Obesity/Overweight as a Meaningful Modifier of Associations Between Gene Polymorphisms Affecting the Sex Hormone-Binding Globulin Content and Uterine Myoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. SNP Selection/Detection
2.3. Statistical/Bioinformatics Genetic Data Analysis
3. Results
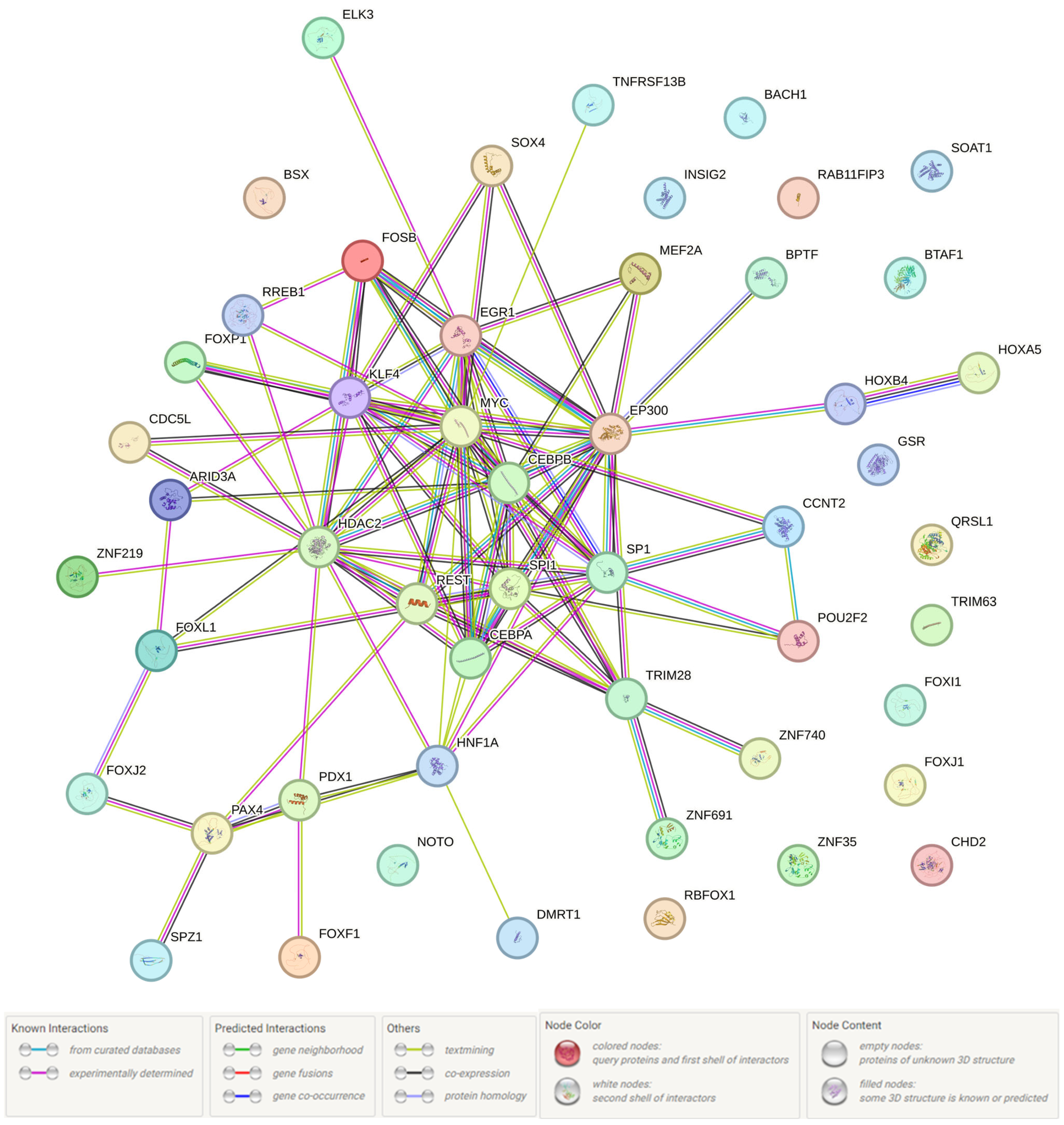
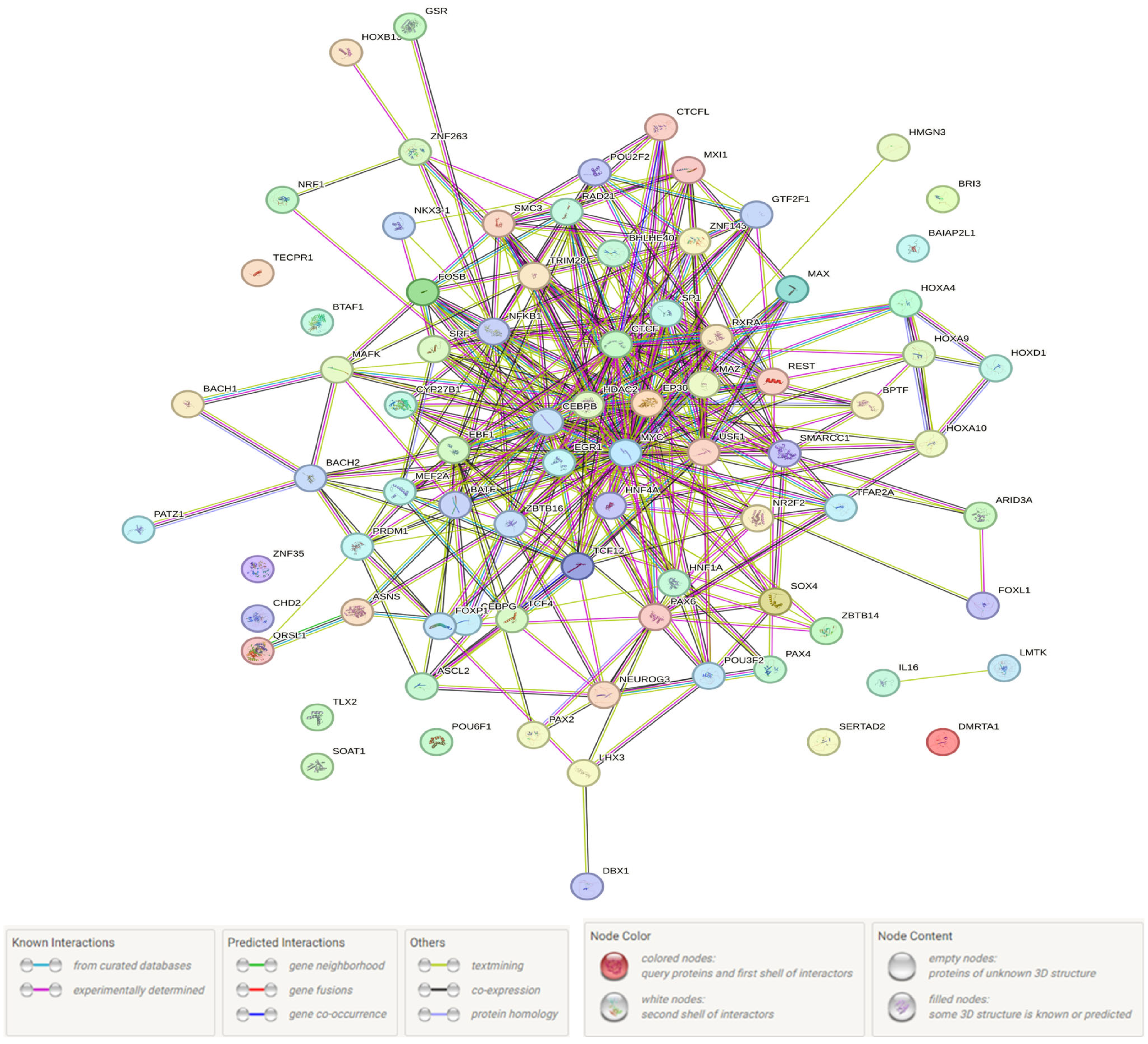
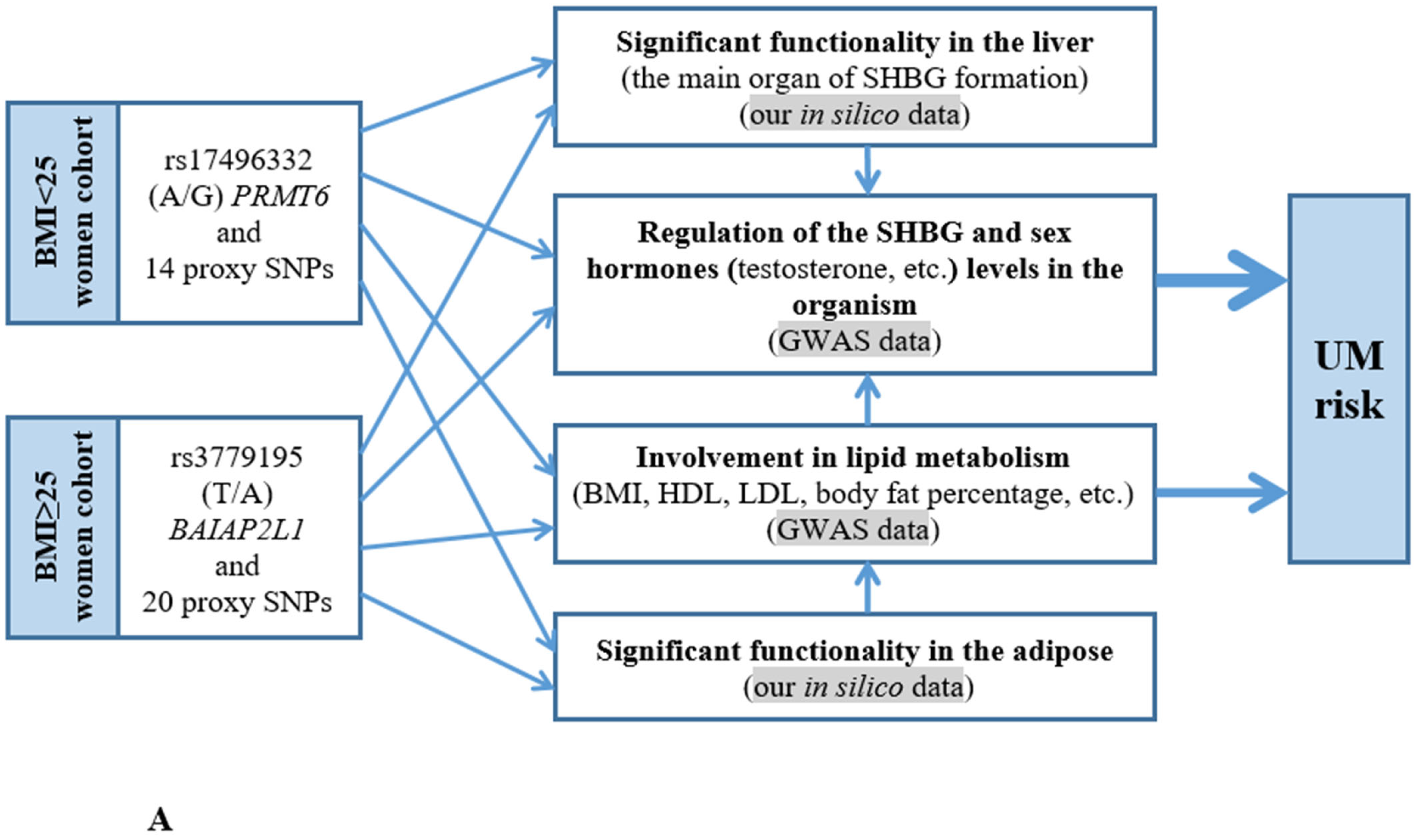
3.1. Probable Functionality of the UM-Associated Loci (In Silico Data)
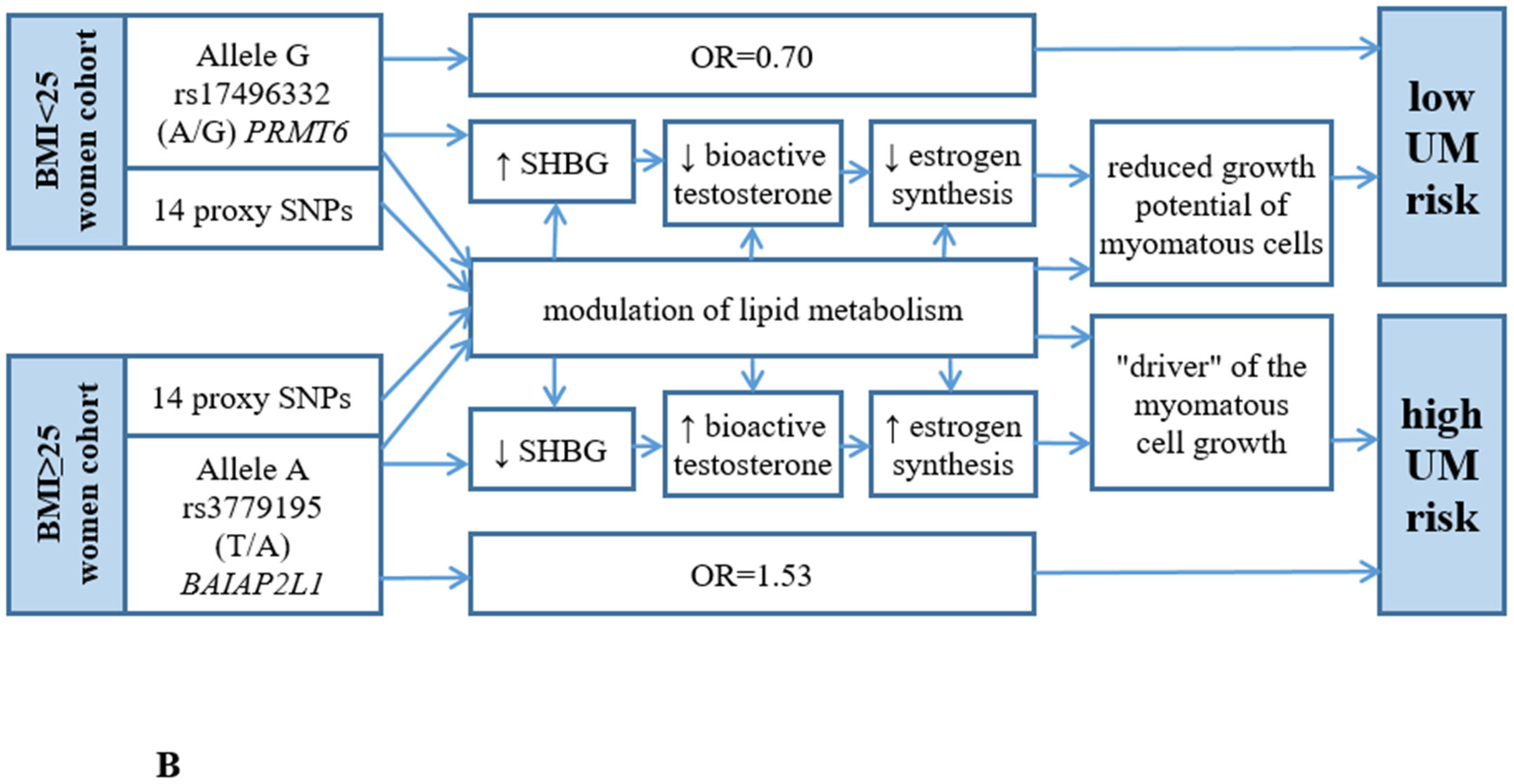
3.2. The Presumed UM-Associated Functionality in the BMI < 25 Women Group rs17496332 (A/G) PRMT6
3.3. The Supposed UM-Associated Functionality in the BMI ≥ 25 Women Group rs3779195 (T/A) BAIAP2L1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| UM | Uterine fibroids |
| SHBG | Sex hormone-binding globulin |
| SHBGlevel | Sex hormone-binding globulin level |
| SNP | Single-nucleotide polymorphism |
| GWAS | Genome-wide association studies |
| BMI | Body mass index |
| DNA | Deoxyribonucleic acid |
| LD | Linkage disequilibrium |
| TFs | Transcription factors |
References
- Bulun, S.E. Uterine fibroids. N. Engl. J. Med. 2013, 369, 1344–1355. [Google Scholar] [CrossRef]
- Yang, Q.; Ciebiera, M.; Bariani, M.V.; Ali, M.; Elkafas, H.; Boyer, T.G.; Al-Hendy, A. Comprehensive Review of Uterine Fibroids: Developmental Origin, Pathogenesis, and Treatment. Endocr. Rev. 2022, 43, 678–719. [Google Scholar] [CrossRef]
- Koltsova, A.S.; Efimova, O.A.; Pendina, A.A. A View on Uterine Leiomyoma Genesis through the Prism of Genetic, Epigenetic and Cellular Heterogeneity. Int. J. Mol. Sci. 2023, 24, 5752. [Google Scholar] [CrossRef]
- Donnez, J.; Dolmans, M.M. Uterine fibroid management: From the present to the future. Hum. Reprod. Update 2016, 22, 665–686. [Google Scholar] [CrossRef] [PubMed]
- Wise, L.A.; Laughlin-Tommaso, S.K. Epidemiology of uterine fibroids: From menarche to menopause. Clin. Obstet. Gynecol. 2016, 59, 2–24. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.A.; Nowak, R.A. Uterine Fibroids: Hiding in Plain Sight. Physiology 2022, 37, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Cianci, S.; Gulino, F.A.; Palmara, V.; La Verde, M.; Ronsini, C.; Romeo, P.; Occhipinti, S.; Incognito, G.G.; Capozzi, V.A.; Restaino, S.; et al. Exploring Surgical Strategies for Uterine Fibroid Treatment: A Comprehensive Review of Literature on Open and Minimally Invasive Approaches. Medicina 2023, 60, 64. [Google Scholar] [CrossRef]
- Wang, T.; Tang, H.; Xie, Z.; Deng, S. Robotic-assisted vs. laparoscopic and abdominal myomectomy for treatment of uterine fibroids: A meta-analysis. Minim. Invasive Ther. Allied Technol. 2018, 27, 249–264. [Google Scholar] [CrossRef]
- Lou, Z.; Huang, Y.; Li, S.; Luo, Z.; Li, C.; Chu, K.; Zhang, T.; Song, P.; Zhou, J. Global, regional, and national time trends in incidence, prevalence, years lived with disability for uterine fibroids, 1990-2019: An age-period-cohort analysis for the global burden of disease 2019 study. BMC Public Health 2023, 23, 916. [Google Scholar] [CrossRef]
- Luoto, R.; Kaprio, J.; Rutanen, E.M.; Taipale, P.; Perola, M.; Koskenvuo, M. Heritability and risk factors of uterine fibroids--the Finnish Twin Cohort study. Maturitas 2000, 37, 15–26. [Google Scholar] [CrossRef]
- Wise, L.A.; Ruiz-Narvaez, E.A.; Palmer, J.R.; Cozier, Y.C.; Tandon, A.; Patterson, N.; Radin, R.G.; Rosenberg, L.; Reich, D. African ancestry and genetic risk for uterine leiomyomata. Am. J. Epidemiol. 2012, 176, 1159–1168. [Google Scholar] [CrossRef]
- Edwards, T.L.; Hartmann, K.E.; Velez Edwards, D.R. Variants in BET1L and TNRC6B associate with increasing fibroid volume and fibroid type among European Americans. Hum. Genet. 2013, 132, 1361–1369. [Google Scholar] [CrossRef]
- Zhang, K.; Wiener, H.; Aissani, B. Admixture mapping of genetic variants for uterine fibroids. J. Hum. Genet. 2015, 60, 533–538. [Google Scholar] [CrossRef][Green Version]
- Liu, B.; Wang, T.; Jiang, J.; Li, M.; Ma, W.; Wu, H.; Zhou, Q. Association of BET1L and TNRC6B with uterine leiomyoma risk and its relevant clinical features in Han Chinese population. Sci. Rep. 2018, 8, 7401. [Google Scholar] [CrossRef]
- Upadhyay, S.; Dubey, P.K. Gene variants polymorphisms and uterine leiomyoma: An updated review. Front. Genet. 2024, 15, 1330807. [Google Scholar] [CrossRef] [PubMed]
- Snieder, H.; MacGregor, A.J.; Spector, T.D. Genes control the cessation of a woman’s reproductive life: A twin study of hysterectomy and age at menopause. J. Clin. Endocrinol. Metab. 1998, 83, 1875–1880. [Google Scholar] [CrossRef] [PubMed]
- Vikhlyaeva, E.M.; Khodzhaeva, Z.S.; Fantschenko, N.D. Familial predisposition to uterine leiomyomas. Int. J. Gynaecol. Obstet. 1995, 51, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Tanikawa, C.; Hirasawa, A.; Chiyoda, T.; Yamagami, W.; Kataoka, F.; Susumu, N.; Terao, C.; Kamatani, Y.; Takahashi, A.; et al. Identification of a novel uterine leiomyoma GWAS locus in a Japanese population. Sci. Rep. 2020, 10, 1197. [Google Scholar] [CrossRef]
- Hellwege, J.N.; Jeff, J.M.; Wise, L.A.; Gallagher, C.S.; Wellons, M.; Hartmann, K.E.; Jones, S.F.; Torstenson, E.S.; Dickinson, S.; Ruiz-Narváez, E.A.; et al. A multi-stage genome-wide association study of uterine fibroids in African Americans. Hum. Genet. 2017, 136, 1363–1373. [Google Scholar] [CrossRef]
- Välimäki, N.; Kuisma, H.; Pasanen, A.; Heikinheimo, O.; Sjöberg, J.; Bützow, R.; Sarvilinna, N.; Heinonen, H.R.; Tolvanen, J.; Bramante, S.; et al. Genetic predisposition to uterine leiomyoma is determined by loci for genitourinary development and genome stability. eLife 2018, 7, e37110. [Google Scholar] [CrossRef]
- Rafnar, T.; Gunnarsson, B.; Stefansson, O.A.; Sulem, P.; Ingason, A.; Frigge, M.L.; Stefansdottir, L.; Sigurdsson, J.K.; Tragante, V.; Steinthorsdottir, V.; et al. Variants associating with uterine leiomyoma highlight genetic background shared by various cancers and hormone-related traits. Nat. Commun. 2018, 9, 3636. [Google Scholar] [CrossRef]
- Gallagher, C.S.; Mäkinen, N.; Harris, H.R.; Rahmioglu, N.; Uimari, O.; Cook, J.P.; Shigesi, N.; Ferreira, T.; Velez-Edwards, D.R.; Edwards, T.L.; et al. Genome-wide association and epidemiological analyses reveal common genetic origins between uterine leiomyomata and endometriosis. Nat. Commun. 2019, 10, 4857. [Google Scholar] [CrossRef]
- Masuda, T.; Low, S.K.; Akiyama, M.; Hirata, M.; Ueda, Y.; Matsuda, K.; Kimura, T.; Murakami, Y.; Kubo, M.; Kamatani, Y.; et al. GWAS of five gynecologic diseases and cross-trait analysis in Japanese. Eur. J. Hum. Genet. 2020, 28, 95–107. [Google Scholar] [CrossRef]
- Cha, P.C.; Takahashi, A.; Hosono, N.; Low, S.K.; Kamatani, N.; Kubo, M.; Nakamura, Y. A genome-wide association study identifies three loci associated with susceptibility to uterine fibroids. Nat. Genet. 2011, 43, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Baranov, V.S.; Osinovskaya, N.S.; Yarmolinskaya, M.I. Pathogenomics of Uterine Fibroids Development. Int. J. Mol. Sci. 2019, 20, 6151. [Google Scholar] [CrossRef] [PubMed]
- Ponomareva, L.; Kobzeva, K.; Bushueva, O. GWAS-Significant Loci and Uterine Fibroids Risk: Analysis of Associations, Gene-Gene and Gene-Environmental Interactions. Front. Biosci. (Sch. Ed.) 2024, 16, 24. [Google Scholar] [CrossRef] [PubMed]
- Edwards, T.L.; Giri, A.; Hellwege, J.N.; Hartmann, K.E.; Stewart, E.A.; Jeff, J.M.; Bray, M.J.; Pendergrass, S.A.; Torstenson, E.S.; Keaton, J.M.; et al. A trans-ethnic genome-wide association study of uterine fibroids. Front. Genet. 2019, 10, 511. [Google Scholar] [CrossRef]
- Lee, S.C.; Chou, Y.H.; Tantoh, D.M.; Hsu, S.Y.; Nfor, O.N.; Tyan, Y.S.; Liaw, Y.P. Risk of uterine leiomyoma based on BET1L rs2280543 single nucleotide polymorphism and vegetarian diet. BMC Women’s Health 2022, 22, 139. [Google Scholar] [CrossRef]
- Qin, H.; Lin, Z.; Vásquez, E.; Luan, X.; Guo, F. Association between obesity and the risk of uterine fibroids: A systematic review and meta-analysis. J. Epidemiol. Community Health 2021, 75, 197–204. [Google Scholar] [CrossRef]
- Venkatesh, S.S.; Ferreira, T.; Benonisdottir, S.; Rahmioglu, N.; Becker, C.M.; Granne, I.; Zondervan, K.T.; Holmes, M.V.; Lindgren, C.M.; Wittemans, L.B.L. Obesity and risk of female reproductive conditions: A Mendelian randomisation study. PLoS Med. 2022, 19, e1003679. [Google Scholar] [CrossRef]
- Ali, M.; Bariani, M.V.; Vafaei, S.; Omran, M.M.; Yang, Q.; Madueke-Laveaux, O.S.; Al-Hendy, A. Prevention of Uterine Fibroids: Molecular mechanisms and potential clinical application. J. Endometr. Uterine Disord. 2023, 1, 100018. [Google Scholar] [CrossRef]
- Qu, Y.; Chen, L.; Guo, S.; Liu, Y.; Wu, H. Genetic liability to multiple factors and uterine leiomyoma risk: A Mendelian randomization study. Front. Endocrinol. 2023, 14, 1133260. [Google Scholar] [CrossRef]
- Harmon, Q.E.; Patchel, S.; Denslow, S.; Wegienka, G.; Baird, D.D. Body Mass Index and Uterine Fibroid Development: A Prospective Study. J. Clin. Endocrinol. Metab. 2024, 109, e2016–e2023. [Google Scholar] [CrossRef] [PubMed]
- Šišljagić, D.; Blažetić, S.; Heffer, M.; Vranješ Delać, M.; Muller, A. The Interplay of Uterine Health and Obesity: A Comprehensive Review. Biomedicines 2024, 12, 2801. [Google Scholar] [CrossRef]
- Chen, Y.W.; Hang, D.; Kværner, A.S.; Giovannucci, E.; Song, M. Associations between body shape across the life course and adulthood concentrations of sex hormones in men and pre- and postmenopausal women: A multicohort study. Br. J. Nutr. 2022, 127, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Szybiak-Skora, W.; Cyna, W.; Lacka, K. New Insights in the Diagnostic Potential of Sex Hormone-Binding Globulin (SHBG)-Clinical Approach. Biomedicines 2025, 13, 1207. [Google Scholar] [CrossRef] [PubMed]
- Alsudairi, H.N.; Alrasheed, A.T.; Dvornyk, V. Estrogens and uterine fibroids: An integrated view. Res. Results Biomed. 2021, 7, 156–163. [Google Scholar] [CrossRef]
- Ohlsson, C.; Wallaschofski, H.; Lunetta, K.L.; Stolk, L.; Perry, J.R.; Koster, A.; Petersen, A.K.; Eriksson, J.; Lehtimäki, T.; Huhtaniemi, I.T.; et al. Genetic determinants of serum testosterone concentrations in men. PLoS Genet. 2011, 7, e1002313. [Google Scholar] [CrossRef]
- Coviello, A.D.; Haring, R.; Wellons, M.; Vaidya, D.; Lehtimäki, T.; Keildson, S.; Lunetta, K.L.; He, C.; Fornage, M.; Lagou, V.; et al. A genome-wide association meta-analysis of circulating sex hormone-binding globulin reveals multiple Loci implicated in sex steroid hormone regulation. PLoS Genet. 2012, 8, e1002805. [Google Scholar] [CrossRef]
- Prescott, J.; Thompson, D.J.; Kraft, P.; Chanock, S.J.; Audley, T.; Brown, J.; Leyland, J.; Folkerd, E.; Doody, D.; Hankinson, S.E.; et al. Genome-wide association study of circulating estradiol, testosterone, and sex hormone-binding globulin in postmenopausal women. PLoS ONE 2012, 7, e37815. [Google Scholar] [CrossRef]
- Ruth, K.S.; Campbell, P.J.; Chew, S.; Lim, E.M.; Hadlow, N.; Stuckey, B.G.; Brown, S.J.; Feenstra, B.; Joseph, J.; Surdulescu, G.L.; et al. Genome-wide association study with 1000 genomes imputation identifies signals for nine sex hormone-related phenotypes. Eur. J. Hum. Genet. 2016, 24, 284–290. [Google Scholar] [CrossRef]
- Ruth, K.S.; Day, F.R.; Tyrrell, J.; Thompson, D.J.; Wood, A.R.; Mahajan, A.; Beaumont, R.N.; Wittemans, L.; Martin, S.; Busch, A.S.; et al. Using human genetics to understand the disease impacts of testosterone in men and women. Nat. Med. 2020, 26, 252–258. [Google Scholar] [CrossRef]
- Harrison, S.; Davies, N.M.; Howe, L.D.; Hughes, A. Testosterone and socioeconomic position: Mendelian randomization in 306,248 men and women in UK Biobank. Sci. Adv. 2021, 7, eabf8257. [Google Scholar] [CrossRef] [PubMed]
- Haas, C.B.; Hsu, L.; Lampe, J.W.; Wernli, K.J.; Lindström, S. Cross-ancestry Genome-wide Association Studies of Sex Hormone Concentrations in Pre- and Postmenopausal Women. Endocrinology 2022, 163, bqac020. [Google Scholar] [CrossRef]
- Churnosov, M.; Abramova, M.; Reshetnikov, E.; Lyashenko, I.V.; Efremova, O.; Churnosova, M.; Ponomarenko, I. Polymorphisms of hypertension susceptibility genes as a risk factors of preeclampsia in the Caucasian population of central Russia. Placenta 2022, 129, 51–61. [Google Scholar] [CrossRef]
- Sergeeva, K.N.; Sorokina, I.N. Assessment of the relationship between marriage and migration characteristics of the population of the Belgorod region in dynamics over 130 years. Res. Results Biomed. 2024, 10, 374–388. (In Russian) [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef]
- Novakov, V.; Novakova, O.; Churnosova, M.; Sorokina, I.; Aristova, I.; Polonikov, A.; Reshetnikov, E.; Churnosov, M. Intergenic Interactions of SBNO1, NFAT5 and GLT8D1 Determine the Susceptibility to Knee Osteoarthritis among Europeans of Russia. Life 2023, 13, 405. [Google Scholar] [CrossRef] [PubMed]
- Golovchenko, O.V. Molecular genetic determinants of pre-eclampsia. Res. Results Biomed. 2019, 5, 139–149. (In Russian) [Google Scholar] [CrossRef]
- Golovchenko, I.; Aizikovich, B.; Golovchenko, O.; Reshetnikov, E.; Churnosova, M.; Aristova, I.; Ponomarenko, I.; Churnosov, M. Sex Hormone Candidate Gene Polymorphisms Are Associated with Endometriosis. Int. J. Mol. Sci. 2022, 23, 13691. [Google Scholar] [CrossRef]
- Ivanova, T.; Churnosova, M.; Abramova, M.; Ponomarenko, I.; Reshetnikov, E.; Aristova, I.; Sorokina, I.; Churnosov, M. Risk Effects of rs1799945 Polymorphism of the HFE Gene and Intergenic Interactions of GWAS-Significant Loci for Arterial Hypertension in the Caucasian Population of Central Russia. Int. J. Mol. Sci. 2023, 24, 8309. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Ivanova, T.; Churnosova, M.; Abramova, M.; Plotnikov, D.; Ponomarenko, I.; Reshetnikov, E.; Aristova, I.; Sorokina, I.; Churnosov, M. Sex-Specific Features of the Correlation between GWAS-Noticeable Polymorphisms and Hypertension in Europeans of Russia. Int. J. Mol. Sci. 2023, 24, 7799. [Google Scholar] [CrossRef]
- Guo, Y.F.; Li, J.; Chen, Y.; Zhang, L.S.; Deng, H.W. A new permutation strategy of pathway-based approach for genome-wide association study. BMC Bioinform. 2009, 10, 429. [Google Scholar] [CrossRef] [PubMed]
- Che, R.; Jack, J.R.; Motsinger-Reif, A.A.; Brown, C.C. An adaptive permutation approach for genome-wide association study: Evaluation and recommendations for use. BioData Min. 2014, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Reshetnikova, Y.; Churnosova, M.; Stepanov, V.; Bocharova, A.; Serebrova, V.; Trifonova, E.; Ponomarenko, I.; Sorokina, I.; Efremova, O.; Orlova, V.; et al. Maternal Age at Menarche Gene Polymorphisms Are Associated with Offspring Birth Weight. Life 2023, 13, 1525. [Google Scholar] [CrossRef] [PubMed]
- Gauderman, W.; Morrison, J. QUANTO 1.1: A Computer Program for Power and Sample Size Calculations Genetic–Epidemiology Studies. 2006. Available online: http://hydra.usc.edu/gxe (accessed on 18 November 2024).
- Butkiewicz, M.; Bush, W.S. In Silico Functional Annotation of Genomic Variation. Curr. Protoc. Hum. Genet. 2016, 88, 6.15.1–6.15.17. [Google Scholar] [CrossRef]
- Fisch, K.M. Biological Interpretation of Complex Genomic Data. Methods Mol. Biol. 2019, 1908, 61–71. [Google Scholar] [CrossRef]
- Minyaylo, O.N. Allele distribution and haploblock structure of matrix metalloproteinase gene polymorphism in patients with H. pylori-negative gastric ulcer and duodenal ulcer. Res. Results Biomed. 2020, 6, 488–502. (In Russian) [Google Scholar] [CrossRef]
- Pavlova, N.; Demin, S.; Churnosov, M.; Reshetnikov, E.; Aristova, I.; Churnosova, M.; Ponomarenko, I. Matrix Metalloproteinase Gene Polymorphisms Are Associated with Breast Cancer in the Caucasian Women of Russia. Int. J. Mol. Sci. 2022, 23, 12638. [Google Scholar] [CrossRef]
- Reshetnikov, E.; Churnosova, M.; Reshetnikova, Y.; Stepanov, V.; Bocharova, A.; Serebrova, V.; Trifonova, E.; Ponomarenko, I.; Sorokina, I.; Efremova, O.; et al. Maternal Age at Menarche Genes Determines Fetal Growth Restriction Risk. Int. J. Mol. Sci. 2024, 25, 2647. [Google Scholar] [CrossRef]
- Ponomarenko, M.; Reshetnikov, E.; Churnosova, M.; Aristova, I.; Abramova, M.; Novakov, V.; Churnosov, V.; Polonikov, A.; Plotnikov, D.; Churnosov, M.; et al. Genetic Variants Linked with the Concentration of Sex Hormone-Binding Globulin Correlate with Uterine Fibroid Risk. Life 2025, 15, 1150. [Google Scholar] [CrossRef] [PubMed]
- Minyaylo, O.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. Functionally significant polymorphisms of the MMP-9 gene are associated with peptic ulcer disease in the Caucasian population of Central Russia. Sci. Rep. 2021, 11, 13515. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Hammond, G.L. Diverse roles for sex hormone-binding globulin in reproduction. Biol. Reprod. 2011, 85, 431–441. [Google Scholar] [CrossRef]
- Hammond, G.L. Plasma steroid-binding proteins: Primary gatekeepers of steroid hormone action. J. Endocrinol. 2016, 230, R13–R25. [Google Scholar] [CrossRef]
- Klarin, D.; Damrauer, S.M.; Cho, K.; Sun, Y.V.; Teslovich, T.M.; Honerlaw, J.; Gagnon, D.R.; DuVall, S.L.; Li, J.; Peloso, G.M.; et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018, 50, 1514–1523. [Google Scholar] [CrossRef]
- Graham, S.E.; Clarke, S.L.; Wu, K.H.; Kanoni, S.; Zajac, G.J.M.; Ramdas, S.; Surakka, I.; Ntalla, I.; Vedantam, S.; Winkler, T.W.; et al. The power of genetic diversity in genome-wide association studies of lipids. Nature 2021, 600, 675–679. [Google Scholar] [CrossRef]
- Hawkes, G.; Beaumont, R.N.; Tyrrell, J.; Power, G.M.; Wood, A.; Laakso, M.; Fernandes Silva, L.; Boehnke, M.; Yin, X.; Richardson, T.G.; et al. Genetic evidence that high BMI in childhood has a protective effect on intermediate diabetes traits, including measures of insulin sensitivity and secretion, after accounting for BMI in adulthood. Diabetologia 2023, 66, 1472–1480. [Google Scholar] [CrossRef]
- Schoeler, T.; Speed, D.; Porcu, E.; Pirastu, N.; Pingault, J.B.; Kutalik, Z. Participation bias in the UK Biobank distorts genetic associations and downstream analyses. Nat. Hum. Behav. 2023, 7, 1216–1227. [Google Scholar] [CrossRef]
- Shi, S.; Rubinacci, S.; Hu, S.; Moutsianas, L.; Stuckey, A.; Need, A.C.; Palamara, P.F.; Caulfield, M.; Marchini, J.; Myers, S. A Genomics England haplotype reference panel and imputation of UK Biobank. Nat. Genet. 2024, 56, 1800–1803. [Google Scholar] [CrossRef]
- Richardson, T.G.; Sanderson, E.; Palmer, T.M.; Ala-Korpela, M.; Ference, B.A.; Davey Smith, G.; Holmes, M.V. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable Mendelian randomisation analysis. PLoS Med. 2020, 17, e1003062. [Google Scholar] [CrossRef]
- Richardson, T.G.; Leyden, G.M.; Wang, Q.; Bell, J.A.; Elsworth, B.; Davey Smith, G.; Holmes, M.V. Characterising metabolomic signatures of lipid-modifying therapies through drug target mendelian randomisation. PLoS Biol. 2022, 20, e3001547. [Google Scholar] [CrossRef]
- Roshandel, D.; Lu, T.; Paterson, A.D.; Dash, S. Beyond apples and pears: Sex-specific genetics of body fat percentage. Front. Endocrinol. 2023, 14, 1274791. [Google Scholar] [CrossRef] [PubMed]
- Goldman, A.L.; Bhasin, S.; Wu, F.C.W.; Krishna, M.; Matsumoto, A.M.; Jasuja, R. A Reappraisal of Testosterone’s Binding in Circulation: Physiological and Clinical Implications. Endocr. Rev. 2017, 38, 302–324. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Yu, J.; Huang, Y.; Ma, J.; Xiang, J.; Wang, Y.; Li, L.; Zhang, Z.; Liao, H. Androgen Signaling in Uterine Diseases: New Insights and New Targets. Biomolecules 2022, 12, 1624. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.Y.; Gold, E.B.; Johnson, W.O.; Lee, J.S. Circulating Sex Hormones and Risk of Uterine Fibroids: Study of Women’s Health Across the Nation (SWAN). J. Clin. Endocrinol. Metab. 2016, 101, 123–130. [Google Scholar] [CrossRef]
- Wang, H.; Li, C.; Chen, L.; Zhang, M.; Ren, T.; Zhang, S. Causal relationship between female reproductive factors, sex hormones and uterine leiomyoma: A Mendelian randomization study. Reprod. Biomed. Online 2024, 48, 103584. [Google Scholar] [CrossRef]
- Sinnott-Armstrong, N.; Naqvi, S.; Rivas, M.; Pritchard, J.K. GWAS of three molecular traits highlights core genes and pathways alongside a highly polygenic background. eLife 2021, 10, e58615. [Google Scholar] [CrossRef]
- Machado-Lopez, A.; Simón, C.; Mas, A. Molecular and Cellular Insights into the Development of Uterine Fibroids. Int. J. Mol. Sci. 2021, 22, 8483. [Google Scholar] [CrossRef]
- Ploumaki, I.; Macri, V.I.; Segars, J.H.; Islam, M.S. Progesterone signaling in uterine fibroids: Molecular mechanisms and therapeutic opportunities. Life Sci. 2025, 362, 123345. [Google Scholar] [CrossRef]
- Ali, M.; Ciebiera, M.; Vafaei, S.; Alkhrait, S.; Chen, H.Y.; Chiang, Y.F.; Huang, K.C.; Feduniw, S.; Hsia, S.M.; Al-Hendy, A. Progesterone Signaling and Uterine Fibroid Pathogenesis; Molecular Mechanisms and Potential Therapeutics. Cells 2023, 12, 1117. [Google Scholar] [CrossRef]
- Szucio, W.; Bernaczyk, P.; Ponikwicka-Tyszko, D.; Milewska, G.; Pawelczyk, A.; Wołczyński, S.; Rahman, N.A. Progesterone signaling in uterine leiomyoma biology: Implications for potential targeted therapy. Adv. Med. Sci. 2024, 69, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Omar, M.; Laknaur, A.; Al-Hendy, A.; Yang, Q. Myometrial progesterone hyper-responsiveness associated with increased risk of human uterine fibroids. BMC Women’s Health 2019, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Sefton, E.C. The role of progesterone signaling in the pathogenesis of uterine leiomyoma. Mol. Cell. Endocrinol. 2012, 358, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Bulun, S.E.; Moravek, M.B.; Yin, P.; Ono, M.; Coon, J.S.; Dyson, M.T.; Navarro, A.; Marsh, E.E.; Zhao, H.; Maruyama, T.; et al. Uterine Leiomyoma Stem Cells: Linking Progesterone to Growth. Semin. Reprod. Med. 2015, 33, 357–365. [Google Scholar] [CrossRef]
- Kovács, K.A.; Lengyel, F.; Környei, J.L.; Vértes, Z.; Szabó, I.; Sümegi, B.; Vértes, M. Differential expression of Akt/protein kinase B, Bcl-2 and Bax proteins in human leiomyoma and myometrium. J. Steroid Biochem. Mol. Biol. 2003, 87, 233–240. [Google Scholar] [CrossRef]
- Mozzachio, K.; Moore, A.B.; Kissling, G.E.; Dixon, D. Immunoexpression of Steroid Hormone Receptors and Proliferation Markers in Uterine Leiomyoma and Normal Myometrial Tissues from the Miniature Pig, Sus scrofa. Toxicol. Pathol. 2016, 44, 450–457. [Google Scholar] [CrossRef]
- Khan, K.N.; Fujishita, A.; Koshiba, A.; Ogawa, K.; Mori, T.; Ogi, H.; Itoh, K.; Teramukai, S.; Kitawaki, J. Expression profiles of E/P receptors and fibrosis in GnRHa-treated and -untreated women with different uterine leiomyomas. PLoS ONE 2020, 15, e0242246. [Google Scholar] [CrossRef]
- Ali, M.; Al-Hendy, A. Selective progesterone receptor modulators for fertility preservation in women with symptomatic uterine fibroids. Biol. Reprod. 2017, 97, 337–352. [Google Scholar] [CrossRef]
- Pavlova, N.; Demin, S.; Churnosov, M.; Reshetnikov, E.; Aristova, I.; Churnosova, M.; Ponomarenko, I. The Modifying Effect of Obesity on the Association of Matrix Metalloproteinase Gene Polymorphisms with Breast Cancer Risk. Biomedicines 2022, 10, 2617. [Google Scholar] [CrossRef]
- Ponomarenko, I.; Pasenov, K.; Churnosova, M.; Sorokina, I.; Aristova, I.; Churnosov, V.; Ponomarenko, M.; Reshetnikova, Y.; Reshetnikov, E.; Churnosov, M. Obesity-Dependent Association of the rs10454142 PPP1R21 with Breast Cancer. Biomedicines 2024, 12, 818. [Google Scholar] [CrossRef]
- Pasenov, K.N. Features of associations of SHBG-related genes with breast cancer in women, depending on the presence of hereditary burden and mutations in the BRCA1/CHEK2 genes. Res. Results Biomed. 2024, 10, 69–88. (In Russian) [Google Scholar] [CrossRef]
- Novakov, V.; Novakova, O.; Churnosova, M.; Aristova, I.; Ponomarenko, M.; Reshetnikova, Y.; Churnosov, V.; Sorokina, I.; Ponomarenko, I.; Efremova, O.; et al. Polymorphism rs143384 GDF5 reduces the risk of knee osteoarthritis development in obese individuals and increases the disease risk in non-obese population. Arthroplasty 2024, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Abramova, M.; Churnosova, M.; Efremova, O.; Aristova, I.; Reshetnikov, E.; Polonikov, A.; Churnosov, M.; Ponomarenko, I. Effects of pre-pregnancy over-weight/obesity on the pattern of association of hypertension susceptibility genes with preeclampsia. Life 2022, 12, 2018. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, I.; Pasenov, K.; Churnosova, M.; Sorokina, I.; Aristova, I.; Churnosov, V.; Ponomarenko, M.; Reshetnikov, E.; Churnosov, M. Sex-Hormone-Binding Globulin Gene Polymorphisms and Breast Cancer Risk in Caucasian Women of Russia. Int. J. Mol. Sci. 2024, 25, 2182. [Google Scholar] [CrossRef]
- Ponomareva, T.A. Genetic variants of sex hormone-binding globulin and hormonal profile in patients with genital endometriosis. Res. Results Biomed. 2025, 11, 75–90. (In Russian) [Google Scholar] [CrossRef]
- Fantus, R.J.; Na, R.; Wei, J.; Shi, Z.; Resurreccion, W.K.; Halpern, J.A.; Franco, O.; Hayward, S.W.; Isaacs, W.B.; Zheng, S.L.; et al. Genetic Susceptibility for Low Testosterone in Men and Its Implications in Biology and Screening: Data from the UK Biobank. Eur. Urol. Open Sci. 2021, 29, 36–46. [Google Scholar] [CrossRef]
| Parameters | BMI ≥ 25 | BMI < 25 | ||||
|---|---|---|---|---|---|---|
| Cases ± SD/% (n) | Controls ± SD/% (n) | p | Cases ± SD/% (n) | Controls ± SD/% (n) | p | |
| N | 379 | 403 | - | 190 | 570 | - |
| Age, years | 45.05 ± 7.78 | 44.07 ± 8.27 | <0.05 | 39.58 ± 8.27 | 35.28 ± 8.13 | <0.001 |
| BMI, kg/m2 | 30.66 ± 4.29 | 28.51 ± 3.88 | <0.001 | 21.99 ± 1.82 | 21.42 ± 1.83 | >0.05 |
| Family history of uterine myoma (mother had uterine leiomyoma) | 35.36 (134) | 19.11 (77) | <0.001 | 34.74 (66) | 15.61 (89) | <0.001 |
| Married | 85.22 (323) | 85.86 (346) | >0.05 | 84.74 (161) | 85.96 (490) | >0.05 |
| Smoker (yes) | 13.72 (52) | 15.14 (61) | >0.05 | 13.68 (26) | 18.42 (105) | >0.05 |
| Drinking alcohol (≥7 drinks per week) | 2.90 (11) | 1.74 (7) | >0.05 | 3.16 (6) | 4.04 (23) | >0.05 |
| Oral contraceptive use | 9.50 (36) | 10.17 (41) | >0.05 | 9.47 (18) | 10.00 (57) | >0.05 |
| Age at first oral contraceptive use (mean, years) | 23.51 ± 2.39 | 23.72 ± 2.37 | >0.05 | 23.32 ± 2.29 | 23.54 ± 2.32 | >0.05 |
| Age at menarche and menstrual cycle | ||||||
| Age at menarche, years | 13.41 ± 1.31 | 13.09 ± 1.23 | >0.05 | 13.57 ± 1.32 | 13.36 ± 1.27 | >0.05 |
| Duration of bleeding menstrual (mean, days) | 5.24 ± 1.68 | 4.94 ± 0.94 | >0.05 | 5.05 ± 1.45 | 4.97 ± 0.96 | >0.05 |
| Menstrual cycle length (mean, days) | 27.94 ± 2.26 | 28.04 ± 2.26 | >0.05 | 28.27 ± 1.80 | 28.20 ± 2.24 | >0.05 |
| Reproductive characteristic | ||||||
| Age at first birth (mean, years) | 21.06 ± 2.35 | 21.57 ± 3.44 | >0.05 | 21.58 ± 3.20 | 21.72 ± 3.42 | >0.05 |
| No of gravidity (mean) | 3.64 ± 2.20 | 2.63 ± 1.56 | <0.001 | 2.73 ± 2.10 | 2.23 ± 1.51 | <0.01 |
| No of births (mean) | 1.58 ± 0.80 | 1.71 ± 0.68 | <0.05 | 1.20 ± 0.90 | 1.41 ± 0.63 | <0.05 |
| No of spontaneous abortions (mean) | 0.29 ± 0.69 | 0.22 ± 0.48 | >0.05 | 0.18 ± 0.49 | 0.23 ± 0.49 | >0.05 |
| No of induced abortions (mean) | 1.73 ± 1.69 | 0.88 ± 0.90 | <0.001 | 1.31 ± 1.50 | 0.48 ± 0.91 | <0.001 |
| No of stillbirths | 0.01 ± 0.08 | 0.02 ± 0.14 | >0.05 | 0.01 ± 0.08 | 0.01 ± 0.11 | >0.05 |
| History of infertility | 13.72 (52) | 5.21 (21) | <0.001 | 13.68 (26) | 5.09 (29) | <0.01 |
| Gynecological pathologies | ||||||
| Cervical disorders | 27.97 (106) | 28.54 (115) | >0.05 | 22.11 (42) | 22.81 (130) | >0.05 |
| History of sexually transmitted disease | 26.91 (102) | 26.55 (107) | >0.05 | 27.37 (52) | 27.19 (155) | >0.05 |
| Chronic endometritis | 11.87 (45) | 7.20 (29) | <0.05 | 6.32 (12) | 4.56 (26) | >0.05 |
| Chronic inflammation of adnexa | 35.88 (136) | 34.24 (138) | >0.05 | 32.11 (61) | 30.35 (173) | >0.05 |
| Endometrial hyperplasia | 47.23 (179) | - | - | 46.84 (89) | - | - |
| Endometriosis | 35.36 (134) | - | - | 38.42 (73) | - | - |
| Adenomyosis | 20.32 (77) | - | - | 23.16 (44) | - | - |
| Chr | SNP | Gene | Nucleotide Sequences of Primers and Probes |
|---|---|---|---|
| 1 | rs17496332 | PRMT6 | F: AGCCTTGAAAGAGTGTATA R: GTGAGAATGTTCCTTGTG FAM-acaaAaCaTaGtAtctgc-BHQ-1 VIC-acaaAaCaCaGtAtctgc-BHQ-2 |
| 2 | rs780093 | GCKR | F: GCCGTTGCTCTCATTCTTA R: CCTTCTTCCACCACCATC FAM-cctGgtTggGggc-BHQ-1 VIC-cctGgtCggGggc-BHQ-2 |
| 2 | rs10454142 | PPP1R21 | F: CCTGCTCTGTATATCTTC R: GTTCCTCTATACATTCATATG FAM-cttacTaaTggCctcc-BHQ-1 VIC-cttacTaaCggCctcc-BHQ-2 |
| 7 | rs3779195 | BAIAP2L1 | F: CGAGAGCACTTTCAACTA R: CCAGGCTTTACTGAGAAA FAM-atttctTgaTttTggggag-BHQ-1 VIC-atttctTgaAttTggggag-BHQ-2 |
| 8 | rs440837 | ZBTB10 | F: CAAGCAAAAATATTGTGAAA R: GAAGGATAGAGTTAATGGA FAM-aattatCtGtTtAgAatttatt-BHQ-1 VIC-aattatCtGtCtAgAatttatt-BHQ-2 |
| 10 | rs7910927 | JMJD1C | F: CACTGACTTCTTAAAAAAG R: TGCAGGTATTTGATATAAC FAM-tgcatAtAaAtTtTctatttta-BHQ-1 VIC-tgcatAtAaCtTtTctatttta-BHQ-2 |
| 12 | rs4149056 | SLCO1B1 | F: ACACCATATTGTCAAAGTTTG R: GCGAAATCATCAATGTAAGAA FAM-tggataTaTgTgTtCatggg-BHQ-1 VIC-tggataTaTgCgTtCatggg-BHQ-2 |
| 15 | rs8023580 | NR2F2 | F: CAAGGAAATATACTTCTTATTCATA R: CCAAGTGGAAATTATTGTC FAM-aagaatTcTaTgTtTagattt-BHQ-1 VIC-aagaatTcTaCgTtTagattt-BHQ-2 |
| 17 | rs12150660 | SHBG | F: GCTGGTCTCAAACTCCTC R: GAGGTAAATTTGTTGGGAACTTA FAM-agccactTcgCccg-BHQ-1 VIC-agccactGcgCccg-BHQ-2 |
| Chr | SNP | Gene | Minor Allele | n | Allelic Model | Additive Model | Dominant Model | Recessive Model | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | p | OR | 95%CI | p | OR | 95%CI | p | OR | 95%CI | p | |||||||||
| L95 | U95 | L95 | U95 | L95 | U95 | L95 | U95 | |||||||||||||
| female with BMI < 25 | ||||||||||||||||||||
| 1 | rs17496332 | PRMT6 | G | 711 | 0.82 | 0.64 | 1.06 | 0.127 | 0.70 | 0.51 | 0.94 | 0.023 | 0.71 | 0.48 | 1.05 | 0.084 | 0.52 | 0.27 | 1.01 | 0.055 |
| 2 | rs780093 | GCKR | T | 727 | 1.16 | 0.91 | 1.47 | 0.232 | 1.13 | 0.85 | 1.50 | 0.384 | 1.25 | 0.82 | 1.90 | 0.303 | 1.08 | 0.65 | 1.81 | 0.764 |
| 2 | rs10454142 | PPP1R21 | C | 717 | 0.92 | 0.71 | 1.19 | 0.520 | 0.94 | 0.71 | 1.26 | 0.699 | 0.90 | 0.61 | 1.33 | 0.602 | 1.00 | 0.54 | 1.85 | 0.999 |
| 7 | rs3779195 | BAIAP2L1 | A | 717 | 0.91 | 0.66 | 1.26 | 0.564 | 1.03 | 0.71 | 1.05 | 0.878 | 1.11 | 0.73 | 1.69 | 0.630 | 0.46 | 0.09 | 2.34 | 0.350 |
| 8 | rs440837 | ZBTB10 | G | 706 | 1.15 | 0.87 | 1.51 | 0.333 | 1.30 | 0.94 | 1.78 | 0.111 | 1.28 | 0.86 | 1.90 | 0.225 | 1.82 | 0.85 | 3.09 | 0.123 |
| 10 | rs7910927 | JMJD1C | T | 726 | 0.89 | 0.70 | 1.13 | 0.329 | 0.87 | 0.66 | 1.14 | 0.303 | 0.89 | 0.59 | 1.36 | 0.591 | 0.75 | 0.46 | 1.21 | 0.239 |
| 12 | rs4149056 | SLCO1B1 | C | 690 | 0.93 | 0.69 | 1.24 | 0.602 | 0.89 | 0.63 | 1.24 | 0.486 | 0.88 | 0.59 | 1.32 | 0.542 | 0.78 | 0.31 | 1.97 | 0.604 |
| 15 | rs8023580 | NR2F2 | C | 720 | 0.94 | 0.72 | 1.23 | 0.663 | 1.03 | 0.76 | 1.39 | 0.846 | 1.13 | 0.76 | 1.67 | 0.542 | 0.80 | 0.39 | 1.62 | 0.536 |
| 17 | rs12150660 | SHBG | T | 731 | 0.97 | 0.74 | 1.29 | 0.846 | 0.98 | 0.71 | 1.33 | 0.875 | 0.99 | 0.67 | 1.46 | 0.952 | 0.90 | 0.41 | 1.98 | 0.784 |
| female with BMI ≥ 25 | ||||||||||||||||||||
| 1 | rs17496332 | PRMT6 | G | 741 | 1.01 | 0.82 | 1.25 | 0.902 | 1.06 | 0.86 | 1.34 | 0.548 | 1.03 | 0.75 | 1.41 | 0.859 | 1.24 | 0.80 | 1.92 | 0.345 |
| 2 | rs780093 | GCKR | T | 743 | 1.06 | 0.86 | 1.31 | 0.559 | 1.06 | 0.85 | 1.33 | 0.586 | 1.04 | 0.75 | 1.44 | 0.801 | 1.16 | 0.76 | 1.77 | 0.483 |
| 2 | rs10454142 | PPP1R21 | C | 728 | 1.09 | 0.87 | 1.36 | 0.461 | 1.07 | 0.83 | 1.37 | 0.604 | 1.04 | 0.76 | 1.43 | 0.802 | 1.24 | 0.70 | 2.20 | 0.457 |
| 7 | rs3779195 | BAIAP2L1 | A | 735 | 1.19 | 0.92 | 1.55 | 0.184 | 1.27 | 0.95 | 1.68 | 0.104 | 1.53 | 1.06 | 2.09 | 0.018 | 0.58 | 0.24 | 1.42 | 0.232 |
| 8 | rs440837 | ZBTB10 | G | 717 | 1.04 | 0.81 | 1.33 | 0.756 | 1.04 | 0.80 | 1.34 | 0.785 | 0.88 | 0.64 | 1.22 | 0.448 | 2.15 | 1.09 | 4.25 | 0.027 |
| 10 | rs7910927 | JMJD1C | T | 745 | 1.14 | 0.93 | 1.39 | 0.216 | 1.12 | 0.89 | 1.40 | 0.321 | 1.00 | 0.69 | 1.44 | 0.996 | 1.35 | 0.94 | 1.95 | 0.106 |
| 12 | rs4149056 | SLCO1B1 | C | 728 | 1.04 | 0.81 | 1.32 | 0.781 | 1.05 | 0.80 | 1.37 | 0.753 | 1.07 | 0.78 | 1.48 | 0.669 | 0.95 | 0.45 | 2.02 | 0.900 |
| 15 | rs8023580 | NR2F2 | C | 731 | 1.07 | 0.85 | 1.34 | 0.576 | 1.00 | 0.78 | 1.28 | 0.983 | 1.04 | 0.76 | 1.42 | 0.823 | 0.90 | 0.50 | 1.61 | 0.717 |
| 17 | rs12150660 | SHBG | T | 755 | 0.97 | 0.77 | 1.22 | 0.775 | 0.92 | 0.72 | 1.19 | 0.533 | 0.95 | 0.69 | 1.29 | 0.721 | 0.76 | 0.41 | 1.44 | 0.404 |
| SNP (Position hg38) (r2, LD) | Haploreg Data | GTE-Portal Data | |||
|---|---|---|---|---|---|
| Transcription Factors | Adipose-Derived Mesenchymal Stem Cell Cultured Cells | Liver | Visceral Adipose | Subcutaneous Adipose | |
| rs113329442 (106996630) (r2 = 0.99, LD = 1.00) | Brachyury, GR, Irf, PU.1, Sox | PRMT6 | PRMT6 | PRMT6 | |
| rs3861909 (107001554) (r2 = 0.97, LD = −0.99) | AP-1, Pdx1, RORalpha1 | H3K4me1_Enh | PRMT6 | PRMT6 | PRMT6 |
| rs17496332 (107003753) | DMRT1, FAC1 | PRMT6 | PRMT6 | PRMT6 | |
| rs2878349 (107006623) (r2 = 0.98, LD = 1.00) | PRMT6 | PRMT6 | PRMT6 | ||
| rs5776878 (107008396) (r2 = 0.98, LD = −1.00) | AP-1, Cart1, HDAC2, Zfp105 | * | * | * | |
| rs72697623 (107011647) (r2 = 0.98, LD = 1.00) | CEBPA, CEBPB, p300 | H3K4me1_Enh | PRMT6 | PRMT6 | PRMT6 |
| rs4914939 (107015739) (r2 = 0.94, LD = 0.99) | Cdc5, Fox, Foxa, Foxf1, Foxi1, Foxj1, Foxj2, Foxl1, Foxp1, HDAC2, Mef2, Pou2f2, TATA, Zfp105, p300 | H3K4me1_Enh | * | * | * |
| rs12406721 (107020621) (r2 = 0.91, LD = 0.96) | EWSR1-FLI1, HDAC2, Hoxa5 | PRMT6 | PRMT6 | PRMT6 | |
| rs61798463 (107023312) (r2 = 0.88, LD = 0.96) | IRC900814 | PRMT6 | PRMT6 | PRMT6 | |
| rs111232683 (107023527) (r2 = 0.85, LD = 0.93) | CACD, CCNT2, CHD2, Ets, Egr-1, GR, Klf4, Myc, NRSF, PU.1, Pax-4, Pou2f2, RREB-1, SP1, SREBP, Spz1, STAT, ZNF219, Zfp281, Zfp740, UF1H3BETA | * | * | * | |
| rs56111229 (107024067) (r2 = 0.85, LD = 0.93) | AP-1, Arid3a, Bach1, Bsx, GATA, GR, KAP1, Zfp691 | PRMT6 | PRMT6 | PRMT6 | |
| rs55924375 (107024068) (r2 = 0.85, LD = 0.93) | AP-1, Arid3a, Bach1, Bsx, GATA, GR, HNF1, Hoxb4, KAP1, Zfp691 | PRMT6 | PRMT6 | PRMT6 | |
| rs61798468 (107026694) (r2 = 0.88, LD = 0.96) | Arid3a, Pou2f2, Sox, Zfp105 | PRMT6 | PRMT6 | PRMT6 | |
| rs200443569 (107028138) (r2 = 0.81, LD = 0.91) | GATA, HDAC2, Ik-2, NF-AT, Sox, TATA | * | * | * | |
| rs72442711 (107028139) (r2 = 0.81, LD = 0.90) | Foxp1, GATA, HDAC2, Irf, Sox, TATA | * | * | * | |
| SNP (Position hg38) (r2, LD) | Haploreg Data | GTE-Portal Data (eQTL/sQTL) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Transcription Factors/Proteins Bound | Liver | Adipocyte Cultured Cells | |||||||
| Mesenchymal Stem Cell-Derived Adipocyte Cultured Cells | Adipose-Derived Mesenchymal Stem Cell Cultured Cells | Adipose Nuclei | Visceral Adipose | Subcutaneous Adipose | Liver | Uterus | |||
| rs6950023 (98286323) (r2 = 0.90, LD = −0.96) | Nkx3/POL24H8, AP2ALPHA, AP2GAMMA, CMYC, GTF2F1, MAX, MXI1, POL2, PRDM1, PU1 | H3K4me1_Enh H3K4me3_Pro H3K27ac_Enh H3K9ac_Pro | H3K4me1_Enh H3K4me3_Pro H3K9ac_Pro | H3K4me1_Enh H3K4me3_Pro H3K9ac_Pro | H3K4me1_Enh H3K4me3_Pro H3K27ac_Enh H3K9ac_Pro | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 |
| rs6967728 (98286325) (r2 = 0.90, LD = −0.96) | Nkx3/POL24H8, AP2ALPHA, AP2GAMMA, CEBPB, CMYC, GTF2F1, MAX, MXI1, POL2, PRDM1, PU1 | H3K4me1_Enh H3K4me3_Pro H3K27ac_Enh H3K9ac_Pro | H3K4me1_Enh H3K4me3_Pro H3K9ac_Pro | H3K4me1_Enh H3K4me3_Pro H3K9ac_Pro | H3K4me1_Enh H3K4me3_Pro H3K27ac_Enh H3K9ac_Pro | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 |
| rs7015 (98291311) (r2 = 0.85, LD = −0.97) | Dbx1, Hoxa10, Hoxa9, Hoxb13, Hoxd10, Ncx, Pou3f2, Sox, Zfp105 | H3K4me1_Enh | H3K9ac_Pro | H3K4me1_Enh H3K27ac_Enh H3K9ac_Pro | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | |
| rs13232861 (98299769) (r2 = 0.81, LD = −0.96) | AP-1, AP-2, BAF155, BATF, Bach1, Bach2, CHD2, E2F, Egr-1, GATA, GR, HMGN3, KAP1, NRSF, Nrf1, PRDM1, SRF, STAT, Sin3Ak-20, TCF4, Zfp161, p300 | H3K27ac_Enh | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | |||
| rs11290747 (98300261) (r2 = 0.95, LD = −0.97) | CHD2, CTCFL, E2F, GR, NF-kappaB, NRSF, Rad21, SP1, UF1H3BETA, ZNF263, Znf143, p300 | H3K4me1_Enh H3K27ac_Enh H3K9ac_Pro | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | |||
| rs2906184 (98310675) (r2 = 0.95, LD = 0.97) | Arid3a, CEBPG, Dbx1, HDAC2, Ncx, PLZF, TATA, Zfp105 | * | * | * | * | ||||
| rs1635609 (98320502) (r2 = 0.96, LD = −0.98) | HNF1, Hoxa4, Pax-4, Pax-6, Pou2f2 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | ||||
| rs1688607 (98322009) (r2 = 0.92, LD = −0.98) | GR, VDR | H3K4me1_Enh | H3K4me1_Enh | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | ||
| rs1688606 (98345539) (r2 = 0.98, LD = −0.99) | GATA | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | ||||
| rs112758337 (98347956) (r2 = 0.98, LD = 0.99) | MAZ, MAZR, MZF1:1–4 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | ||||
| rs77032872 (98355009) (r2 = 0.98, LD = 0.99) | Foxl1, HNF1, Mef2, Nkx2, Pax-2, TATA | H3K4me1_Enh H3K27ac_Enh | H3K4me1_Enh H3K9ac_Pro | H3K4me1_Enh | H3K4me1_Enh H3K27ac_Enh | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 |
| rs12704986 (98357118) (r2 = 0.97, LD = −0.99) | EBF, HNF4, Nr2f2, RXRA | H3K27ac_Enh | * | * | * | * | |||
| rs3779196 (98360794) (r2 = 0.98, LD = −0.99) | Ascl2, BHLHE40, CEBPB, CTCF, Lmo2-complex, TCF12/USF1, CTCF, RAD21, SMC3, GABP, HDAC2, MAFK, POL2 | H3K4me1_Enh H3K27ac_Enh H3K9ac_Pro | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | |||
| rs6965424 (98361813) (r2 = 0.98, LD = −0.99) | H3K4me1_Enh | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | ||||
| rs3779195 (98364050) | Foxp1 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | ||||
| rs4268041 (98376226) (r2 = 0.91, LD = −0.98) | Rad21 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | ||||
| rs201244010 (98378225) (r2 = 0.95, LD = −0.99) | * | * | * | * | |||||
| rs5886063 (98378229) (r2 = 0.95, LD = −0.98) | BATF, FAC1, MAZ, Myc, Pax-2 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | ||||
| rs10953259 (98383795) (r2 = 0.95, LD = −0.98) | DMRT4, Lhx3, Pou6f1 | H3K4me1_Enh H3K9ac_Pro | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | |||
| rs13310668 (98393841) (r2 = 0.88, LD = −0.94) | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | |||||
| rs10953260 (98404180) (r2 = 0.93, LD = −0.96) | PU1 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3/BRI3 | RP11-307C18.1, BRI3 | RP11-307C18.1 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ponomarenko, M.; Reshetnikov, E.; Churnosova, M.; Aristova, I.; Abramova, M.; Novakov, V.; Churnosov, V.; Polonikov, A.; Churnosov, M.; Ponomarenko, I. Obesity/Overweight as a Meaningful Modifier of Associations Between Gene Polymorphisms Affecting the Sex Hormone-Binding Globulin Content and Uterine Myoma. Life 2025, 15, 1459. https://doi.org/10.3390/life15091459
Ponomarenko M, Reshetnikov E, Churnosova M, Aristova I, Abramova M, Novakov V, Churnosov V, Polonikov A, Churnosov M, Ponomarenko I. Obesity/Overweight as a Meaningful Modifier of Associations Between Gene Polymorphisms Affecting the Sex Hormone-Binding Globulin Content and Uterine Myoma. Life. 2025; 15(9):1459. https://doi.org/10.3390/life15091459
Chicago/Turabian StylePonomarenko, Marina, Evgeny Reshetnikov, Maria Churnosova, Inna Aristova, Maria Abramova, Vitaly Novakov, Vladimir Churnosov, Alexey Polonikov, Mikhail Churnosov, and Irina Ponomarenko. 2025. "Obesity/Overweight as a Meaningful Modifier of Associations Between Gene Polymorphisms Affecting the Sex Hormone-Binding Globulin Content and Uterine Myoma" Life 15, no. 9: 1459. https://doi.org/10.3390/life15091459
APA StylePonomarenko, M., Reshetnikov, E., Churnosova, M., Aristova, I., Abramova, M., Novakov, V., Churnosov, V., Polonikov, A., Churnosov, M., & Ponomarenko, I. (2025). Obesity/Overweight as a Meaningful Modifier of Associations Between Gene Polymorphisms Affecting the Sex Hormone-Binding Globulin Content and Uterine Myoma. Life, 15(9), 1459. https://doi.org/10.3390/life15091459









