6733 | 0.6733 | 0.6733 | 0.6733 | 0.0000 | 4 | 0.7935 | 0.9984 | 0.9250 | 0.9540 | 0.0948 |
| 2 | 14 | 0.4390 | 1.0000 | 0.8432 | 0.9330 | 0.1813 | 18 | 0.5406 | 0.9997 | 0.8794 | 0.9420 | 0.1423 |
| 3 | 13 | 0.5118 | 0.9866 | 0.8424 | 0.8855 | 0.1332 | 6 | 0.5114 | 0.9965 | 0.7682 | 0.8096 | 0.2134 |
| 4 | 8 | 0.6453 | 0.9993 | 0.8231 | 0.8198 | 0.1417 | 5 | 0.4572 | 0.8498 | 0.6560 | 0.6431 | 0.1709 |
|
Messidor-2 | 0 | 819 | 0.3463 | 1.0000 | 0.9308 | 0.9931 | 0.1291 | 198 | 0.3577 | 0.9995 | 0.8039 | 0.8493 | 0.1718 |
| 1 | 2 | 0.3396 | 0.4064 | 0.3730 | 0.3730 | 0.0472 | 268 | 0.2867 | 1.0000 | 0.8680 | 0.9346 | 0.1602 |
| 2 | 153 | 0.3798 | 0.9992 | 0.8226 | 0.8694 | 0.1586 | 194 | 0.3337 | 0.9999 | 0.8437 | 0.9468 | 0.1847 |
| 3 | 40 | 0.4838 | 0.9991 | 0.8317 | 0.9486 | 0.1870 | 35 | 0.4035 | 0.9994 | 0.7386 | 0.7425 | 0.1873 |
| 4 | 2 | 0.4735 | 0.7559 | 0.6147 | 0.6147 | 0.1997 | 33 | 0.4775 | 0.9967 | 0.7858 | 0.8327 | 0.1733 |
Table 7.
Mean Intersection-over-Union (mIoU) scores of our segmentation models. In Round 1, the models were trained on the training set and evaluated on the test set. In Round 2, the models were retrained on the combined training and test sets. In both rounds, the same validation set was used. Here, MA: microaneurysms, EX: hard exudates, HE: hemorrhage, SE: soft exudates, OD: optic disc, MC: macula.
Table 7.
Mean Intersection-over-Union (mIoU) scores of our segmentation models. In Round 1, the models were trained on the training set and evaluated on the test set. In Round 2, the models were retrained on the combined training and test sets. In both rounds, the same validation set was used. Here, MA: microaneurysms, EX: hard exudates, HE: hemorrhage, SE: soft exudates, OD: optic disc, MC: macula.
| Segmentation Category | Round 1 | Round 2 |
|---|
| Val. Set | Test Set | Val. Set |
| MA | 0.6042 | 0.5800 | 0.5996 |
| EX | 0.7904 | 0.7608 | 0.8240 |
| HE | 0.7169 | 0.6942 | 0.7344 |
| SE | 0.7513 | 0.6411 | 0.7659 |
| OD | 0.9162 | 0.9229 | 0.9382 |
| MC | 0.8159 | 0.8237 | 0.8229 |
Table 8.
Manual analysis of the highlighted regions in Grad-CAM and IG heatmaps for our ADRC trained using the Kaggle EyePACS dataset. The analysis was performed on the IDRiD-DR test set, showing the number of images for which the ADRC paid attention to various DR lesions across the dataset. Here, # Images: total images per DR grade, OD: optic disc, MC: macula, HE: hemorrhage, EX: exudate, MA: microaneurysms, NV: neovascularization, OT: other areas of the fundus image.
Table 8.
Manual analysis of the highlighted regions in Grad-CAM and IG heatmaps for our ADRC trained using the Kaggle EyePACS dataset. The analysis was performed on the IDRiD-DR test set, showing the number of images for which the ADRC paid attention to various DR lesions across the dataset. Here, # Images: total images per DR grade, OD: optic disc, MC: macula, HE: hemorrhage, EX: exudate, MA: microaneurysms, NV: neovascularization, OT: other areas of the fundus image.
| Method | DR | # | Correct Prediction | Incorrect Prediction |
|---|
| Grade | Images | Total | MA | EX | HE | NV | MC | OD | OT | Total | MA | EX | HE | NV | MC | OD | OT |
| Grad-CAM | 0 | 34 | 22 | 0 | 0 | 0 | 0 | 19 | 11 | 22 | 12 | 0 | 0 | 0 | 0 | 8 | 2 | 12 |
| 1 | 5 | 0 | - | - | - | - | - | - | - | 5 | 4 | 0 | 0 | 0 | 0 | 0 | 1 |
| 2 | 32 | 6 | 5 | 3 | 0 | 0 | 0 | 0 | 1 | 26 | 18 | 18 | 9 | 0 | 0 | 1 | 10 |
| 3 | 19 | 16 | 16 | 14 | 10 | 0 | 1 | 6 | 11 | 3 | 2 | 1 | 1 | 0 | 0 | 0 | 1 |
| 4 | 13 | 7 | 5 | 6 | 6 | 4 | 0 | 0 | 1 | 6 | 3 | 3 | 6 | 4 | 0 | 1 | 3 |
| IG | 0 | 34 | 22 | 0 | 0 | 0 | 0 | 9 | 13 | 20 | 12 | 0 | 0 | 0 | 0 | 7 | 1 | 12 |
| 1 | 5 | 0 | - | - | - | - | - | - | - | 5 | 3 | 0 | 0 | 0 | 2 | 1 | 5 |
| 2 | 32 | 6 | 5 | 4 | 0 | 0 | 5 | 4 | 6 | 26 | 25 | 19 | 9 | 0 | 13 | 22 | 24 |
| 3 | 19 | 16 | 16 | 14 | 12 | 0 | 9 | 16 | 16 | 3 | 3 | 1 | 1 | 0 | 2 | 2 | 3 |
| 4 | 13 | 7 | 6 | 5 | 7 | 2 | 5 | 7 | 6 | 6 | 6 | 4 | 5 | 1 | 4 | 6 | 6 |
Table 9.
Automatic analysis of the ADRC highlighted areas in Grad-CAM and IG heatmaps of the IDRiD-DR test set using automatic segmentation models. It displays the number of IDRiD-DR images where the ADRC focused on various DR lesions and landmarks, as identified by masks generated using six trained segmentation models. Here, # Images: total images per DR grade, MA: microaneurysm, SE: soft exudate, EX: hard exudate, HE: hemorrhage, MC: macula, and OD: optic disc.
Table 9.
Automatic analysis of the ADRC highlighted areas in Grad-CAM and IG heatmaps of the IDRiD-DR test set using automatic segmentation models. It displays the number of IDRiD-DR images where the ADRC focused on various DR lesions and landmarks, as identified by masks generated using six trained segmentation models. Here, # Images: total images per DR grade, MA: microaneurysm, SE: soft exudate, EX: hard exudate, HE: hemorrhage, MC: macula, and OD: optic disc.
| Method | Class ID | # Images | Correct Prediction | Incorrect Prediction |
|---|
| Total | MA | SE | EX | HE | MC | OD | Total | MA | SE | EX | HE | MC | OD |
| Grad-CAM | 0 | 34 | 22 | 8 | 11 | 20 | 16 | 21 | 16 | 12 | 7 | 2 | 9 | 7 | 11 | 5 |
| 1 | 5 | 0 | - | - | - | - | - | - | 5 | 3 | 0 | 4 | 4 | 2 | 5 |
| 2 | 32 | 6 | 4 | 3 | 6 | 6 | 5 | 6 | 26 | 18 | 14 | 25 | 21 | 14 | 26 |
| 3 | 19 | 16 | 13 | 8 | 16 | 16 | 2 | 15 | 3 | 2 | 1 | 2 | 1 | 2 | 3 |
| 4 | 13 | 7 | 5 | 5 | 6 | 7 | 3 | 7 | 6 | 5 | 4 | 6 | 6 | 1 | 4 |
| IG | 0 | 34 | 22 | 14 | 12 | 17 | 18 | 22 | 22 | 12 | 11 | 6 | 8 | 11 | 11 | 12 |
| 1 | 5 | 0 | - | - | - | - | - | - | 5 | 4 | 0 | 4 | 5 | 5 | 5 |
| 2 | 32 | 6 | 5 | 2 | 6 | 5 | 6 | 6 | 26 | 23 | 17 | 24 | 25 | 22 | 26 |
| 3 | 19 | 16 | 16 | 12 | 16 | 16 | 12 | 16 | 3 | 3 | 0 | 2 | 2 | 2 | 3 |
| 4 | 13 | 7 | 7 | 5 | 6 | 7 | 7 | 7 | 6 | 5 | 6 | 6 | 6 | 3 | 6 |
Table 10.
Automatic analysis of the highlighted areas in Grad-CAM and IG heatmaps of the Retinal-Lesions (B) dataset. It shows the number of Retinal-Lesions (B) images where the ADRC focused on various DR lesions and landmarks located by masks generated using six trained segmentation models. Here, # Images: total images per DR grade, MA: microaneurysm, SE: soft exudate, EX: hard exudate, HE: hemorrhage, MC: macula, and OD: optic disc.
Table 10.
Automatic analysis of the highlighted areas in Grad-CAM and IG heatmaps of the Retinal-Lesions (B) dataset. It shows the number of Retinal-Lesions (B) images where the ADRC focused on various DR lesions and landmarks located by masks generated using six trained segmentation models. Here, # Images: total images per DR grade, MA: microaneurysm, SE: soft exudate, EX: hard exudate, HE: hemorrhage, MC: macula, and OD: optic disc.
| Method | Class ID | # Images | Correct Prediction | Incorrect Prediction |
|---|
| Total | MA | SE | EX | HE | MC | OD | Total | MA | SE | EX | HE | MC | OD |
| Grad-CAM | 2 | 415 | 238 | 190 | 157 | 206 | 189 | 111 | 229 | 177 | 132 | 117 | 147 | 117 | 83 | 168 |
| 3 | 136 | 74 | 72 | 56 | 71 | 71 | 16 | 30 | 62 | 53 | 44 | 56 | 53 | 21 | 38 |
| 4 | 65 | 21 | 17 | 17 | 18 | 20 | 10 | 14 | 44 | 41 | 36 | 42 | 41 | 23 | 20 |
| IG | 2 | 415 | 238 | 224 | 200 | 228 | 216 | 230 | 234 | 177 | 144 | 135 | 156 | 137 | 168 | 177 |
| 3 | 136 | 74 | 74 | 64 | 72 | 74 | 69 | 73 | 62 | 59 | 50 | 59 | 58 | 60 | 62 |
| 4 | 65 | 21 | 19 | 18 | 20 | 21 | 16 | 21 | 44 | 41 | 36 | 44 | 42 | 40 | 44 |
Table 11.
Semi-automatic analysis of the highlighted areas in Grad-CAM and IG heatmaps of the Retinal-Lesions dataset. It shows the number of images of the Retinal-Lesions dataset for which our ADRC (trained on the Kaggle EyePACS dataset) paid attention to various DR lesions and landmarks located by manually prepared ground truth binary masks. Here, # Images: total images per DR grade, MA: microaneurysm, SE: soft exudates, EX: hard exudate, rHE: retinal hemorrhage, pHE: preretinal hemorrhage, vHE: vitreous hemorrhage, FP: fibrous proliferation, NV: neovascularization, Subset: subset of Retinal-Lesion dataset.
Table 11.
Semi-automatic analysis of the highlighted areas in Grad-CAM and IG heatmaps of the Retinal-Lesions dataset. It shows the number of images of the Retinal-Lesions dataset for which our ADRC (trained on the Kaggle EyePACS dataset) paid attention to various DR lesions and landmarks located by manually prepared ground truth binary masks. Here, # Images: total images per DR grade, MA: microaneurysm, SE: soft exudates, EX: hard exudate, rHE: retinal hemorrhage, pHE: preretinal hemorrhage, vHE: vitreous hemorrhage, FP: fibrous proliferation, NV: neovascularization, Subset: subset of Retinal-Lesion dataset.
| Subset | Method | DR | # | Correct Prediction | Incorrect Prediction |
|---|
| Grade | Images | Total | MA | SE | EX | rHE | vHE | pHE | FP | NV | Total | MA | SE | EX | rHE | vHE | pHE | FP | NV |
| (A) | Grad-CAM | 0 | 67 | 57 | 9 | 0 | 4 | 3 | 0 | 0 | 1 | 0 | 10 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| 1 | 184 | 33 | 26 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 151 | 75 | 2 | 6 | 13 | 0 | 0 | 0 | 0 |
| 2 | 561 | 320 | 254 | 80 | 165 | 235 | 0 | 0 | 0 | 2 | 241 | 140 | 46 | 65 | 112 | 0 | 0 | 2 | 0 |
| 3 | 120 | 94 | 80 | 44 | 70 | 92 | 0 | 0 | 0 | 0 | 26 | 23 | 8 | 15 | 20 | 0 | 0 | 0 | 0 |
| 4 | 45 | 29 | 18 | 6 | 15 | 24 | 8 | 6 | 10 | 16 | 16 | 13 | 5 | 10 | 15 | 0 | 0 | 0 | 2 |
| IG | 0 | 67 | 57 | 24 | 3 | 9 | 5 | 0 | 0 | 1 | 0 | 10 | 8 | 2 | 3 | 3 | 0 | 0 | 0 | 0 |
| 1 | 184 | 33 | 32 | 0 | 3 | 4 | 0 | 0 | 0 | 0 | 151 | 106 | 6 | 28 | 17 | 0 | 0 | 0 | 0 |
| 2 | 516 | 320 | 288 | 91 | 197 | 251 | 0 | 0 | 0 | 2 | 241 | 166 | 52 | 79 | 118 | 0 | 0 | 2 | 0 |
| 3 | 120 | 94 | 87 | 52 | 76 | 92 | 0 | 0 | 0 | 0 | 26 | 24 | 10 | 16 | 22 | 0 | 0 | 0 | 0 |
| 4 | 45 | 29 | 23 | 8 | 20 | 26 | 8 | 6 | 12 | 16 | 16 | 16 | 6 | 11 | 15 | 0 | 0 | 0 | 2 |
| (B) | Grad-CAM | 2 | 415 | 238 | 194 | 64 | 127 | 201 | 0 | 0 | 0 | 0 | 177 | 106 | 40 | 46 | 97 | 0 | 0 | 0 | 1 |
| 3 | 136 | 74 | 57 | 45 | 55 | 72 | 0 | 0 | 0 | 0 | 62 | 53 | 24 | 29 | 53 | 0 | 0 | 0 | 1 |
| 4 | 65 | 21 | 12 | 5 | 8 | 16 | 2 | 4 | 9 | 13 | 44 | 31 | 14 | 32 | 39 | 2 | 1 | 2 | 11 |
| IG | 2 | 415 | 238 | 222 | 81 | 150 | 215 | 0 | 0 | 1 | 0 | 177 | 131 | 50 | 68 | 100 | 0 | 0 | 1 | 1 |
| 3 | 136 | 74 | 69 | 48 | 59 | 73 | 0 | 0 | 0 | 1 | 62 | 59 | 26 | 33 | 54 | 0 | 0 | 0 | 1 |
| 4 | 65 | 21 | 15 | 6 | 9 | 18 | 2 | 4 | 8 | 14 | 44 | 36 | 21 | 33 | 41 | 2 | 1 | 2 | 13 |
Table 12.
Kappa scores for lesion detection agreement between our manual and automatic analyses of the highlighted areas in Grad-CAM and IG heatmaps across DR grades of the IDRiD-DR test set. Here, positive values indicate better-than-chance agreement, negative values indicate disagreement, zeros represent no agreement beyond chance, dashes (“– ”) denote missing cases, and “N/A” indicates that the kappa score is undefined due to no variability in the labels for that category.
Table 12.
Kappa scores for lesion detection agreement between our manual and automatic analyses of the highlighted areas in Grad-CAM and IG heatmaps across DR grades of the IDRiD-DR test set. Here, positive values indicate better-than-chance agreement, negative values indicate disagreement, zeros represent no agreement beyond chance, dashes (“– ”) denote missing cases, and “N/A” indicates that the kappa score is undefined due to no variability in the labels for that category.
| Method | DR Grade | Correct Prediction | Incorrect Prediction |
|---|
| MA | EX | HE | OD | MC | MA | EX | HE | OD | MC |
| Grad-CAM | 0 | 0.0000 | 0.0000 | 0.0000 | 0.3636 | 0.4634 | 0.0000 | 0.0000 | 0.0000 | 0.0625 | 0.3077 |
| 1 | – | – | – | – | – | −0.3636 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| 2 | 0.5714 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | −0.2639 | 0.0000 | 0.2239 | 0.0000 | 0.0000 |
| 3 | 0.0000 | 0.0000 | 0.0000 | 0.0769 | −0.0909 | −0.5000 | 0.0000 | 1.000 | 0.0000 | 0.0000 |
| 4 | 0.3000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | −0.3333 | 0.0000 | “N/A” | 0.1818 | 0.0000 |
| IG | 0 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.258 |
| 1 | – | – | – | – | – | 0.5455 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| 2 | 1.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.4694 | 0.1959 | 0.0415 | 0.0000 | 0.0000 |
| 3 | “N/A” | 0.0000 | 0.0000 | “N/A” | 0.3333 | “N/A” | 0.4000 | 0.4000 | 0.0000 | −0.5000 |
| 4 | 0.0000 | 0.0000 | “N/A” | “N/A” | 0.0000 | 0.0000 | 0.0000 | 0.0000 | “N/A” | 0.6667 |